Abstract
Understanding tissue complexity requires spatially resolved multi-omics data at single-cell resolution. Here, we present a workflow integrating high-resolution matrix-assisted laser desorption ionization mass spectrometry imaging (MALDI-MSI) with Xenium spatial transcriptomics (SPT) on a single tissue section. This strategy ensures pixel-scale spatial correspondence between metabolic and transcriptomic features, avoiding misalignment issues of serial sections, where even minor offsets result in sampling different cells. We investigated MALDI-MSI compatibility with downstream SPT revealing that the number of transcripts per cell decreased by ~ 30% after MSI, whilst cell recovery and cell-type assignments are preserved. Validated using mouse brain and demonstrated using human glioblastoma tissues, we achieved pixel-scale modality co-registration, enabling per-cell MALDI spectra extraction aligned with gene expression. Integrated clustering revealed enhanced cell-type resolution and identified metabolic heterogeneity within transcriptionally defined populations. This facilitates precise correlations of a cell’s function and its biochemical state, providing a holistic view of cellular function, heterogeneity, and interaction in health and disease. Our workflow provides a scalable path to multi-omic atlases, advancing both data integration and translational research.
Similar content being viewed by others
Introduction
Understanding the spatial distribution of biomolecules and their interactions is key for unravelling the complexities of biological systems, particularly in complex systems such as brain1. Modalities such as matrix-assisted laser desorption ionization-mass spectrometry imaging (MALDI-MSI) and spatial transcriptomics (SPT) have revolutionized spatial biology by enabling the visualization of metabolic and transcriptomic information directly within tissue section2,3. However, the integration of these modalities at a single cell level within the same tissue section remains a challenging yet promising area, offering unique insights into the molecular and cellular landscape of tissues.
MALDI-MSI enables the spatial detection of a wide range of molecular species, including lipids, metabolites, and proteins at a single cell level4,5. Although, MALDI-MSI is not inherently suitable for the detection of larger RNA panels due to the technical challenges associated with ionizing and detecting nucleic acids using MALDI-MSI6,7. RNA molecules are chemically distinct and more prone to degradation during sample preparation, and their relatively low abundance and complexity of ionization profiles further limit their detection with MALDI8. SPT, particularly with cutting-edge platforms like Xenium, complements this approach by providing single-cell resolution maps of RNA expressions9. The integration of these two modalities provides a comprehensive view of the molecular and transcriptomic states of individual cells, creating a deeper understanding of the interplay between molecular and genomic layers10. One of the advantages of integrating MALDI-MSI and SPT on the same tissue section lies in the removal of serial-section variability. When both modalities are performed on the same section, the spatial registration of data from both modalities is essentially the same, reducing artifacts introduced by tissue heterogeneity and sectioning. This approach can also ensure that molecular and transcriptomic data originate from the exact same cells and microenvironmental niches, preserving spatial fidelity and increasing confidence in biological inferences. Nevertheless, despite the clear advantages of single-section multi-omics, the potential impact of MALDI-MSI on subsequent spatial transcriptomics quality and performance has not been systematically evaluated. Research by Godfrey et al., has shown that combining DESI-MSI and Visium allow for understanding the relationship between gene expression and the metabolic phenotype of cells in the complex tumour microenvironment11. It needs to be mentioned that Visium lacks true single-cell resolution, it instead captures transcriptomes from spatially defined spots encompassing multiple cells. Consequently, Visium does not enable true single cell spatial multi-omics. Therefore, we have chosen Xenium for our spatial single cell approach.
Achieving single-cell resolution is essential for understanding tissue architecture and cellular dynamics in complex tissues. While previous studies have already integrated spatial transcriptomics and molecular imaging, they often lack the resolution required to resolve individual cells, instead providing data averaged over larger regions or tissue compartments11,12,13. This hinders accurate mapping of molecular features to specific cell types. In contrast, our approach enables single-cell co-mapping of metabolic and transcriptomic states and provides the basis for hypothesis-driven analyses of spatial metabolism at the individual cellular environment. This capability is critical for uncovering spatial relationships, identifying functional cell states, and detecting previously unresolved subpopulations14. By preserving both the spatial and cellular integrity, single-section integration of MALDI-MSI and spatial transcriptomics enhances the reliability of downstream analyses, such as cell type-specific molecular profiling and multimodal machine learning, while extensively reducing, tissue misalignment and variability relative to serial sections.
In this study, we assess the effect of MALDI-MSI prior to Xenium SPT and demonstrate the feasibility of integrating 5 µm spatial resolution MALDI-MSI measurements and the single-cell Xenium spatial transcriptomics platform on the same tissue section. By leveraging the complementary strengths of these technologies, we established a workflow that combines single-cell MALDI-MSI with single-cell SPT data. To our knowledge, this is the first demonstrations of true single-section MALDI-MSI and SPT analysis at cellular resolution, representing a significant technical advancement in spatial biology. As a proof of concept, we applied this approach to sagittal mouse brain sections, to provide a robust model to explore the integration of MALDI-MSI and SPT. The effect of MALDI-MSI on tissue integrity and the quality of spatial transcriptomics were evaluated to assess the impact of one technology on the other. Using human glioblastoma (GBM) tissue we established methods to spatially align and integrate data from MALDI-MSI and Xenium at single-cell resolution. We show the ability to obtain cell-specific MALDI-MSI spectra, linking molecular profiles to defined cell types. This work establishes a foundational framework for future studies that aim to link metabolic and transcriptomic data within single tissue sections on a per-cell basis providing powerful new insights into health and disease.
Results
Assess effects of prior MALDI-MSI on spatial transcriptomics tissue integrity and data quality
We first performed a series of validation experiments on adult mouse brain tissue to establish the feasibility of sequentially combining high-resolution MALDI-MSI with SPT. These preliminary studies served two purposes: to verify that the MALDI laser ablation and matrix application would not irreversibly damage tissue morphology, and to confirm that the semi-destructive MALDI step would not compromise downstream RNA capture or hybridization performance. Fresh-frozen mouse brain sections were mounted on Xenium-compatible slides, sublimated with 2,5-DHB matrix, and subjected to MALDI-MSI at 5 µm pixel size. Post-MSI, the same section underwent the Xenium SPT workflow. Cell boundaries were defined using Xenium Ranger. We observed that the MALDI-laser ablation produced a regular grid of micron-scale fluorescent spots within the intracellular matrix (Fig. 1-A), corresponding precisely to the MALDI-MSI pixel array. Fluorescent ablation fiducials were visible on-tissue but not off-tissue. These ablation marks did not obscure nuclear outlines, nor did they interfere with subsequent cell segmentation or identification. To assess whether ablation produced spatially periodic depletion or delocalization, we inspected transcript localizations relative to the 5 µm ablation grid and did not detect any grid-aligned patterns. RNA spots remained distributed with a uniform reduction in counts, cell segmentation and identification were preserved (Fig. 2). To assess whether MALDI-MSI would fully ablate tissue or remain limited to the surface, scanning electron microscopy (SEM) of a GBM-derived cell after MALDI-MSI demonstrates only surface-limited etching (Fig. 1-B). This is consistent with the observed preservation of nuclear outlines and segmentation. We directly compared counts in MALDI-MSI prior to SPT versus no MALDI-MSI prior to SPT (control) sections of mouse brain to quantify the impact of the MALDI laser on the transcript counts. Figure 1-C, shows the transcript counts per cell within the defined cell boundaries per condition. In Fig. 1-D, we aggregated transcript counts across five anatomically matched 1 mm2 regions in MALDI-MSI versus control sections. Boxplots of total RNA counts per cell and detected transcript count per mm2 reveal statistically significant reductions in both RNA yield and transcript detection following MALDI-MSI. Although MALDI measured regions exhibited moderate statistically significant reductions in total RNA counts (-20%, p < 0.05) and detected transcripts per cell (-30%, p < 0.005) compared to control tissues, more than 90% of cells in ablated zones still surpassed 250 detected transcripts. To visualize the approximately 30% count loss, we compared per-gene RNA abundances between MALDI-MSI measured and control regions, normalizing by tissue area and number of segmented cells. Across two similar ROI, the per-gene profiles were broadly similar; genes that were most abundant in control remained among the most abundant after MALDI-MSI as well as the proportional decreases rather than complete gene-dropouts (sup. Figure 1). This supports a predominantly uniform decrease of signal, consistent with the results of Fig. 1C-D and Fig. 2. Despite this decrease, overall transcriptome complexity remained high, confirming that the semi-destructive MALDI step imposes only minor impacts on the cell and its spatial transcriptome.
Impact of MALDI-MSI on tissue and spatial transcriptomics data quality using control mouse brain tissue and GBM-derived cells. (A) DAPI-based cell segmentation (nucleus + 5 µm) control versus MALDI-MSI regions. Maximum-intensity projections of DAPI fluorescence in control (left) and MALDI-MSI (right) measured mouse brain. White outlines indicate cell boundaries defined by DAPI segmentation with a 5 µm radial expansion. The MALDI spot grid appears as a regular array of fluorescent micro-dots on the MALDI-MSI measured section but does not interfere with nuclear detection or boundary detection. Scale bar: 50 µm. (B) Scanning electron micrograph of a GBM-derived cell on ITO after MALDI-MSI showing superficial surface modification. Boxplots compare (C) total transcript counts per cell and (D) number of detected transcripts per area (mm2) between MALDI-MSI measured and non-MALDI-MSI measured (Control) mouse brain sections (≥ 40,000 cells per section). For the transcripts per area, 5 anatomical similar regions of 1mm2 were analysed on both tissues. Statistical significance was assessed by paired student T-test (** p < 0.005; * p < 0.05).
Xenium transcript localizations and cell identifications in mouse brain with and without prior MALDI-MSI. Left: section (control) not measured with MALDI-MSI. Right: consecutive section after MALDI-MSI. Top row: transcript localizations over tissue background. Each coloured dot represents a detected transcript corresponding to the target RNA. Bottom row: Xenium cell segmentation and identifications; grayscale shows tissue background. Legend at right maps colours to RNA-based cell type labels. No grid-aligned pattern is observed after ablation. RNA spots remain distributed with a uniform reduction in counts. Nuclear outlines and downstream cell segmentation and identification are preserved. Scale bar: 100 µm.
Integration of MALDI-MSI with xenium spatial transcriptomics
The integration of MALDI-MSI and SPT provides comprehensive multimodal insights into tissue biology at single-cell resolution using human glioblastoma tissue. Here, we present a single sample proof-of-concept demonstration of the detailed overview of the workflow designed for integrating these two distinct, but complementary datasets obtained from the same tissue section (Fig. 3A). Firstly, MALDI-MSI and SPT measurements were sequentially performed on the same tissue section. The section was stained with a panel of fluorescent markers to enable cell segmentation: DAPI for nuclear identification, ATP1A1/CD45/E-Cadherin to define cell boundaries, 18S ribosomal RNA for interior RNA distribution, and αSMA/Vimentin for interior protein architecture. Following data acquisition, computational integration of both datasets is performed using an in-house MATLAB-based software platform we named ‘ESCDAT’ (sup. Figure 2). ESCDAT enables data handling and seamless spatial alignment between MALDI-MSI ion maps and spatial transcriptomics fluorescence images. Within ESCDAT, MALDI-MSI data is first binned into a lower resolution dataset (using a bin width of 1 Da), significantly enhancing computational efficiency and facilitating rapid preliminary analyses. Simultaneously, a fluorescence image from the spatial transcriptomics measurement is selected as the spatial reference for co-registration (Fig. 3B). Figure 3B. illustrates the practical approach to accurately overlay MALDI-MSI data with corresponding spatial transcriptomics data. In panel I, a representative m/z channel from the MALDI-MSI analysis is visualized, providing spatial distribution of the selected molecule across the entire tissue section. Panel II shows a fluorescence image of the same tissue section, displaying cellular and molecular structures through labelled fluorescence markers which serve as reference data for integration. A focused examination of the region indicated by the red boxes in panels I and II is presented in panels III through VI. Panel III demonstrates a zoomed-in MALDI-MSI image revealing a sharp corner of a deliberately created fiducial marker region. The corresponding fluorescence image (panel IV) distinctly shows tissue ablation dots created by the MALDI laser for each pixel, which were strategically utilized as fiducial markers for precise pixel co-registration. In panel V, the overlay of MALDI-MSI sampling points (red circles) onto the fluorescence image demonstrates accurate spatial alignment achieved by the co-registration method. Panel VI further highlights this alignment by presenting the MALDI-MSI data combined with cell boundary segmentation derived from SPT. A direct fluorescence-MALDI-MSI overlay (panel VII) demonstrates that the MALDI signal follows the fiducial grid per pixel, supporting pixel-scale alignment where fiducials are clear. Small local offsets may occur at edges with reduced fiducial contrast. The alignment of these modalities leveraged fiducial markers identifiable in both MALDI-MSI and fluorescence images. This step ensured precise spatial co-registration, essential for accurate downstream biological interpretation. Following spatial alignment, cell boundary coordinates obtained via Xenium Ranger using the cell boundary segmentation staining were imported into ESCDAT, allowing for extraction of high-resolution per-cell m/z spectra from the MALDI-MSI data. The resulting integrated multimodal dataset, combined high-resolution mass spectra (m/z interval of 0.01 Da) and spatially resolved gene expression profiles, was then analysed using Seurat v515,16,17. This integrated visualization clearly demonstrates how molecular data from MALDI-MSI align spatially with single-cell transcriptomic data, thereby enabling detailed multi-omic cellular profiling. Additional observations of interest include the precision and spatial resolution attainable through this method, confirming the potential in correlating distinct metabolic profiles identified by MALDI-MSI directly with specific cell types and their transcriptional states.
Overview of overlaying and combining MALDI-MSI and spatial transcriptomics data on human glioblastoma tissue. (A) Shows the schematic workflow used using ESCDAT. First MALDI-MSI (timsTOF fleX) and spatial transcriptomics (Xenium) are sequentially conducted on the same tissue section. Afterwards, using ESCDAT, a low mass resolution dataset is spatially aligned with the spatial transcriptomics data. Using fiducial markers and visible laser spots in the tissue generated via the MALDI laser. Using the cell boundaries from the cell segmentation staining, pixels per cell are averaged and a high-resolution m/z spectrum is created per cell. These data modalities are afterwards combined, and downstream analysis are performed using Seurat V5 and Python. (B) Visualization of co-registration MALDI-MSI with spatial transcriptomics. (I) A representative m/z ion map from MALDI-MSI showing the spatial distribution of a selected molecule across the tissue. (II) Fluorescence image of the same section with labelled markers as reference. (III-IV) Zoom into the red-boxed region: (III) MALDI-MSI view of a sharp fiducial marker corner; (IV) matching fluorescence view with the laser-ablation dots used as fiducials. (V) Overlay of MALDI sampling points (red circles) on the fluorescence image after co-registration. (VI) Final overlay of MALDI-MSI data with cell-boundary segmentation from spatial transcriptomics, demonstrating precise multimodal alignment. (VII) Direct overlay of fluorescence fiducials and MALD-MSI intensity map. The MALDI signal recapitulates the per-pixel fluorescence pattern, confirming grid-anchored, pixel-scale alignment in fully gridded regions.
Cell identification and cell-specific MALDI-MSI spectra
We profiled 8,318 cells across the human glioblastoma tissue that overlapped between MALDI-MSI and SPT, after applying filters to remove low quality cells (qv < 20). Initial dataset quality was assessed by examining total RNA counts (nCount_RNA) and detected transcript numbers (nFeature_RNA) across all predicted cell-type clusters (sup. Figure 3). The similar shapes and ranges of these distributions confirm consistent capture efficiency and sequencing depth amongst the cell types. Accurate cell-type identification was performed based on spatial transcriptomic profiles using Seurat v5 and log-normalization, leveraging predefined marker genes for distinct cell populations (sup. table 1)18,19,20,21,22,23,24,25,26,27,28,29,30. The cell identifications were set as a ground-truth for the cell identities throughout. These marker genes, selected based on prior biological knowledge, demonstrated high specificity and robust expression patterns within targeted cell types (sup. Figure 4). In every cell type panel, the distribution of scores peaks sharply in the matching cell-type group and remains low in all others. For example, the astrocyte signature has its highest values exclusively in cells predicted as astrocytes, and the endothelial cell signature peaks only in endothelial-predicted cells. Minor low-level RNA-expression signals in non-target groups reflect baseline expression and technical background. Further validation at the single-gene level confirms that each marker’s normalized expression is tightly confined to its predicted lineage. Analysis of individual marker-gene expression distributions (sup. Figure 5) confirms that each gene is predominantly expressed in its assigned cell-type cluster, with negligible off-target expression. For instance, NRP1 and PECAM1 localize exclusively to endothelial cells, and MOG and CAPN3 peaks are restricted to oligodendrocytes. These plots validate both our marker lists and the prediction strategy: each marker panel cleanly identifies its intended lineage with minimal crosstalk, further substantiating the accuracy of our annotations. To further resolve the transcriptional heterogeneity underlying our spatially defined cell-type assignments, we used Seurat to identify the top ten positively enriched genes per predicted cell type (ranked by average log2-fold change). We subsequently visualized the normalized expression of these 100 marker genes across all cells in a heatmap (sup. Figure 6), with cells ordered by their predicted cell type. Across all clusters, marker-gene expression is sharply confined to its associated cell-type segment, with minimal off-target signals. Similar analysis was performed for the MALDI-MSI markers. Using Seurat on the TIC-normalized MALDI assay, we generated a heatmap of the top 100 m/z features based on log-fold-change across all cell types for all cells (sup. Figure 7). Cells remain ordered by their predicted cell type. Unlike the binary on/off patterns seen in transcript data, MALDI-MSI data exhibits graded enrichment: certain m/z values are elevated in one cell type yet maintain low-level expression elsewhere. These subtler, non-binary distributions reflect the shared metabolic milieu of neighbouring cell types yet still show clear molecular signatures that can complement the RNA-based annotations. To illustrate within-lineage metabolic heterogeneity that is undetected by spatial transcriptomics alone, we examined m/z 546.52 within RNA-defined astrocytes in human GBM. Despite near-uniform expression of astrocytic marker transcripts, MALDI-MSI intensities at m/z 546.52 segregated astrocytes into high and low m/z 546.52 intensity subpopulations, with the high-intensity subgroup enriched in pathological different regions. A heat map of astrocytes for this feature shows a clear pattern, and a spatial overlay of astrocyte centroids reveals enrichment of the high-intensity cells within certain pathological areas (sup. Figure 8).
Combined molecular profiling of single cell data
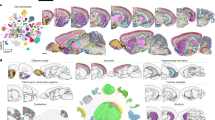
To compare modality structure before integration, we computed modality-specific UMAPs and plotted their spatial projections (sup. Figure 9). The RNA UMAP resolves multiple cell types. In contrast, the MALDI-only UMAP, reveals multiple metabolomic clusters that segregate regionally on the glioblastoma tissue. Within astrocytes, cells occupy more than one MALDI cluster and these astrocyte subclusters show distinct localization, whereas RNA-defined astrocytes remain broadly distributed on the tissue. Therefore, RNA emphasizes cell identity, while MALDI captures regional metabolic state and within-lineage heterogeneity. When both modalities are integrated per cell (Fig. 4-A), the embedding displays additional substructure relative to RNA alone. Distinct cell populations, including oligodendrocytes, astrocytes, endothelial cells, TAMs, and neuronal subpopulations, were clearly delineated. The integrated modalities can capture complementary molecular information, thus greatly refining the spatial resolution and biological interpretation compared to either modality alone. It also showed the value and necessity of integrating these complementary data modalities to enhance biological relevance. While RNA markers provide clear cell-type identification, MALDI-MSI data enriched the molecular profiling, particularly evident from the combined UMAP clusters. The modality contribution analysis (Fig. 4-B) reveals distinct patterns in the reliance on RNA versus MALDI-MSI data for clustering per cell type. Certain cell types, including astrocytes, neuronal populations, endothelial cells, GSCs and MES cells showed balanced modality contributions (close to a 50/50 ratio), suggesting equally important roles of transcriptomic and metabolic signatures for clustering. In contrast, other populations, such as oligodendrocytes, OPCs, TAMs and T-cells predominantly relied on RNA for accurate clustering. The increase of information by integrating SPT and MALDI-MSI is supported by visualizing all transcripts and all m/z values (sup. Figure 10) in a correlation heatmap. Here we saw several coherent clusters that show either positive or negative correlation. By systematically exploring the associations between top RNA marker genes characteristic of specific cell types and top MALDI-MSI ions (Fig. 4-C). To aid interpretation, we annotated ions in Fig. 4C where possible. LC–MS/MS supported lipids are shown without asterisk, and MS1-level, putative lipid assignments are marked with an asterisk (*) using common positive-mode adducts. Here we notably saw moderate correlations emerge between distinct cell-type markers and particular ions, suggesting functional and metabolic specificity for these cell populations. This correlation heatmap further suggests precise associations between specific transcripts and molecular ions, offering insights into cell-type-specific biological processes or metabolic pathways. As an illustration of strong gene-ion correlation, we visualize spatial co-localization of the oligodendrocyte marker MOBP with CE (18:2) (sup. Figure 11). These findings highlight variability in the molecular information content contributed by each modality across cell types and emphasize the complementary strengths of integrating both RNA and MALDI-MSI data to achieve additional biological information and understanding.
Integrated multimodal single-cell molecular profiling reveals distinct cell clusters in human glioblastoma. (A) Combined UMAP visualization of integrated spatial transcriptomics (RNA) and MALDI-MSI data. Distinct cell-type clusters, labelled according to RNA-based ground-truth. (B) Violin plots showing the contribution of each modality (RNA vs. MALDI-MSI) to cell-type clustering. Notably, some cell populations, such as astrocytes, endothelial cells, excitatory and inhibitory neurons, GSCs and MES cells, exhibit balanced contributions (~ 50% RNA and 50% MALDI-MSI) to the UMAP. whereas oligodendrocytes, OPCs, T-cells and TAMs predominantly rely on RNA data, indicating variability in modality-specific information content across cell types. (C) Cell-wise Pearson correlation matrix between the top 80 RNA marker genes and the top 80 MALDI-MSI ions (features selected from FindAllMarkers() and ranked by average log2 fold-change). Where available, ions are annotated to lipid classes. Labels without an asterisk are supported by LC–MS/MS from our published dataset on the same GBM samples, and labels with an asterisk (*) denote MS1-level, putative assignments based on adducts [M + H]+ and [M + Na]+ 31. Correlation patterns between RNA expression levels and molecular ions, hint at underlying biological or metabolic pathways concentrated within spatially localized cellular niches.
In this study, we established and demonstrate a workflow for integrating high-resolution MALDI-MSI with Xenium single cell spatial transcriptomics on one single tissue section, overcoming the limitations of prior approaches that relied on adjacent sections or lower-resolution co-registration11,13,32. This approach is motivated by the need to achieve a comprehensive, spatially precise, and cell-specific understanding of tissue biology. This cannot be reliably achieved using either modality alone or by analysing adjacent sections. First, we validated our approach in mouse brain, then applied it to human glioblastoma. We used 10 µm thick sections for this workflow. Although some applications report improved MALDI-MSI signal-to-noise at 5 µm, 10 µm is recommended for Xenium measurements on fresh-frozen tissue33. Because MALDI-MSI is surface-confined, the signal arises predominantly from the uppermost cell layer, preventing oversampling of multiple layers in the z-direction34,35. Consistent with this surface-confined effect, SEM of a GBM-derived cell imaged after MALDI-MSI shows superficial surface etching, aligning with the preserved nuclear outlines and the observed ~ 30% reduction in transcript counts rather than a complete loss of cells. We did not observe any grid-aligned patterns in Xenium transcript localizations after MALDI-MSI. Instead, the decrease was uniform across the field and cell segmentation and identification were preserved. This argues that MALDI-MSI sampling does not imprint periodic bias on downstream RNA measurements and supports the validity of per-cell analyses on the same section.
Sublimation of 2,5‐DHB and laser ablation at a 5 µm pixel pitch produced a characteristic fluorescent grid of ablation spots, yet nuclear morphology and cell segmentation remained entirely intact. Recent reports demonstrate matrix-dependent fluorescence enhancement through matrix-fluorophore cocrystallization, with significant differences across matrices and wavelengths, which could directly impact the visibility of optical fiducials used for registration36. Consistent with these observations, our 2,5-DHB matrix resulted sufficient ablation-spot contrast for alignment. Though our approach should, in principle, work with any matrix as long as fiducials are visible in both modalities and are compatible with Xenium chemistry. Importantly, we performed MALDI-MSI directly on Xenium slides, without requiring conventional conductive ITO-coated substrates, demonstrating that high-resolution MALDI imaging is feasible on transcriptomics compatible surfaces using a timsTOF fleX. This removes a key compatibility barrier between platforms, simplifies multimodal sample preparation, and enables broader adoption of integrated workflows. While the integration was successful, we observed moderate reductions in transcript counts following MALDI-MSI without any significant effect on cell type identifications37. Per-gene abundance profiles showed a uniform 30% attenuation without gene-specific dropouts, indicating that overall transcriptome complexity remains sufficient for stable labelling despite the MALDI step. This indicates that the transcriptomic data remains biologically informative, supporting reliable cell-type classification and downstream analyses, making the trade-off acceptable for integrated multimodal workflows. By co-registering MALDI-MSI and SPT on the same section, our maps permit within-cell-type comparisons across spatial niches (e.g., tumor core, perivascular, margin) at single-cell resolution.
Using MALDI-MSI and Xenium on the same section captures the identical tissue plane, enabling pixel-scale co-registration via fluorescent ablation fiducials and preserving per-cell boundaries. The main trade-off is a ~ 30% reduction in Xenium transcripts per cell, with all cells retained and labels unchanged. The count loss likely reflects local loss at ablation sites rather than complete cell loss; practical improvements could include laser fluency, fewer shots per pixel, and possibly RNA-preserving washes, with QC stratified by MALDI-exposed vs. unexposed areas. By contrast, consecutive sections could maximize RNA yield and protocol flexibility but introduce deformation and true 3D heterogeneity that does not allow one-to-one mapping. When precise per-cell correspondence is necessary, the same-section strategy is preferable; when maximal RNA coverage is the priority, consecutive sections are appropriate, with analyses emphasizing region-level or neighbourhood statistics.
Using the regular grid of MALDI-MSI laser ablation spots, ESCDAT enabled pixel-scale alignment of MSI ion maps with fluorescence-based cell segmentation, allowing per-cell mass spectra to be paired with spatial transcriptomes. While single cell RNA markers formed discrete, lineage-specific clusters, MALDI-MSI features showed graded biochemical enrichments. Cell-type specific masses were markedly elevated in their respective types but also showed low-level expression elsewhere, reflecting shared metabolic contexts38. RNA and MALDI-MSI separate different, yet complementary microenvironmental states. RNA primarily captures cell identity, resulting in separated lineage clusters that are spatially mixed across the section. In contrast, MALDI-MSI highlights regional metabolic states, producing broad clusters that segregate into distinct tissue areas. Same-section integration therefore enables within-cell-type comparisons across microenvironments, providing a way to interrogate microenvironmental reprogramming of GBM. While this single-case methods study stops short of pathway claims, these findings motivate cohort-level tests linking regional metabolite patterns to transcriptomic programs and pathology. Correlation heatmaps linking gene expression to m/z features revealed that specific cell types exhibit distinct combinations of transcripts and molecular ions, effectively defining unique cellular ‘metabotypes’. These gene-metabolite associations suggest that transcriptomic identity is tightly coupled to underlying biochemical states, enabling functional characterization of cells beyond RNA alone and suggests candidate gene-metabolite regulatory circuits for further investigation.
This methods study is limited in sample size; mouse brain validation and a single human GBM case. Our primary goal is to present a reproducible and open workflow for same-section MALDI-MSI and SPT co-registration. Consequently, our goal is technical validation and any biological insights given are illustrative and hypothesis-generating. Even so, the resulting multimodal maps point to potential new biological insights. The combined MALDI signals with RNA-defined clusters point to metabolic heterogeneity even among transcriptionally similar cells, hinting at subpopulations within the section. Unlike traditional 2D or 3D cultures, our single tissue-based workflow maintains cells in their native spatial and microenvironmental context, enabling the study of molecular and transcriptomic phenotypes as they exist in situ39. This allows not only for more biologically relevant single cell profiling but also supports future analysis of cell-to-cell interactions, by examining how neighbouring cells with distinct molecular identities could influence one another through spatial proximity. Looking forward, expanding our workflow to include targeted lipidomic, protein markers, and advanced statistical integration frameworks will further extend the understanding of cellular states in health and disease. As transcriptomic panels expand toward full-transcriptome coverage, our integrated modalities can enable a more refined classification. However, sensitivity for very low-abundance transcripts may be constrained, especially given the transcript reduction observed after MSI, so we suggest markers that remain robust under these conditions. By linking gene expression to cell-type-specific metabolic or signalling profiles, this integration enhances our ability to dissect disease mechanisms, identify biomarkers or drug targets, and distinguish cell states that are transcriptionally similar but functionally distinct. It also aligns with the goals of atlas-scale efforts, such as the Human Cell Atlas, by enabling the generation of spatially resolved, multi-omic reference maps that preserve both cellular identity and microenvironmental context40,41. Ultimately, this integrated MALDI-MSI/SPT platform lays the groundwork for comprehensive, single-cell resolved multi-omic atlases.
To enable broader adoption, key steps in our workflow such as matrix deposition, imaging, and registration are already compatible with high-throughput instrumentation. Our integration software (ESCDAT) is designed to be modular and will be further developed for other modalities and scalable deployment, such as multiple tissue sections. We also view future compatibility with open-MSI-formats, such as imzML, beneficial to the community. Also, the ability to generate ROIs using QuPath would be beneficial to the software. Compared with existing multimodal platforms, our method uniquely achieves subcellular co-registration of lipidomic and transcriptomic data without sacrificing either modality’s resolution. By generating truly cell-specific mass spectra that directly correspond to Xenium derived transcriptomes, our workflow sets a new standard for comprehensive multi-omic tissue mapping, enabling more precise investigation of metabolic reprogramming in tumour microenvironments and delivering a superior tool for both fundamental research and translational pathology.
Materials and methods
Sample preparation – MALDI-MSI
Ethical approval was obtained from the UZ/KU Leuven Ethical Review Committee of University Hospitals Leuven (Gasthuisberg, Leuven, Belgium) with reference number S60290 and the project was registered under the UZ Leuven Tissue Bank. The patients gave consent for the resected tissue samples to be included in the research. All experiments using human patient samples were conducted in compliance with the respective institutional guidelines. Fresh frozen mouse brain (n = 3) was obtained from the Department of Pathology at Maastricht University. All animal experiments were performed with appropriate approval granted by IvD (Instantie voor Dierenwelzijn, University Maastricht) and in compliance with the respective institutional guidelines. GBM tissue (n = 1) was resected and collected at the Department of Neurosurgery at KU Leuven. GBM tissue was embedded in 10% gelatin 2% CMC (SigmaAldrich). Afterwards mouse brain and GBM tissues were cryo-sectioned at 10 µm sections using a cryostat (CM1850, Leica Biosystems) according to Protocol CG000579 published by 10X Genomics. The sections were stored at − 80 °C until further analysis. Upon retrieval, they were immediately transferred to boxes filled with hygroscopic desiccant beads and subsequently vacuum-dried in a desiccation chamber for 20 min. Before MALD-MSI experiments, sublimation of 50 mg of 2,5-dihydroxybenzoic acid (Sigma-Aldrich) matrix at 160 °C for 160 s was performed using an HTX Sublimator (HTX Technologies, USA). As for the GBM patient-derived cell lines, fresh GBM tumor tissue was obtained from patients undergoing surgical resection at UZ Gasthuisberg after written informed consent (S59804). On receipt at the LPCM laboratory, specimens were immediately processed to establish patient-derived GBM stem-cell cultures (S61081) as described previously. For the data presented here, we highlight one donor (female, age 45). Approximately 100 cells (~ 1.5 × 105 cells/mL) were seeded onto poly-L-lysine coated ITO-glass slides suitable for MALDI-MSI, following published protocols (15). Cells were snap-frozen in liquid nitrogen and stored at -80 °C until measurement.
MALDI-MSI analysis
Whilst setting up the imaging-experiment, to create fiducial markers, several sharp corners and features on the tissue were created during the ROI selection (Fig. 2. B-III). MALDI-MSI analysis was performed on a timsTOF fleX equipped with microGRID (Bruker Daltonics GmbH, Germany). Spectra were obtained in positive ion mode with a mass range of m/z 250 – 1200 with a pixel size of 5 × 5 µm and laser spot size of approximately 4 µm. The laser frequency was set to 1 kHz, and 25 shots were accumulated at each pixel. The MALDI laser consists of a Nd:YAG 355 nm SmartBeam 3D laser (Ekspla, Lithuania). Time-of-flight calibration was performed before imaging experiments using red phosphorus. Transfer settings were 200 V peak-to-peak (Vpp; funnel 1 RF), 350 Vpp (funnel 2 RF), and 450 Vpp (multipole RF). Focus pre-time-of-flight (TOF) transfer time was set at 100 μs. The quadrupole ion energy was 5.0 eV with a low mass of m/z 200. Collision cell energy was 10.0 eV with collision RF at 1500 Vpp. MALDI-MSI features were annotated by cross-referencing LC–MS/MS identifications as previous analysed by us on the same human GBM sample (supplementary text 1)31,42. MS1-level matches to LIPID MAPS using positive-mode adducts [M + H] + and [M + Na] + within the instrument mass tolerance (0.01 Da)43.
SEM analysis of single cell
Following MALDI-MSI of the GBM patient-derived cell line, slides were dried in a desiccator at room temperature. To ensure surface conductivity for high-vacuum imaging, samples were gold coated by sputter deposition (~ 5 – 10 nm Au) SEM-imaging was performed on a field-emission scanning electron microscope (ETD secondary-electron detector) (Aquilos 2, Thermo Fisher) under high vacuum. Instrument settings for the representative micrograph were: accelerating voltage 10.0 kV, working distance 6.3 mm, beam current 1.6 nA, magnification 5,000x (horizontal field width 41.4 µm), chamber pressure 7.88 × 10–4 Pa.
Sample preparation – spatial transcriptomics
After MALDI-MSI, to remove any excess 2,5-DHB, slides were washed for two times 1 min in 100% EtOH (BioSolve). Slides were then dried under a steady N2 stream. The Xenium slides were then fixed and permeabilized following Protocol CG000581. In addition to this protocol, slides were fully submerged in PBS at 37 °C for 2 min. Afterwards, any gelatin covering the Xenium slide fiducials was manually removed using a tissue (KimTech wipes, Sigma Aldrich). Probe hybridization, ligation, and amplification were carried out using the pre-designed Mouse Brain and Human Brain reagent in accordance with Protocol CG000749. Reagents for the Xenium measurements were bought according to Protocol CG000601. Finally, the processed slides were loaded into the Xenium Analyzer as per Protocol CG000584. After the Xenium analysis, raw FASTQ files were analysed using Xenium Ranger (v2.0.0.10) using the Xenium multimodal cell segmentation method. The sagittal mouse brain sections were processed using the Xenium nuclear cell expansion (5 µm). Data was then visualized in Xenium Explorer (v3.2.0) and processed using the Seurat R package (v5.3.0).
Integration of MALDI-MSI and spatial transcriptomics
Single cells detected via the Xenium multimodal cell segmentation were converted to a GeoJSON format containing the cell-identifier, cell cluster, individual transcript count and spatial coordinates using our custom python script. Afterwards, Tims-TOF MALDI-MSI and Xenium spatial transcriptomics datasets were aligned using our custom co-registration software (Version P1 04–06-2025) compiled in MATLAB (v2022b). First, the Bruker Tims-TOF binary (.tsf) files were imported via Python, and a low-resolution 1 Da mass-spectrometry image was generated. We then created either a total ion current (TIC) image or a single-mass-channel image and matched it to the fluorescence microscope image (.tiff, .ome.tiff, .png, .jpg or .svs) produced during Xenium’s multi-modal cell-segmentation staining. MALDI-laser ablation spots are readily visible in the fluorescence channel, providing precise markers for pixel-scale alignment. Tissue fiducials placed before MALDI-MSI were co-localized with these laser spots to match at least four corresponding registration points. A piecewise-linear (PWL) transformation was then computed, producing a transformation matrix that links coordinates across modalities. Finally, we applied this matrix to the GeoJSON file exported from the Xenium run containing cell IDs, spatial coordinates, and transcript counts to extract high-resolution mass spectra for each cell-id within the combined coordinate frame. MALDI-MSI peaks were extracted based on TIC-normalized intensity ≥ 5. To account for partial-volume effects and varying cell-pixel overlap, we weighted each MALDI pixel’s intensity by the fraction of its area overlapping the cell segmentation mask, as demonstrated before44.
Data processing and cell type identification
Xenium data was processed using the Seurat R package (v5.3.0). Filtering of cells excluded all low-quality cells with a quality value (qv) of < 20. Also, any empty cells with an nCount lower than 0 were removed. Further analysis was only conducted on common cells between the MALDI-MSI and Xenium measurement. The MALDI-MSI data was imported as a new assay in the Xenium Seurat object. Data for both modalities were normalized and scaled. Afterwards, PCA was conducted on both for the optimal number of PCs, found via elbow plots and PC expression heatmaps. Dimensionality and visualization were performed using UMAP. Any clusters with overall low nCount were removed, and PCA and UMAP visualization were performed again on the filtered object. Cell identifications in the clusters were characterized and visualized using expressed marker genes from the Human Brain panel. Cell-type scores were computed using Seurat AddModuleScore() on log-normalized counts. We did not apply expression cutoffs. Low-level scores denote near-zero module scores in non-target groups relative to the cell type distribution.
Differential marker analysis per cell type
To identify both transcriptomic and m/z features that are distinguishable for each predicted cell type, we performed cluster-wise differential testing using Seurat v5. First, for RNA markers, the RNA expression values were log normalized. We ran a FindAllMarkers() test which employs a Wilcoxon rank-sum test and controls the false discovery rate via Bonferroni correction, top markers were ranked by log2 fold-change. From the resulting table, we selected the top ten genes per cluster ranked by average log2-fold change and visualized in a grouped heatmap. For the m/z features, we used TIC-normalized MALDI data and performed similar tests. To jointly leverage transcriptomic and mass spectrometric modalities, we employed Seurat v5’s weighted nearest-neighbour (WNN) framework. First, we computed independent principal component analyses on the RNA and TIC-normalized MALDI assays, retaining the first 30 PCs from each modality. We constructed a WNN graph from both RNA and MALDI-MSI PC spaces and generated a UMAP for visualization. For feature-pair analysis, per-cell scaled matrices were correlated using Pearson’s r (cor, “pairwise.complete.obs”). For visualizations we displayed an 80 × 80 subset ranked by average log2 foldchange.
Data availability
Raw and processed MALDI-MSI and Xenium spatial transcriptomics data from the mouse measurements, including Seurat objects, co-registration overlays, and associated analysis scripts, are available from the corresponding author upon reasonable request. Please note that glioblastoma data cannot be shared directly, as this falls outside the scope of the approved ethical consents. Access to this data requires additional institutional review and adherence to the original patient consent.
Code availability
We used ESCDAT Version P1 04-06-2025 for the overlay in this manuscript. The software packages such as R, Seurat and Python are publicly available and well documented. The Xenium to GeoJSON package and MALDI-MSI – Xenium SPT software ‘ESCDAT’ underlying these analysis are deposited at GitHub (https://github.com/M4i-Imaging-Mass-Spectrometry/MALDI-MSI---Spatial-Transcriptomics-Overlay).
References
Fangma, Y., Liu, M., Liao, J., Chen, Z. & Zheng, Y. Dissecting the brain with spatially resolved multi-omics. J. Pharm. Anal. 13(7), 694–710. https://doi.org/10.1016/j.jpha.2023.04.003FromNLMPubMed-not-MEDLINE (2023).
Janesick, A. et al. High resolution mapping of the tumor microenvironment using integrated single-cell, spatial and in situ analysis. Nat. Commun. 14(1), 8353. https://doi.org/10.1038/s41467-023-43458-xFromNLMMedline (2023).
Mirzazadeh, R. et al. Spatially resolved transcriptomic profiling of degraded and challenging fresh frozen samples. Nat. Commun. 14(1), 509. https://doi.org/10.1038/s41467-023-36071-5FromNLMMedline (2023).
Sekera, E. R. et al. Single cell MALDI-MSI analysis of lipids and proteins within a replicative senescence fibroblast model. J. Am. Soc. Mass. Spectrom. 35(12), 2815–2823. https://doi.org/10.1021/jasms.4c00095FromNLMMedline (2024).
McKinnon, J. C. et al. Enhancing metabolite coverage in MALDI-MSI using laser post-ionisation (MALDI-2). Anal. Methods 15(34), 4311–4320. https://doi.org/10.1039/d3ay01046eFromNLMMedline (2023).
Zhang, H. et al. Mass spectrometry imaging for spatially resolved multi-omics molecular mapping. Npj Imaging 2(1), 20. https://doi.org/10.1038/s44303-024-00025-3FromNLMPubMed-not-MEDLINE (2024).
Vanderschoot, K.A. et al. Spatial multiomics lipids and gene expression using MALDI ISH MSI. BioRxiv. https://doi.org/10.1101/2024.06.01.596997 (2024).
Emanuelson, C., Ankenbruck, N., Deiters, A. & Yu, M. S. High-throughput amenable MALDI-MS detection of RNA and DNA with on-surface analyte enrichment using fluorous partitioning. SLAS Discov. 26(1), 58–66. https://doi.org/10.1177/2472555220958391FromNLMMedline (2021).
Liu, Q. et al. Single-cell, single-nucleus and xenium-based spatial transcriptomics analyses reveal inflammatory activation and altered cell interactions in the hippocampus in mice with temporal lobe epilepsy. Biomark. Res. 12(1), 103. https://doi.org/10.1186/s40364-024-00636-3FromNLMPubMed-not-MEDLINE (2024).
Zheng, P. et al. Integrated spatial transcriptome and metabolism study reveals metabolic heterogeneity in human injured brain. Cell Rep. Med. 4(6), 101057. https://doi.org/10.1016/j.xcrm.2023.101057FromNLMMedline (2023).
Godfrey, T. M. et al. Integrating ambient mass spectrometry imaging and spatial transcriptomics on the same cancer tissues to identify gene-metabolite correlations. BioRxiv. https://doi.org/10.1101/2024.12.20.626670 (2024).
Kreutzer, L. et al. Simultaneous metabolite MALDI-MSI, whole exome and transcriptome analysis from formalin-fixed paraffin-embedded tissue sections. Lab. Invest. 102(12), 1400–1405. https://doi.org/10.1038/s41374-022-00829-0FromNLMMedline (2022).
Vicari, M. et al. Spatial multimodal analysis of transcriptomes and metabolomes in tissues. Nat. Biotechnol. 42(7), 1046–1050. https://doi.org/10.1038/s41587-023-01937-yFromNLMMedline (2024).
Huynh, K. L. A. et al. Spatial deconvolution of cell types and cell states at scale utilizing TACIT. bioRxiv. https://doi.org/10.1101/2024.05.31.596861 (2024) (from NLM PubMed-not-MEDLINE).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33(5), 495–502. https://doi.org/10.1038/nbt.3192FromNLMMedline (2015).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36(5), 411–420. https://doi.org/10.1038/nbt.4096FromNLMMedline (2018).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42(2), 293–304. https://doi.org/10.1038/s41587-023-01767-yFromNLMMedline (2024).
Li, J. et al. Redistribution of the astrocyte phenotypes in the medial vestibular nuclei after unilateral labyrinthectomy. Front. Neurosci. 17, 1146147. https://doi.org/10.3389/fnins.2023.1146147FromNLMPubMed-not-MEDLINE (2023).
Yang, Y. et al. Four specific biomarkers associated with the progression of glioblastoma multiforme in older adults identified using weighted gene co-expression network analysis. Bioengineered 12(1), 6643–6654. https://doi.org/10.1080/21655979.2021.1975980FromNLMMedline (2021).
Amin, M. et al. Myelin oligodendrocyte glycoprotein (MOG) antibodies in a patient with glioblastoma: Red flags for false positivity. J. Neuroimmunol. 361, 577743. https://doi.org/10.1016/j.jneuroim.2021.577743FromNLMMedline (2021).
Ye, D. et al. Identifying genes that affect differentiation of human neural stem cells and myelination of mature oligodendrocytes. Cell Mol. Neurobiol. 43(5), 2337–2358. https://doi.org/10.1007/s10571-022-01313-5FromNLMMedline (2023).
Hayashida, S. et al. Distinct microglial and macrophage distribution patterns in the concentric and lamellar lesions in Balo’s disease and neuromyelitis optica spectrum disorders. Brain Pathol. 30(6), 1144–1157. https://doi.org/10.1111/bpa.12898FromNLMMedline (2020).
Mecca, C., Giambanco, I., Donato, R. & Arcuri, C. Microglia and aging: The role of the TREM2-DAP12 and CX3CL1-CX3CR1 axes. Int. J. Mol. Sci. 19(1). https://doi.org/10.3390/ijms19010318 (2018) (from NLM Medline).
Ravi, V. M. et al. T-cell dysfunction in the glioblastoma microenvironment is mediated by myeloid cells releasing interleukin-10. Nat. Commun. 13(1), 925. https://doi.org/10.1038/s41467-022-28523-1FromNLMMedline (2022).
Lichtenberger, B. M. et al. Autocrine VEGF signaling synergizes with EGFR in tumor cells to promote epithelial cancer development. Cell 140(2), 268–279. https://doi.org/10.1016/j.cell.2009.12.046FromNLMMedline (2010).
Tasic, B. et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 563(7729), 72–78. https://doi.org/10.1038/s41586-018-0654-5FromNLMMedline (2018).
Kim, M. H. et al. Target cell-specific synaptic dynamics of excitatory to inhibitory neuron connections in supragranular layers of human neocortex. Elife 12. https://doi.org/10.7554/eLife.81863 (2023) (from NLM Medline).
Ooki, A. et al. Epigenetically regulated PAX6 drives cancer cells toward a stem-like state via GLI-SOX2 signaling axis in lung adenocarcinoma. Oncogene 37(45), 5967–5981. https://doi.org/10.1038/s41388-018-0373-2 (2018).
Krepela, E. et al. Regulation of fibroblast activation protein by transforming growth factor Beta-1 in glioblastoma microenvironment. Int. J. Mol. Sci. 22 (3). https://doi.org/10.3390/ijms22031046 (2021) (from NLM Medline).
Li, Y. et al. MGST1 expression is associated with poor prognosis, enhancing the Wnt/beta-catenin pathway via regulating AKT and inhibiting ferroptosis in gastric cancer. ACS Omega 8(26), 23683–23694. https://doi.org/10.1021/acsomega.3c01782FromNLMPubMed-not-MEDLINE (2023).
Hendriks, T. F. E. et al. MALDI-MSI-LC-MS/MS workflow for single-section single step combined proteomics and quantitative lipidomics. Anal. Chem. 96(10), 4266–4274. https://doi.org/10.1021/acs.analchem.3c05850 (2024).
Wess, M. et al. Spatial integration of multi-omics data from serial sections using the novel multi-omics imaging integration toolset. Gigascience 14. https://doi.org/10.1093/gigascience/giaf035 (2025) (from NLM medline).
Wang, X., Zhang, L., Xiang, Y., Ye, N. & Liu, K. Systematic study of tissue section thickness for MALDI MS profiling and imaging. Analyst 148(4), 888–897. https://doi.org/10.1039/d2an01739cFromNLMMedline (2023).
Joignant, A. N., Knizner, K. T., Xi, Y. & Muddiman, D. C. Evaluating the optimal tissue thickness for mass spectrometry imaging using infrared matrix-assisted laser desorption electrospray ionization. Rapid Commun. Mass Spectrom. 37(22), e9638. https://doi.org/10.1002/rcm.9638FromNLMMedline (2023).
Dreisewerd, K. Recent methodological advances in MALDI mass spectrometry. Anal. Bioanal. Chem. 406(9–10), 2261–2278. https://doi.org/10.1007/s00216-014-7646-6FromNLMMedline (2014).
Yang, E. et al. FluoMALDI microscopy: Matrix co-crystallization simultaneously enhances fluorescence and MALDI imaging. Adv. Sci. (Weinh) 10(35), e2304343. https://doi.org/10.1002/advs.202304343FromNLMMedline (2023).
Tuck, M., Grelard, F., Blanc, L. & Desbenoit, N. MALDI-MSI towards multimodal imaging: Challenges and perspectives. Front. Chem. 10, 904688. https://doi.org/10.3389/fchem.2022.904688FromNLMPubMed-not-MEDLINE (2022).
Ali, A. et al. Single cell metabolism: Current and future trends. Metabolomics 18(10), 77. https://doi.org/10.1007/s11306-022-01934-3FromNLMMedline (2022).
Kapalczynska, M. et al. 2D and 3D cell cultures - A comparison of different types of cancer cell cultures. Arch. Med. Sci. 14(4), 910–919. https://doi.org/10.5114/aoms.2016.63743FromNLMPubMed-not-MEDLINE (2018).
Rood, J. E. et al. The Human Cell Atlas from a cell census to a unified foundation model. Nature 637(8048), 1065–1071. https://doi.org/10.1038/s41586-024-08338-4FromNLMMedline (2025).
Wang, L. et al. A single-cell atlas of glioblastoma evolution under therapy reveals cell-intrinsic and cell-extrinsic therapeutic targets. Nat. Cancer 3(12), 1534–1552. https://doi.org/10.1038/s43018-022-00475-xFromNLMMedline (2022).
Hendriks, T. F. E., Birmpili, A., de Vleeschouwer, S., Heeren, R. M. A. & Cuypers, E. Integrating rapid evaporative ionization mass spectrometry classification with matrix-assisted laser desorption ionization mass spectrometry imaging and liquid chromatography-tandem mass spectrometry to unveil glioblastoma overall survival prediction. ACS Chem. Neurosci. 16(6), 1021–1033. https://doi.org/10.1021/acschemneuro.4c00463FromNLMMedline (2025).
Conroy, M. J. et al. LIPID MAPS: Update to databases and tools for the lipidomics community. Nucleic Acids Res. 52(D1), D1677–D1682. https://doi.org/10.1093/nar/gkad896FromNLMMedline (2024).
Scupakova, K., Dewez, F., Walch, A. K., Heeren, R. M. A. & Balluff, B. Morphometric cell classification for single-cell MALDI-mass spectrometry imaging. Angew. Chem. Int. Ed. Engl. 59(40), 17447–17450. https://doi.org/10.1002/anie.202007315FromNLMMedline (2020).
Acknowledgements
The authors would like to thank Dr. Veerle Melotte and Lisa Mayrginther for the mouse brain tissue (Department of Pathology, Maastricht University). Dr. Morgane Rouault, Henk Buermans and Najiba Mammadova (10X Genomics) for performing the mouse Xenium Spatial Transcriptomics experiment. Dr. Frederik de Smet (KU Leuven) for the GBM patient-derived cell line. Hans Duimel (Department of Nanoscopy, Maastricht University) for assisting with the SEM. Ruben Jacobs (Maastricht University) for performing the human glioblastoma Xenium Spatial Transcriptomics experiment and Ian G.M. Anthony (Maastricht University) for helping with the python code. The authors would also like to thank the Department of Neurosurgery at KU Leuven, especially Prof. Dr. Steven de Vleeschouwer, for providing the GBM sample.
Funding
This research was supported by the LINK program funded through the Netherlands Organization for Scientific Research (NWO) and NWO-STEM (Project Number 19013 to E.C.). We also gratefully acknowledge the support of the FWO Research Foundation, Belgium (TBM T001919N) and the Interreg Flanders-Netherlands Molecular Brain Tumor Detector.
Author information
Authors and Affiliations
Contributions
T.F.E.H. contributed to the investigation, formal analysis, conceptualization, writing – review, and editing; G.B.E. contributed to software design and development, conceptualization and review; T.V. contributed to conceptualization, writing and review; B.B. contributed to software design and review; R.M.A.H. contributed to review and editing, funding acquisition; E.C. contributed to conceptualization, supervision, writing—review and editing, funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hendriks, T.F.E., Eijkel, G.B., Visvikis, T. et al. One section, two worlds: single-cell integration of MALDI-MSI and spatial transcriptomics on the same single tissue section. Sci Rep 15, 42660 (2025). https://doi.org/10.1038/s41598-025-26735-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-26735-1