Abstract
This study aims to preliminarily explore the impact of super-enhancer (SE)-associated genes on breast cancer (BC) and their potential regulatory mechanisms. We first identified differentially expressed SE-associated genes between BC patients and healthy controls. Subsequently, Cox regression analysis and the Least Absolute Shrinkage and Selection Operator (LASSO) algorithm were applied to evacuate SE-associated genes that related to the overall survival (OS) of patients. The Super-Enhancer-Related Score (SERS) was then calculated based on the LASSO coefficients and incorporated with clinical features to construct a clinical prediction model. Besides, we applied multiple bioinformatic approaches to investigate the potential regulatory mechanism of SE-associated genes using bulk RNA and single-cell RNA sequencing data. Finally, the hub gene was identified using various machine learning methods and Immunohistochemistry (IHC) assay. We found that SERS was negatively correlated with OS of BC patients. Both time-dependent ROC analysis and calibration curves demonstrated the strong predictive ability and high accuracy of the prediction model we built. Significant differences were observed between the two groups in terms of tumor mutation burden, tumor immune microenvironment, and drug sensitivity. Importantly, we identified TFF1 as the core gene, and the immunohistochemical (IHC) assay using the tissue microarray revealed that TFF1-positive patients had significantly better OS than their counterparts. We constructed a highly precise clinical prediction model based on SERS, demonstrating the impact of SE-associated genes on the prognosis of BC patients and their regulatory effects on somatic mutations, tumor immune microenvironment, and drug sensitivity in BC.
Similar content being viewed by others
Introduction
Breast cancer (BC) is one of the most prevalent malignant tumors among women worldwide, ranking first in both incidence and mortality rates among female cancers1. Despite significant advancements in early screening, molecular subtyping, and targeted therapies2, BC continues to pose challenges such as tumor recurrence, metastasis, and treatment resistance3,4,5. Thus, there is an urgent need to identify novel therapeutic targets and strategies for BC.
With advancements in genomics and epigenetics, there has been a renewed understanding of the mechanisms governing gene expression regulation6,7,8. Previous studies primarily focused on promoter regions and classical enhancers. In recent years, however, super-enhancers (SEs) have emerged as a focal point in cancer research9,10,11. SEs, which are large regulatory regions composed of multiple enhancer elements and typically spanning thousands of base pairs, can regulate the efficient transcription of target genes through physical interactions with gene promoters. In contrast to classical enhancers, SEs exhibit significant structural and functional differences, characterized by higher densities of transcription factors and greater transcriptional activation capabilities, playing a crucial role in tumorigenesis and cancer progression9.
In the field of BC, accumulating evidence suggests that alterations in SE activity may serve as a critical factor in tumorigenesis and progression. Numerous BC-associated oncogenes, such as MYC and ESR1, are regulated by SEs12. The aberrant activation of these SEs may drive tumor aggressiveness, metastatic potential, and chemotherapy resistance13. Consequently, targeting SEs or their key components has emerged as a novel potential therapeutic strategy.
Therefore, this study aims to elucidate the prognostic predictive capability of SE-associated genes in BC, as well as their correlations with somatic mutations, tumor immune microenvironment, and drug sensitivity. Furthermore, we seek to investigate the role of the SE-associated core gene in BC, thereby providing novel insights for early diagnosis and targeted therapeutic strategies against SEs in BC.
Materials and methods
Data source
The gene expression profile and corresponding clinicopathological data for the training cohort were obtained from The Cancer Genome Atlas (TCGA) database14. The external validation cohort was sourced from the METABRIC dataset15,16. All enrolled patients were diagnosed with primary BC without distant metastasis (M0). The clinicopathological characteristics are summarized in Table S1. SE-associated genes were extracted from Sample_02_0667, Sample_02_0670, Sample_02_0671, and Sample_02_1517 in the SEdb 2.0 database17. Duplicate genes were subsequently removed to ensure a non-redundant gene list. Single-cell RNA sequencing (scRNA-seq) data and theraptic response date were obtained from the GSE161529 dataset and GSE194040 in the Gene Expression Omnibus (GEO) database, respectively18.
Development of SE-related score and clinical prediction model
Firstly, differential expression analysis was performed in the counts data of BC patients and healthy controls from the TCGA-BRCA dataset using the DESeq2 package in R software19. Differentially expressed genes (DEGs) were identified with a threshold of |log2FoldChange|> 2 and an adjusted p-value < 0.05. These DEGs were then intersected with SE-associated genes to obtain SE-related DEGs in BC. Subsequently, the Cox proportional hazards regression model from the survival package was applied to evaluate the association between the expression of each SE-related DEG and overall survival (OS) in the TCGA-BRCA cohort. Genes with statistically significant differences (p < 0.05) were included in the LASSO algorithm to ensure model simplicity and minimize overfitting during model training. The risk score was constructed using the regression coefficients derived from LASSO Cox regression analysis:
SERS means the super-enhancer-related score. Coefi and Expi represent the coefficient and expression level of the corresponding gene, respectively. All samples were stratified into two groups based on the median SERS value: the high-risk group (SERS > median) and the low-risk group (SERS < median). Kaplan–Meier (KM) analysis was then employed to compare the differences in OS between the two groups. Time-dependent receiver operating characteristic (tROC) curves were utilized to evaluate the prognostic predictive performance of SERS. Furthermore, clinicopathological features with statistical significance (p < 0.05) in univariate and multivariate Cox regression analyses were integrated, and a nomogram was constructed using the rms package to predict 1-years, 3-years, 5-years, and 10-years OS. The discriminative ability of the nomogram was quantitatively assessed using the area under the curve (AUC) of the tROC curves, while the calibration curves were used to evaluate the calibration performance of the nomogram.
Functional enrichment analysis
DEGs between the high-risk and low-risk groups were identified using the DESeq2 package in R19. Functional enrichment analysis of the identified DEGs was performed based on the Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), and Reactome databases using the clusterProfiler20 and ReactomePA packages21. Additionally, a Bayesian network was constructed using the CBNplot package to explore interactions among pathways22.
Somatic mutation analysis
Somatic mutation data were retrieved from TCGA and analyzed using the maftools package in R23. The tumor mutational burden (TMB) of each patient between the high-risk and low-risk groups was calculated and compared using the Wilcoxon rank-sum test.
Tumor immune microenvironment analysis
The CIBERSORT algorithm was applied to predict the proportions of 22 types of tumor-infiltrating immune cells (TIICs) in each sample between the high-risk and low-risk groups. Single-sample Gene Set Enrichment Analysis (ssGSEA) was utilized to estimate the abundance of 28 TIICs in individual tissue samples. Additionally, the xCell package was employed to assess the infiltration levels of 64 immune and stromal cell types24. The ESTIMATE package was used to calculate the ESTIMATE score and tumor purity for each sample. Immunophenoscore data for each sample were collected from The Cancer Immunome Atlas (TCIA) database, and tumor immune exclusion scores were computed using the Tumor Immune Dysfunction and Exclusion (TIDE) database. Furthermore, the Wilcoxon rank-sum test was applied to compare the expression levels of several key co-stimulatory molecules and human leukocyte antigen (HLA) family genes between the high-risk and low-risk groups.
Drug Sensitivity prediction
The R package oncoPredict was utilized to calculate the half-maximal inhibitory concentration (IC50) of cancer therapeutic drugs for each sample in the high-risk and low-risk groups based on the GDSC2 dataset of the Genomics of Drug Sensitivity in Cancer (GDSC) database25.
Single-cell data analysis
Single-cell RNA sequencing (scRNA-seq) data were quality-controlled and processed using the Seurat package (v4.3.0)26. Cell types were annotated based on the CellMarker2.0 database and relevant literature27,28. Subsequently, the single-cell identification of subpopulations with bulk sample phenotype correlation (Scissor) package was employed to identify phenotype-related cell subpopulations29. Input data included TCGA-BRCA bulk-seq data, GSE161529 scRNA-seq data, and SE-related risk group data obtained from LASSO analysis. The single-cell dataset was ultimately divided into Scissor-positive (Scissor+) and Scissor-negative (Scissor -) cells, corresponding to the high-risk and low-risk groups in bulk-seq, respectively. The CellChat package was used to calculate differential information flow between Scissor-positive and Scissor-negative cells and to compare the number and strength of cell–cell communication across different cell subtypes30.
Screening of SE-associated core genes
The random forest (RF) algorithm was applied to analyze the importance of each SE-associated gene used to calculate the SERS. The eXtreme Gradient Boosting (XGBoost) algorithm was further employed to assess the importance of each gene31. Finally, the SE-associated core gene was determined by integrating the results from LASSO, RF, and XGBoost analyses.
Experimental validation of the core gene
The expression of the core gene and its impact on prognosis were validated using immunohistochemistry (IHC) on tissue microarrays (TMA). The TMA was made using wax blocks of 133 BC and 45 adjacent breast tissue. All samples were obtained from primary BC patients without metastasis at the time of diagnosis in the Department of Breast Surgery at the Second Affiliated Hospital of Harbin Medical University. The inclusion criteria are: (i) pathological diagnosis of invasive carcinoma; (ii) operable; (iii) no prior treatment before surgery; (iv) OS longer than 3 months; (v) complete clinical data and postoperative follow-up records. The exclusion criteria include: (i) concurrent immune system diseases; (ii) presence of distant metastasis; (iii) surgical complications or infections; (iv) history of antibiotic use within three weeks before surgery. After deparaffinization and rehydration, tissue sections were incubated in antigen retrieval buffer and heated in a steamer at > 97 °C for 20 min. IHC staining was performed using the Ventana Discovery XT automated slide stainer, which automated the processes of dewaxing, antigen retrieval, blocking, DAB detection, counterstaining, post-counterstaining, and slide washing. The trefoil factor 1 (TFF1) antibody (Abclonal, A1789) was applied to the TMA at a 1:200 dilution and incubated overnight at 4 °C in a constant temperature and humidity chamber. After washing in TBS, antigen–antibody binding was detected using the Envision + system and DAB + chromogen (DAKO). TMA was briefly immersed in hematoxylin for counterstaining, rinsed with water, and cover slipped. The Fiji Image-J software was used for semi-quantitative analysis of the average optical density (AOD) to determine TFF1 expression levels.
Statistical analysis
All statistical analyses were performed using R v4.2.2 and SPSS 26.0. A p-value < 0.05 was considered statistically significant unless otherwise specified.
Results
Identification of super-enhancer-associated genes in BC
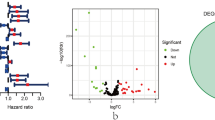
In this study, the TCGA-BRCA dataset was utilized as the training set. Through differentially expressed gene (DEG) analysis, we identified 1846 DEGs between breast cancer (BC) patients and healthy controls. By intersecting these DEGs with 1688 super-enhancer (SE)-associated genes, 150 SE-associated DEGs were obtained and subjected to univariate Cox regression analysis. Ultimately, 8 genes significantly associated with overall survival (OS) were identified (Fig. 1A). These 8 genes were subsequently used for LASSO regression analysis (Fig. 1B,C), and the super-enhancer-related score (SERS) was calculated based on the LASSO regression coefficients. The formula for SERS is as follows:
Identification of SE-associated genes in BC. (A) Forest plot of statistically significant genes in the univariate Cox regression analysis; (B) Coefficient profiles of SE-associated genes in the least absolute shrinkage and selection operator (LASSO); (C) Identification of the best parameter (lambda) in LASSO; (D) Violin plot of the expression of SE-associated genes in high and low SERS groups; (E) Kaplan–Meier (KM) analysis based on SERS in the training cohort; (F) KM analysis based on SERS in the validation cohort. HR: hazard ratio; CI: confidence interval; high: high-risk group; low: low-risk group; ****:p < 0.0001.
To further elucidate the potential mechanisms of SE-associated genes in BC, patients were stratified into a high-risk group and a low-risk group based on the SERS. The expression levels of the 8 SE-associated genes used to construct the model were significantly different between the high-risk and low-risk groups (Fig. 1D). Notably, Kaplan–Meier (KM) curves revealed that patients in the high-risk group had significantly worse OS compared to those in the low-risk group (Fig. 1E). In external validation, the results from the METABRIC dataset were consistent with those from the TCGA-BRCA training set, further confirming the prognostic predictive value of SERS for BC patients (Fig. 1F).
Construction and validation of the clinical prediction model
After initially confirming the prognostic predictive value of the SERS for BC patients, we aimed to establish a clinical prediction model by integrating SERS with other clinical features. First, the time-dependent receiver operating characteristic (tROC) curve was applied to evaluate the predictive performance of SERS. As shown in Fig. 2A, the AUC values of SERS for predicting 1-years, 3-years, 5-years, and 10-years OS were 0.627, 0.721, 0.695, and 0.682, respectively. Next, the impact of SERS, age, hormone receptor (HR) status, T stage, lymph node status, ethnicity and prior treatment on prognosis was assessed using univariate Cox regression analysis (Fig. 2B). Statistically significant factors were subsequently included in multivariate Cox regression analysis (Fig. 2C). Based on the results of the multivariate Cox regression analysis, age, lymph node status, and SERS were selected to construct the final model. Considering the complexity of the risk score formula, a nomogram was developed to predict 1-years, 3-years, 5-years, and 10-years OS in BC patients (Fig. 2D). In the nomogram model, higher OS was associated with lower SERS, younger age, HR-positive status, and absence of lymph node metastasis. The AUC values of the nomogram for predicting 1-years, 3-years, 5-years, and 10-years survival rates were 0.876, 0.746, 0.737, and 0.798, respectively (Fig. 2E). The calibration curves demonstrated that the predicted survival rates from the nomogram closely aligned with the actual survival rates of BC patients, indicating strong concordance between predicted and observed outcomes at 1, 3, 5, and 10 years (Fig. 2F). Furthermore, external validation using the METABRIC dataset confirmed the robust predictive performance of the model, as evidenced by the tROC and calibration curves (Fig. 2G,H).
Construction and validation of a clinical nomogram. (A) Time-dependent receiver operating characteristic (tROC) curves of the SERS in predicting the 1-, 3-, 5- and 10-year OS; (B) Univariate Cox regression analysis of SERS and clinical characteristics; (C) Multivariate Cox regression analysis of SERS and clinical characteristics; (D) Nomogram based on the SERS, age and lymph node status; (E) Time-dependent ROC curves of the nomogram in predicting the 1-, 3-, 5- and 10-years OS in the training cohort; (F) Calibration curves of the nomogram in predicting the 1-, 3-, 5- and 10-years OS in the training cohort; (G) Time-dependent ROC curves of the nomogram in predicting the 1-, 3-, 5- and 10-years OS in the validation cohort; (H) Calibration curves of the nomogram in predicting the 1-, 3-, 5- and 10-years OS in the validation cohort. AUC, area under curve; HR: hazard ratio; CI: confidence interval; SERS, super enhancer-related score; OS, overall survival.
Identification of SE-related signaling pathways
After establishing a clinical prediction model with high predictive performance using SE-associated genes and clinical features, we further explored the potential mechanisms by which SE-associated genes influence the prognosis of BC patients. A total of 718 DEGs were identified between the high-risk and low-risk groups, including 578 upregulated genes and 140 downregulated genes in the high-risk group (Fig. 3A). In the KEGG functional enrichment analysis, the DEGs were primarily enriched in pathways related to molecular interactions and signal transduction within the cellular microenvironment, such as neuroactive ligand-receptor interaction and the estrogen signaling pathway (Fig. 3B). Gene Ontology (GO) analysis revealed that the DEGs were significantly enriched in pathways associated with transmembrane protein activity and ion channel activity (Fig. 3C). Enrichment analysis based on the Reactome database further indicated that the DEGs were involved in G protein-coupled receptor ligand binding, synaptic signaling, and ion transport (Fig. 3D). Additionally, a Bayesian network (BN) demonstrated close interactions among the enriched pathways (Figs. 3E,F).
Identification of SE-related signaling pathways. (A) Volcano plot of DEGs based on SERS; (B) Dot plot of KEGG enrichment analysis; (C) Dot plot of GO enrichment analysis; (D) Dot plot of Reactome enrichment analysis; (E) Bayesian network based on GO analysis; (F) Bayesian network based on Reactome analysis.
Somatic mutation analysis
Given that genetic mutations are a critical factor in tumorigenesis, we evaluated the landscape of somatic mutations between the high- and low-risk groups. In the high-risk group, the somatic mutation rate was 87.89% (341 out of 388 samples), with missense mutations being the most prevalent. The most frequently mutated gene was TP53 (47%) (Fig. 4A). In the low-risk group, the mutation rate was 87.1% (324 out of 372 samples), also dominated by missense mutations, with PIK3CA being the most frequently mutated gene (41%) (Fig. 4B). Furthermore, quantitative analysis of tumor mutation burden (TMB) revealed that the high-risk group had a significantly higher TMB compared to the low-risk group (Fig. 4C–E).
Tumor mutation analysis. (A) Waterfall plot of somatic mutation features established with high SERS; (B) Waterfall plot of somatic mutation features established with low SERS; (C) TMB in the high-risk group; (D) TMB in the low-risk group; (E) Violin plot of TMB in the high- and low-risk groups. high: high-risk group; low: low-risk group; ****p < 0.0001.
Tumor immune microenvironment analysis
To characterize the status of the tumor immune microenvironment, we first employed the CIBERSORT algorithm to calculate the proportions of immune cells in the high-risk and low-risk groups. The results revealed that the high-risk group exhibited significantly reduced levels of CD8 + T cells, dendritic cells, and monocytes compared to the low-risk group (Fig. 5A). Single-sample Gene Set Enrichment Analysis (ssGSEA) further demonstrated that the expression of most tumor-infiltrating immune cells (TIICs) was lower in the high-risk group, including activated B cells, CD8 + T cells, NK-T cells, Th1 cells, and Th17 cells (Fig. 5B). Subsequently, the xCell package was used to assess the infiltration levels of 64 immune and stromal cell types, yielding similar results (Fig. 5C). These findings suggested that the heterogeneity of TIICs in the tumor microenvironment might contribute to the prognostic differences between patients with high and low SERS.
Comprehensive analysis of tumor immune microenvironment. (A) Boxplot of immune cell proportion in the high- and low-risk groups calculated by CIBERSORT algorithm; (B) Boxplot of immune cell expression in the high- and low-risk groups calculated by ssGSEA algorithm; (C) Boxplot of infiltration levels of 64 immune cells and stromal cells in the high- and low-risk groups calculated by xCell; (D)Violin plot of ESTIMATE score in the high- and low-risk groups; (E) Violin plot of Tumor Purity in the high- and low-risk groups; (F) Violin plot of TIDE exclusion score in the high- and low-risk groups; (G) Violin plot of IPS in the high- and low-risk groups; (H) Violin plot of the expression levels of co-stimulators; (I) Violin plot of the expression levels of HLA molecules. high: high-risk group; low: low-risk group; IPS, Immunophenoscore; ns, non-significant;*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Additionally, the ESTIMATE algorithm showed that the immune score was significantly lower in the high-risk group compared to the low-risk group (Fig. 5D), while the tumor purity was significantly higher (Fig. 5E). The tumor immune exclusion score, calculated using the Tumor Immune Dysfunction and Exclusion (TIDE) database, also indicated higher scores in the high-risk group, suggesting a more pronounced degree of immune suppression in the tumor microenvironment (Fig. 5F). Comparative analysis using the Cancer Immunome Atlas (TCIA) database revealed that the relative probability of immunotherapy response was lower in the high-risk group, regardless of CTLA4 and PD1 status (Fig. 5G). Furthermore, the expression levels of most co-stimulatory molecules (Fig. 5H) and human leukocyte antigen (HLA) family genes (Fig. 5I) were significantly higher in the low-risk group.
Drug sensitivity analysis
We then explored the relationship between the SERS and sensitivity to anti-cancer drugs using the Genomics of Drug Sensitivity in Cancer (GDSC) database. Notably, it was found that several commonly used anti-cancer drugs in BC treatment, including Alpelisib (Fig. 6A), Cisplatin (Fig. 6B), Epirubicin (Fig. 6C), Fulvestrant (Fig. 6D), Olaparib (Fig. 6E), Palbociclib (Fig. 6F), Ribociclib (Fig. 6G), Temozolomide (Fig. 6H), Vinorelbine (Fig. 6I), and Zoledronate (Fig. 6J), exhibited lower half-maximal inhibitory concentration (IC50) values in the low-risk group, which suggested that patients with low SERS were more sensitive to these anti-cancer drugs. Moreover, we found that patients who achieved pathological complete response (pCR) under Paclitaxel + Ganitumab therapy shared significantly lower SERS than those non-pCR patients, further validated SERS’s discrimination ability in therapeutic response (Fig. 6K).
Drug sensitivity analysis. Sensitivity analysis for Alpelisib (A), Cisplatin (B), Epirubicin (C), Fulvestrant (D), Olaparib (E), Palbociclib (F), Ribociclib (G), Temozolomide (H), Vinorelbine (I), and Zoledronate (J). (K) SERS of patients who received pCR and non-PCR under Paclitaxel + Ganitumab therapy. IC50, half-maximal inhibitory concentration; ns, non-significant; high: high-risk group; low: low-risk group; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
SE-related single-cell analysis
In this study, we employed the standard Seurat pipeline to analyze single-cell RNA sequencing (scRNA-seq) data. Through dimension reduction, cell clustering, and annotation, 7 cell types were identified (Fig. 7A–C). Subsequently, the Scissor algorithm was applied to identify SE-related cell subpopulations. We identified 1379 Scissor-positive cells (Scissor+, corresponding to the high-risk group) and 1300 Scissor-negative cells (Scissor-, corresponding to the low-risk group) based on the SERS from bulk RNA sequencing (bulk-seq) data (Fig. 7G). Notably, the proportions of T cells, macrophages, and fibroblasts were higher in the Scissor- group, while the proportions of epithelial cells, B cells, and mast cells were lower compared to the Scissor + group (Fig. 7H).
SE-associated TME analysis at single-cell resolution. (A) UMAP visualization of cell clusters; (B) Visualization of marker gene expression; (C) UMAP visualization of cell-type-specific annotation; (D) UMAP visualization of cell clusters among extracted immune cells; (E) Visualization of marker gene expression among extracted immune cells; (F) UMAP visualization of cell-type-specific annotation among extracted immune cells; (G) UMAP visualization of Scissor + and Scissor- cells; (H) Proportional fractions of identified cell types across Scissor + /- condition; (I) UMAP visualization of Scissor + and Scissor- cells among extracted immune cells; (J) Proportional fractions of identified cell types across Scissor + /-condition among extracted immune cells; (K) Relative information flow in Scissor + and Scissor- cells; (L) Changes in cell interaction number and strength in Scissor + and Scissor- cells. Left: Differential numbers of interactions in Scissor + and Scissor- cells; right: Differential interaction strength in Scissor + and Scissor- cells.
Subsequently, we extracted T cells, B cells, and macrophages to evaluate the relationship between SERS and the tumor immune microenvironment. In subgroup analysis, immune cells were categorized into 10 subtypes (Fig. 7D–F), and the Scissor algorithm identified 556 Scissor + cells and 559 Scissor- cells (Fig. 7I). Concordantly, we found that the Scissor- group had a higher proportion of effector memory CD8 + T cells but lower proportions of regulatory T cells and M2 macrophages compared to the Scissor + group (Fig. 7J). These results align with the immune infiltration analysis using bulk data, further supporting the notion that the tumor immune microenvironment in the high-risk group has weaker anti-tumor capabilities.
Additionally, cell–cell communication analysis indicated that pathways such as VEGF, CALCR, and EGF exhibited significantly increased information flow in Scissor + cells, while pathways like COMPLEMENT showed enhanced information flow in Scissor- cells (Fig. 7K). Besides, we compared the number and strength of communications among different cell types to explore differences in intercellular interactions between the high- and low-risk groups. The results demonstrated stronger communication between T cells, macrophages, and epithelial cells in the Scissor- group (Fig. 7L).
Screening and experimental validation of the core gene
To identify the most critical core gene from the 8 SE-associated genes, we integrated the results of LASSO, random forest (RF), and eXtreme Gradient Boosting (XGBoost) algorithms, ultimately determining Trefoil Factor 1 (TFF1) as the core gene (Fig. 8A, B). Subsequently, IHC analysis of TFF1 was performed using tissue microarrays (TMA) composed of 133 breast cancer tissues and 45 adjacent normal tissues. Based on the average optical density (AOD) of IHC staining, all tissues were classified as TFF1-positive (AOD ≥ 0.1) or TFF1-negative (AOD < 0.1), with representative microscopic images shown in Fig. 8C. The results indicated no significant difference in TFF1 expression levels between tumor tissues and adjacent normal tissues (Table 1).
Experimental validation using IHC analysis. (A) The mean decrease accuracy of each SE-associated gene calculated using random forest; (B) The importance score of each SE-associated gene calculated using XGBoost; (C) Representative images of TFF1 expression in adjacent (left panel) and tumor (right panel) tissue; (D) KM survival analysis between patients in TFF1-negative and TFF1-positive groups. IHC, immunohistochemistry; KM, Kaplan–Meier.
BC patients were then divided into TFF1-positive and TFF1-negative groups. No significant differences were observed in clinicopathological features, including age, lymph node status, tumor size, molecular subtype, and Ki-67 levels, between the two groups (Table 2). However, Kaplan–Meier (KM) curves demonstrated that patients in the TFF1-positive group had significantly better prognosis compared to those in the TFF1-negative group (Fig. 8D), confirming the protective role of the SE-associated core gene TFF1 in BC patients.
Discussion
Despite significant advancements in basic research and clinical treatment of BC, it remains the most commonly diagnosed malignancy and the leading cause of cancer-related deaths among women globally, with incidence and mortality rates continuing to rise1. This may be attributed to the high genetic heterogeneity of BC, underscoring the urgent need for a more comprehensive and in-depth exploration of the non-coding regions of the genome.
One of the most prominent regulatory elements in the genome is the enhancer, which is often hijacked by cancer cells. Although enhancers were initially described as DNA sequences that increase the transcription of associated genes, recent research has focused on their properties beyond binding sequence-specific transcription factors, which may provide clues to their mechanisms of action and aid in their identification. For example, the enrichment of histone modifications (e.g., H3K4me1, H3K27ac), coactivators (e.g., p300/CBP, mediator complex), RNA polymerase II, open chromatin structure (e.g., chromatin accessibility via ATAC-seq), and “loop-mediated” interactions with target gene promoters have been identified as genomic features marking or identifying enhancers32,33,34.
Studies have shown that SEs regulate key cell identity genes and drive oncogene expression in several forms of cancer10,35,36. Increasing evidence suggests that mutations and structural rearrangements can alter the function of SE in cancer, leading to abnormally high levels of oncogene expression. For instance, structural rearrangements within the genomes of malignant cells can result in enhancer hijacking, placing previously unrelated enhancers near oncogenes and causing aberrant overexpression37,38. Super enhancers are particularly susceptible to hijacking, and studies have confirmed that certain cancers arise from the unintended binding of SEs to oncogenes, including medulloblastoma, leukemia, and adenoid cystic carcinoma39,40,41,42. Additionally, beyond their oncogenic roles in cancers, SEs also actively promote the expression of tumor suppressor genes43. For example, RCAN1.4, a potential tumor suppressor in BC located on human chromosome 21 (HSA21), is driven by a SE approximately 23 kb in length and is sensitive to BET inhibitors (BETi). Deletion of RCAN1.4 reduces its expression by over 90% and induces a malignant phenotype in BC cells43. Therefore, identifying SE-associated genes and investigating their roles are crucial for exploring novel therapeutic targets in cancer.
Other researchers have conducted similar studies, developing prognostic models based on SE-associated genes in liver cancer and head and neck cancer44,45. In this study, we utilized BC-related SE data from the SEdb2.0 database and employed a combination of Cox regression and LASSO algorithms to screen for SE-associated DEGs. Moreover, we constructed a highly accurate clinical prognostic model using SE-associated genes and clinical features, and its predictive value was further validated through external validation.
To further explore the potential mechanisms by which SE-associated genes influence the prognosis of BC, functional enrichment analysis of DEGs between the high-risk and low-risk groups was performed. The results revealed significant enrichment in pathways related to signal transduction (e.g., neuroactive ligand-receptor interaction, estrogen signaling pathways), ion transport, and transmembrane transport (e.g., G protein-coupled receptor ligand binding). Previous studies have shown that the neuroactive ligand-receptor system, specific G protein-coupled receptors, and trace amine-associated receptors (TAARs) are co-expressed in BC and are associated with subtypes of BC with poorer prognosis46. Additionally, estrogen-related signaling pathways are closely linked to hormone-dependent BC, but the complex resistance mechanisms to anti-estrogen therapies are a major contributor to worse patient outcomes47.
Beyond differences in enriched pathways, significant disparities were also observed in TMB, tumor immune microenvironment, and drug sensitivity between the high-risk and low-risk groups. The high-risk group exhibited a higher TMB and lower expression of tumor-infiltrating immune cells, along with higher tumor purity and lower immune scores. The Scissor algorithm further confirmed these differences at the single-cell level, revealing a significantly lower proportion of effector memory CD8 + T cells in the high-risk group. Consistent with previous findings, the tumor microenvironment can disrupt immune cell trafficking and function, leading to immune dysfunction. For example, in pancreatic and ovarian cancers48, common KRAS mutations impair T cell infiltration and anti-tumor immune responses49,50. Additionally, overexpression of VEGF in various tumors results in abnormal vasculature, limiting immune cell trafficking and recruitment, ultimately leading to dysregulation of anti-tumor immune cells51,52. This dysregulated state of immune cells in the tumor microenvironment is increasingly recognized as a major barrier to anti-tumor immunity. Furthermore, the high-risk group exhibited lower sensitivity to several commonly used anti-cancer drugs (e.g., cisplatin, epirubicin, fulvestrant, palbociclib, ribociclib), which may contribute to the poorer prognosis observed in high-SERS patients53,54,55.
Finally, by integrating the results of LASSO, random forest, and XGBoost algorithms, we identified the SE-associated gene TFF1 as the core gene. Studies on TFF1 in various malignancies have revealed its dual role as both an oncogene and a tumor suppressor. For example, TFF1 is overexpressed in prostate cancer and promoting tumor growth56. Besides, it can also promote cell survival in colonic carcinoma cell lines, and its enforced expression induces anchorage-independent growth and drives malignant transformation of premalignant colonic adenoma cells derived from familial adenomatous polyposis (FAP) patients57,58,59. However, in gastric cancer, TFF1 functions as a tumor suppressor; its expression is significantly downregulated in tumor tissues compared to normal mucosa, and mechanistically, it reduces cellular proliferation by inducing G1/S phase arrest in gastric carcinoma cells60,61,62. Furthermore, TFF1 mRNA expression is associated with pancreatic carcinogenesis and was identified as the most significantly upregulated gene in intraductal papillary mucinous neoplasms (IPMNs)59,63,64. Paradoxically, researchers have also found that TFF1 can suppress epithelial-mesenchymal transition (EMT), inhibit Wnt pathway activation, and reduce cancer stem cell properties, thereby promoting apoptosis in pancreatic carcinoma cells65.
In breast cancer, studies indicated that elevated TFF1 expression inversely correlates with tumor size and histological grade, while positively associating with estrogen receptor (ER) status and improved survival outcomes66,67,68,69. Mechanistically, TFF1’s promoter harbors estrogen response elements (EREs), consistent with its ER-positive association70,71. However, there is also in vitro evidence demonstrating that TFF1 stimulates BC cell migration, suggesting hormonal therapies may exert efficacy partialy by suppressing TFF1 expression to inhibit tumor cell motility72. Collectively, TFF1’s expression and functions in breast cancer exhibit multifaceted complexity beyond estrogen regulation. It may trigger diverse cellular responses, acting as both a morphogen and motogen, while exerting context-dependent anti-proliferative and anti-apoptotic effects73. This functional duality parallels the paradoxical behaviors of certain tumor suppressors involved in cellular differentiation62. In this study, IHC analysis of tissue microarrays revealed no significant difference in TFF1 expression between the tumor and adjacent tissues. However, KM curves demonstrated that patients in the TFF1-positive group had significantly better prognoses than those in the TFF1-negative group, confirming the protective role of TFF1 in BC patients.
Despite these significant findings, several limitations of this study should be acknowledged. First, the limited repertoire of clinicopathological features in the training and validation datasets constrains both the accuracy and generalizability of the model. Future iterations should incorporate additional clinical variables to enhance model’s utility when richer data become available. Moreover, the clinical prediction model was developed and validated using retrospective data. Therefore, prospective studies in real-world settings are essential to validate its clinical applicability. Additionally, although IHC analysis based on tissue microarrays was performed for experimental validation, comprehensive and precise mechanistic studies are needed to elucidate the specific regulatory mechanisms of SE-associated genes in BC.
In conclusion, our study utilized bioinformatics approaches to explore the predictive potential of super-enhancer-associated genes in breast cancer, established a highly accurate clinical prediction model, and uncovered their roles in regulating somatic mutations, tumor immune microenvironment, and drug sensitivity. Furthermore, we demonstrated the protective impact of the super enhancer-associated core gene TFF1 on breast cancer prognosis, suggesting its potential as a novel therapeutic target.
Data availability
Publicly available datasets analyzed in this study can be found here: https://xenabrowser.net/, https://www.cbioportal.org/, https://www.ncbi.nlm.nih.gov/geo/, and http://www.licpathway.net/sedb/.
Abbreviations
- AUC:
-
Area under curve
- AOD:
-
Average optical density
- ATAC-seq:
-
Assay for transposase-accessible chromatin with high throughput sequencing
- BC:
-
Breast cancer
- DEGs:
-
Differentially expressed genes
- EMT:
-
Epithelial-mesenchymal transition
- ER:
-
Estrogen receptor
- EREs:
-
Estrogen response elements
- FAP:
-
Familial adenomatous polyposis
- GO:
-
Gene ontology
- GEO:
-
Gene expression omnibus
- GDSC:
-
Genomics of drug sensitivity in cancer
- H3K4me1:
-
Histone H3 lysine 4 monomethylation
- H3K27ac:
-
Histone H3 lysine 27 acetylation
- H3K4me3:
-
Histone H3 lysine 4 trimethylation
- HSA21:
-
Human chromosome 21
- HLA:
-
Human leukocyte antigen
- HR:
-
Hormone receptor
- IHC:
-
Immunohistochemistry
- IC50:
-
Half-maximal inhibitory concentration
- IPMNs:
-
Intraductal papillary mucinous neoplasms
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- LASSO:
-
Least absolute shrinkage and selection operator
- OS:
-
Overall survival
- pCR:
-
Pathological complete response
- RF:
-
Random forest
- SEs:
-
Super enhancers
- SERS:
-
Super-enhancer-related score
- ssGSEA:
-
Single sample gene set enrichment analysis
- scRNA-seq:
-
Single cell RNA sequencing
- TIME:
-
Tumor immune microenvironment
- TCGA:
-
The cancer genome atlas
- TMB:
-
Tumor mutational burden
- TIICs:
-
Tumor-infiltrating immune cells
- TCIA:
-
The cancer immunome atlas
- TIDE:
-
Tumor immune dysfunction and exclusion
- TAARs:
-
Trace amine-associated receptors
- timeROC:
-
Time-dependent receiver operating characteristic curve
- XGBoost:
-
EXtreme gradient boosting
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263. https://doi.org/10.3322/caac.21834 (2024).
Zubair, M., Wang, S. & Ali, N. Advanced approaches to breast cancer classification and diagnosis. Front. Pharmacol. 11, 632079. https://doi.org/10.3389/fphar.2020.632079 (2020).
Herzog, S. K. & Fuqua, S. A. W. ESR1 mutations and therapeutic resistance in metastatic breast cancer: Progress and remaining challenges. Br. J. Cancer 126, 174–186. https://doi.org/10.1038/s41416-021-01564-x (2022).
Bai, X., Ni, J., Beretov, J., Graham, P. & Li, Y. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat Rev. 69, 152–163. https://doi.org/10.1016/j.ctrv.2018.07.004 (2018).
Kumar, H. et al. A review of biological targets and therapeutic approaches in the management of triple-negative breast cancer. J. Adv. Res. 54, 271–292. https://doi.org/10.1016/j.jare.2023.02.005 (2023).
Signor, S. A. & Nuzhdin, S. V. The evolution of gene expression in cis and trans. Trends Genet 34, 532–544. https://doi.org/10.1016/j.tig.2018.03.007 (2018).
Brown, D. D. Gene expression in eukaryotes. Oncodev. Biol. Med. 4, 9–29 (1982).
Wu, Y., Sarkissyan, M. & Vadgama, J. V. Epigenetics in breast and prostate cancer. Methods Mol. Biol. 1238, 425–466. https://doi.org/10.1007/978-1-4939-1804-1_23 (2015).
Thandapani, P. Super-enhancers in cancer. Pharmacol. Ther. 199, 129–138. https://doi.org/10.1016/j.pharmthera.2019.02.014 (2019).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947. https://doi.org/10.1016/j.cell.2013.09.053 (2013).
Tang, F., Yang, Z., Tan, Y. & Li, Y. Super-enhancer function and its application in cancer targeted therapy. NPJ. Precis. Oncol. 4, 2. https://doi.org/10.1038/s41698-020-0108-z (2020).
Jahan, R. et al. Odyssey of trefoil factors in cancer: Diagnostic and therapeutic implications. Biochim. Biophys. Acta Rev. Cancer 1873, 188362. https://doi.org/10.1016/j.bbcan.2020.188362 (2020).
Zhou, R. W. & Parsons, R. E. Etiology of super-enhancer reprogramming and activation in cancer. Epigenetics Chromatin 16, 29. https://doi.org/10.1186/s13072-023-00502-w (2023).
Weinstein, J. N. et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120. https://doi.org/10.1038/ng.2764 (2013).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, pl1. https://doi.org/10.1126/scisignal.2004088 (2013).
Cerami, E. et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. https://doi.org/10.1158/2159-8290.Cd-12-0095 (2012).
Wang, Y. et al. SEdb 2.0: A comprehensive super-enhancer database of human and mouse. Nucleic Acids Res. 51, D280–D290. https://doi.org/10.1093/nar/gkac968 (2023).
Pal, B. et al. A single-cell RNA expression atlas of normal, preneoplastic and tumorigenic states in the human breast. Embo J. 40, e107333. https://doi.org/10.15252/embj.2020107333 (2021).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Wu, T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. (Camb.) 2, 100141. https://doi.org/10.1016/j.xinn.2021.100141 (2021).
Yu, G. & He, Q. Y. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 12, 477–479. https://doi.org/10.1039/c5mb00663e (2016).
Sato, N., Tamada, Y., Yu, G. & Okuno, Y. CBNplot: Bayesian network plots for enrichment analysis. Bioinformatics 38, 2959–2960. https://doi.org/10.1093/bioinformatics/btac175 (2022).
Mayakonda, A., Lin, D. C., Assenov, Y., Plass, C. & Koeffler, H. P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 28, 1747–1756. https://doi.org/10.1101/gr.239244.118 (2018).
Aran, D., Hu, Z. & Butte, A. J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, 220. https://doi.org/10.1186/s13059-017-1349-1 (2017).
Yang, W. et al. Genomics of drug sensitivity in cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955-961. https://doi.org/10.1093/nar/gks1111 (2013).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573-3587.e3529. https://doi.org/10.1016/j.cell.2021.04.048 (2021).
Hu, C. et al. Cell Marker 2.0: An updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 51, 870–876. https://doi.org/10.1093/nar/gkac947 (2023).
Azizi, E. et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 174, 1293-1308.e1236. https://doi.org/10.1016/j.cell.2018.05.060 (2018).
Sun, D. et al. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat. Biotechnol. 40, 527–538. https://doi.org/10.1038/s41587-021-01091-3 (2022).
Jin, S., Plikus, M. V. & Nie, Q. Cell Chat for systematic analysis of cell-cell communication from single-cell transcriptomics. Nat. Protoc. 20, 180–219. https://doi.org/10.1038/s41596-024-01045-4 (2025).
Yuan, K. C. et al. The development an artificial intelligence algorithm for early sepsis diagnosis in the intensive care unit. Int. J. Med. Inform. 141, 104176. https://doi.org/10.1016/j.ijmedinf.2020.104176 (2020).
Franco, H. L. et al. Enhancer transcription reveals subtype-specific gene expression programs controlling breast cancer pathogenesis. Genome Res. 28, 159–170. https://doi.org/10.1101/gr.226019.117 (2018).
Chen, H. et al. A pan-cancer analysis of enhancer expression in nearly 9000 patient samples. Cell 173, 386-399.e312. https://doi.org/10.1016/j.cell.2018.03.027 (2018).
Li, W., Notani, D. & Rosenfeld, M. G. Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat. Rev. Genet. 17, 207–223. https://doi.org/10.1038/nrg.2016.4 (2016).
Kelly, M. R. et al. A multi-omic dissection of super-enhancer driven oncogenic gene expression programs in ovarian cancer. Nat. Commun. 13, 4247. https://doi.org/10.1038/s41467-022-31919-8 (2022).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319. https://doi.org/10.1016/j.cell.2013.03.035 (2013).
Weischenfeldt, J. et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 49, 65–74. https://doi.org/10.1038/ng.3722 (2017).
Spielmann, M., Lupiáñez, D. G. & Mundlos, S. Structural variation in the 3D genome. Nat. Rev. Genet. 19, 453–467. https://doi.org/10.1038/s41576-018-0007-0 (2018).
Northcott, P. A. et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434. https://doi.org/10.1038/nature13379 (2014).
Gröschel, S. et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 157, 369–381. https://doi.org/10.1016/j.cell.2014.02.019 (2014).
Yamazaki, H. et al. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell 25, 415–427. https://doi.org/10.1016/j.ccr.2014.02.008 (2014).
Drier, Y. et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat. Genet. 48, 265–272. https://doi.org/10.1038/ng.3502 (2016).
Deng, R. et al. Disruption of super-enhancer-driven tumor suppressor gene RCAN1.4 expression promotes the malignancy of breast carcinoma. Mol. Cancer 19, 122. https://doi.org/10.1186/s12943-020-01236-z (2020).
Wei, X. et al. A novel signature constructed by super-enhancer-related genes for the prediction of prognosis in hepatocellular carcinoma and associated with immune infiltration. Front. Oncol. 13, 1043203. https://doi.org/10.3389/fonc.2023.1043203 (2023).
Wang, A., Xia, H., Li, J., Diao, P. & Cheng, J. Development of a novel prognostic signature derived from super-enhancer-associated gene by machine learning in head and neck squamous cell carcinoma. Oral Oncol. 159, 107016. https://doi.org/10.1016/j.oraloncology.2024.107016 (2024).
Vaganova, A. N., Maslennikova, D. D., Konstantinova, V. V., Kanov, E. V. & Gainetdinov, R. R. The expression of trace amine-associated receptors (TAARs) in breast cancer is coincident with the expression of neuroactive ligand-receptor systems and depends on tumor intrinsic subtype. Biomolecules https://doi.org/10.3390/biom13091361 (2023).
Clusan, L., Ferrière, F., Flouriot, G. & Pakdel, F. A basic review on estrogen receptor signaling pathways in breast cancer. Int. J. Mol. Sci. https://doi.org/10.3390/ijms24076834 (2023).
Forbes, S. A. et al. COSMIC: Mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res. 39, D945-950. https://doi.org/10.1093/nar/gkq929 (2011).
Bayne, L. J. et al. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell 21, 822–835. https://doi.org/10.1016/j.ccr.2012.04.025 (2012).
Pylayeva-Gupta, Y., Lee, K. E., Hajdu, C. H., Miller, G. & Bar-Sagi, D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell 21, 836–847. https://doi.org/10.1016/j.ccr.2012.04.024 (2012).
Nagy, J. A., Chang, S. H., Dvorak, A. M. & Dvorak, H. F. Why are tumour blood vessels abnormal and why is it important to know?. Br. J. Cancer 100, 865–869. https://doi.org/10.1038/sj.bjc.6604929 (2009).
Slaney, C. Y., Kershaw, M. H. & Darcy, P. K. Trafficking of T cells into tumors. Cancer Res. 74, 7168–7174. https://doi.org/10.1158/0008-5472.Can-14-2458 (2014).
Koual, M. et al. Environmental chemicals, breast cancer progression and drug resistance. Environ. Health 19, 117. https://doi.org/10.1186/s12940-020-00670-2 (2020).
Hanker, A. B., Sudhan, D. R. & Arteaga, C. L. Overcoming endocrine resistance in breast cancer. Cancer Cell 37, 496–513. https://doi.org/10.1016/j.ccell.2020.03.009 (2020).
Dong, X. et al. Exosomes and breast cancer drug resistance. Cell Death Dis. 11, 987. https://doi.org/10.1038/s41419-020-03189-z (2020).
Soutto, M. et al. Loss of TFF1 is associated with activation of NF-κB-mediated inflammation and gastric neoplasia in mice and humans. J. Clin. Invest. 121, 1753–1767. https://doi.org/10.1172/jci43922 (2011).
Welter, C. et al. Expression pattern of breast-cancer-associated protein pS2/BCEI in colorectal tumors. Int. J. Cancer 56, 52–55. https://doi.org/10.1002/ijc.2910560110 (1994).
Taupin, D. & Podolsky, D. K. Trefoil factors: Initiators of mucosal healing. Nat. Rev. Mol. Cell Biol. 4, 721–732. https://doi.org/10.1038/nrm1203 (2003).
Emami, S. et al. Trefoil factor family (TFF) peptides and cancer progression. Peptides 25, 885–898. https://doi.org/10.1016/j.peptides.2003.10.019 (2004).
Qian, Z. et al. Validation of the DNA methylation landscape of TFF1/TFF2 in gastric cancer. Cancers (Basel) https://doi.org/10.3390/cancers14225474 (2022).
Ge, Y. et al. TFF1 inhibits proliferation and induces apoptosis of gastric cancer cells in vitro. Bosn. J. Basic Med. Sci. 12, 74–81. https://doi.org/10.17305/bjbms.2012.2499 (2012).
Bossenmeyer-Pourié, C. et al. The trefoil factor 1 participates in gastrointestinal cell differentiation by delaying G1-S phase transition and reducing apoptosis. J. Cell Biol. 157, 761–770. https://doi.org/10.1083/jcb200108056 (2002).
Rodrigues, S. et al. Induction of the adenoma-carcinoma progression and Cdc25A-B phosphatases by the trefoil factor TFF1 in human colon epithelial cells. Oncogene 25, 6628–6636. https://doi.org/10.1038/sj.onc.1209665 (2006).
Regalo, G., Wright, N. A. & Machado, J. C. Trefoil factors: From ulceration to neoplasia. Cell Mol. Life Sci. 62, 2910–2915. https://doi.org/10.1007/s00018-005-5478-4 (2005).
Yamaguchi, J. et al. Trefoil factor 1 suppresses stemness and enhances chemosensitivity of pancreatic cancer. Cancer Med. 13, e7395. https://doi.org/10.1002/cam4.7395 (2024).
Soubeyran, I. et al. Immunohistochemical determination of pS2 in invasive breast carcinomas: A study on 942 cases. Breast Cancer Res. Treat. 34, 119–128. https://doi.org/10.1007/bf00665784 (1995).
Corte, M. D. et al. Cytosolic levels of TFF1/pS2 in breast cancer: Their relationship with clinical-pathological parameters and their prognostic significance. Breast Cancer Res. Treat. 96, 63–72. https://doi.org/10.1007/s10549-005-9041-7 (2006).
Foekens, J. A. et al. Prognostic value of PS2 and cathepsin D in 710 human primary breast tumors: Multivariate analysis. J. Clin. Oncol. 11, 899–908. https://doi.org/10.1200/jco.1993.11.5.899 (1993).
Gion, M. et al. PS2 in breast cancer–alternative or complementary tool to steroid receptor status? Evaluation of 446 cases. Br. J. Cancer 68, 374–379. https://doi.org/10.1038/bjc.1993.343 (1993).
Henry, J. A. et al. Expression of the pNR-2/pS2 protein in diverse human epithelial tumours. Br. J. Cancer 64, 677–682. https://doi.org/10.1038/bjc.1991.380 (1991).
Longman, R. J., Thomas, M. G. & Poulsom, R. Trefoil peptides and surgical disease. Br. J. Surg. 86, 740–748. https://doi.org/10.1046/j.1365-2168.1999.01131.x (1999).
Prest, S. J., May, F. E. & Westley, B. R. The estrogen-regulated protein, TFF1, stimulates migration of human breast cancer cells. Faseb J. 16, 592–594. https://doi.org/10.1096/fj.01-0498fje (2002).
Williams, R., Stamp, G. W., Gilbert, C., Pignatelli, M. & Lalani, E. N. pS2 transfection of murine adenocarcinoma cell line 410.4 enhances dispersed growth pattern in a 3-D collagen gel. J. Cell Sci. 109(1), 63–71. https://doi.org/10.1242/jcs.109.1.63 (1996).
Funding
This research was supported by grants from the National Natural Science Foundation of China (81872135, 82002791) and the Funds for Distinguished Young Scientists of the Second Affiliated Hospital of Harbin Medical University.
Author information
Authors and Affiliations
Contributions
MCL, FM, and BLG designed the study. ZBF, YHJ, YLL, WLC, YQD, XLW, YHS, FJK, JWL, DLC and YLC collected the data. MCL, TSY, JYF, YSL, JRZ,TW, ABH, HYZ, ZYR, and SSS conducted the statistical analyses and visualized the results. MCL, ZBF, and YHJ collectively conceptualized the manuscript. FM, BLG and YSL edited the manuscript and provided critical comments. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki and approved by the Institutional Review Board of The Second Affiliated Hospital of Harbin Medical University.
Consent to participate
Informed consent was obtained from all subjects involved in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, M., Fang, Z., Ji, Y. et al. Identification of a super enhancer associated gene signature for the prognosis prediction and regulatory mechanism exploration in breast cancer. Sci Rep 15, 43517 (2025). https://doi.org/10.1038/s41598-025-26694-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-26694-7