Abstract
De novo genome assembly allows for the genome reconstruction of an organism without using a reference sequence. Assembly results depend on various sequencing technologies that generate data with differing fidelity, read lengths, and coverage levels, as well as on performance of a wide variety of algorithms. These attributes generate a diversity of assemblies for each single genome, which collectively defines the pan-assembly. In this study, we aimed to benchmark pan-assemblies of the prokaryotic models Bartonella henselae, Escherichia coli, Pseudomonas aeruginosa, and Xylella fastidiosa, using different attributes and their impact on metrics of the 3 C (contiguity, correctness, and completeness) criterion for selecting the best conditions for de novo genome assembly. Results showed that short-read assembly strategies presented higher accuracy with fewer errors (high correctness) and a high degree of completeness but lower contiguity due to fragmented assemblies. In contrast, long-read-based strategies showed high contiguity but lower completeness and accuracy. The hybrid strategy yielded the best overall results across all parameters by leveraging the strengths of both types of technology. Regarding assembly algorithms, Unicycler was the top assembler in 3 C metrics, using any of the short-read (compared to Megahit), long-read (compared to Canu), or hybrid strategies (compared to Wengan). Overall, the hybrid approach with Unicycler proved to be the best general approach for genome assembly of the four bacterial models. Finally, regarding coverage depth, increasing depth did not significantly affect assembly quality results if a minimum data level was maintained, indicating that high-quality assemblies can be achieved using moderate coverage levels. Jointly, the results of the pan-assembly provide working conditions for de novo genome assembly that can be applied to bacterial models of interest, guiding the selection of optimized experimental and bioinformatics conditions while reducing sequencing costs for generating high-quality sequences.
Similar content being viewed by others
Introduction
Whole genome assembly is the reconstruction of a nucleotide sequence that represents the actual genome from an organism, in which fragmented DNA pieces on average covered multiple times (or raw sequencing read data) are used1,2. When no prior knowledge of the source DNA sequence is assumed, a strategy called de novo genome assembly is considered. This approach is essential for investigating species in which a reference genome is unavailable or is not considered representative of the target genome due to a spectrum of genetic variations3.
In de novo assembly, sequencing reads are assembled into consensus sequences named “contigs”, that together represents most of the genome at a first level of assembly2. Several graph-based algorithms are available for this purpose, including Overlap-Layout-Consensus, de Bruijn and greedy approaches4,5. A subsequent level is the assembly of scaffolds. Scaffolding aims to bridge the gaps between the contigs using experimental data (for example mate pairs and paired end sequencing) or a reference genome1, in which nucleotides of unsolved regions are represented with letter “N”. When all gaps are solved (gap closing) by generating longer contigs6 or Polymerase Chain Reaction7 for example, a complete genome is assumed as a final level of assembly.
Regarding the generation of reads by sequencing technology, strategies can be separated based on the read length in short- or long-read sequencing. In short-read sequencing, such as Illumina’s instruments (second-generation sequencing), reads can be produced with a length of up to 600 bases, accuracy > 99.9% and cost-effective8. In contrast, classical long-reads or third-generation strategies, such as Pacific Biosciences’ (PacBio) single-molecule real-time sequencing and Oxford Nanopore Technologies’ (ONT) sequencing, reads are obtained with > 10 kb and 85–90% accuracy with a higher cost3,8. Advances in PacBio technology have consolidated the HiFi sequencing method, which yields highly long-read sequencing with accuracy > 99.5%9 but remains comparatively expensive. Recent advances in ONT sequencing technology have significantly enhanced both its hardware and data analysis pipelines, reducing error rates to approximately 6%10. Continuous methodological improvements are further increasing ONT accuracy; however, certain applications—especially those requiring single-nucleotide resolution—still benefit from the complementary use of short-read sequencing10,11.
Notwithstanding these technologies have been widely used to determine the DNA sequence of thousands of genomes, a diversity of candidate genome sequences can be obtained depending not only on the experimental procedures but also on the bioinformatic pipelines5,12. These parameters include genome complexity (repeats, number of chromosomes, mobilome), sample conditions, DNA extraction protocol, reads length and sequencing technology, sequencing depth (number of times that a given nucleotide has been read in an experiment), algorithm to assemble sequence (assemblers), databases, and others1,13. In addition, criteria to benchmark assemblies and selecting a winner are also complicated, which depends on the aim of the study14,15. Due to all this, to de novo genome assembly is not straightforward and is still a very classical and challenging problem in bioinformatics2,6.
To provide some insights to continue overcoming these challenges, we previously proposed the 3 C criterion to compare and assess de novo assemblies based on Contiguity (pieces of the assembled genome), Correctness (fidelity of the assembly) and Completeness (ability to assemble expected genes) using different assemblers with short- and long-reads for a bacterial model14,16. In line with the features of each technology and previous works1,5,6,9,17, assemblies only using short reads (Illumina) resulted with a high correctness and completeness (not considering HiFi sequencing or improved ONT), while best values for contiguity were obtained for long-reads. Best values for all metrics were obtained when both strategies were used at the same time in hybrid assemblies.
As a proof of concept, in this current work we extend the assessment of a collection of assemblies for a specific organism, a concept that we here termed “pan-assembly”, to select the best assembly based on the 3 C criterion. For this purpose, we selected four prokaryotic models in which short- and long-reads were used to assemble the genome with six assemblers (two short-reads only, two long-reads only, and two hybrid assemblers), and different levels of sequencing depth of coverage. The impact of these conditions on several 3 C parameters was quantified. Thus, the aim of the study was to benchmark pan-assemblies of the prokaryotic models Bartonella henselae, Escherichia coli, Pseudomonas aeruginosa, and Xylella fastidiosa, using different attributes and their impact on metrics of the 3 C (contiguity, correctness, and completeness) criterion for selecting the best conditions for de novo genome assembly.
Methods
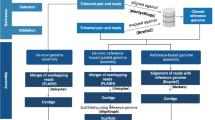
With the aim of providing conditions for generating a high-quality assembly for five bacterial models, a comparative assembly approach was implemented with three strategies (data source: short-reads, long-reads, or both), with two algorithms per strategy and different sequencing depth levels.
Data source and pre-processing
Sequencing data were retrieved from Sequence Read Archive database (SRA-NCBI, https://www.ncbi.nlm.nih.gov/sra) for four bacterial models: Bartonella henselae, Xylella fastidiosa, Escherichia coli, and Pseudomonas aeruginosa (Table 1). Data were selected based on the FDA-ARGOS framework (https://www.fda.gov/medical-devices/science-and-research-medical-devices/ database-reference-grade-microbial-sequences-fda-argos) or similar projects.
Quality control was performed with FASTQC v.0.11.9 tool18. Trimmomatic v.0.3919 was used for eliminating low-quality bases (Q < 30) and adapters.
Estimation of sequencing depth (also called depth or coverage, or simply coverage) was achieved by first mapping reads to the corresponding a reference sequence (from NCBI) with the BWA-MEM 0.7.5a-r405 software20. Then, alignment was examined with Qualimap 2.3 platform21 to obtain several metrics, including sequencing depth.
Pan-assembly of bacterial genomes
A standardized bioinformatics protocol was developed to assemble and annotate genomes for each bacterium. AnalysEs were run using the High-Performance Computing Cluster of the Center for Research in Materials Science and Engineering (CICIMA-HPC), University of Costa Rica. Raw sequencing data were subsampled based on sequencing depth (Table 1) using Seqtk 1.4 software (https://github.com/lh3/seqtk). Thus, trimmed reads subsets were generated with a sequencing depth of 12.5X, 25X, 50X, 100X, 200X* and 400X* (*: when possible). Depth was classified as low when < 100X, medium for 100X and high for > 100X.
Using trimmed data in each level of sequencing depth, two algorithms were implemented for each of the approaches using short-reads only, long-reads only, and hybrid (both short- and long-reads). The short-reads were processed with Unicycler v0.4.722 and Megahit v1.1.323. Assemblers for long-reads were Unicycler v0.4.7, and Canu v1.824. Finally, Wengan25 and Unicycler v0.4.7 were implemented for hybrid assemblies. Jointly, all assemblies in the different conditions for a single genome were considered the pan-assembly.
Metrics related to assembly quality were calculated with QUAST 5.2.0 tool26. Gene prediction (structural genome annotation) was performed using Prokka v1.13.327, and results (GFF files) were used to compare assemblies based on gene content (similar to pan-genome analysis) using Roary v3.12.028.
Benchmarking of pan-assembly using the 3 C criterion
The 3 C criterion (Contiguity, Completeness and Correctness), defined previously in14 was used to compare genome sequences in each pan-assembly. For this purpose, a variety of metrics were selected, as follows:
-
Contiguity or number of assembled segments: the total number of fragments or contigs, N50 value (shortest contig length that needs to be included for covering 50% of the genome and other metrics were obtained using QUAST 5.2.0 tool26.
-
Completeness: the ability to assemble expected genes was assessed using the number of predicted genes by Prokka v1.13.327, as well as the completeness score based on analysis of orthologs with BUSCO v5.4.729 within the gVolante platform30.
-
Correctness: the fidelity of the assembly was estimated based on rates of mismatches and insertions/deletions of the assembly with respect to the reference sequence. Calculations were obtained for the analysis with QUAST 5.2.0 tool26.
In addition, assessed conditions for each pan-assembly (depth level, assembly approach and algorithm/assembler) were compared and used to select the parameters for the reconstruction of each genome with the best quality possible. For this purpose, tests of statistical significance were performed with R v4.3.1 software (www.r-project.org/) using RStudio interface (www.rstudio.com). Statistical analyzes were run with parametric and non-parametric tests, as appropriate, including ANOVA tests or Kruskal–Wallis tests, T or U tests, multiple linear regression or generalized linear models (see Results). Additionally, a Hierarchical clustering and Principal Component Analysis were performed in R software to study each pan-assembly based on all metrics of the 3 C criterion.
Results
Sequencing data obtained by short- and long-reads for four bacterial models were used to generate de novo genome assemblies. The pan-assembly was established for each model using three strategies (based on a single technology separately or in a hybrid mode), two algorithms per strategy, and several levels of sequencing depth. The number of assemblies obtained for the organisms was 67 for B. henselae, 76 for E. coli, 69 for P. aeruginosa, 89 for X. fastidiosa.
Metrics of the 3 C criterion were calculated for each assembly, including those obtained from the comparison against the correspondent reference (consensus) sequence. Distribution of values for all assemblies evidenced a non-gaussian pattern, in which the median was used as measure of central tendency, as well as non-parametric tests (including multivariable models) for comparisons. Six metrics (two for each 3 C category) were used as key parameters for in-depth comparisons: number of contigs, N50, mismatches/100kbp, indels/100kbp, BUSCO score, and number of CDS. Using the whole set or selected metrics, pan-assembly was then described depending on sequencing strategy, assembler, and sequencing depth.
Regarding sequencing strategy, clear patterns of segregation depending on technology were found for all the models. This assessment of each pan-assembly was first done using clustering, based on the similarity among the whole set of 15 metrics of the 3 C criterion (details in Supplementary file). For X. fastidiosa, shown in Fig. 1-A-B, hierarchical clustering and PCA were implemented. These analyses not only assess the variation of metrics among assemblies, but also which sequences were more or less similar to each other. It is observed that clusters are defined by the sequencing strategy and then by algorithm for the assembly (assembler), rather than depth sequencing. In this line, distribution of values for the six key 3 C metrics defines a particular pattern depending on the strategy, as shown in Table 2; Fig. 2. Fragmented genome and lower N50 (lower contiguity) are reported for short-reads only, with median values of 90 and 101 890 respectively, in contrast to long-reads only (2 contigs and N50 = 2 513 184) or hybrid approaches (4 contigs and N50 = 1 441 411). In correctness, mismatches and indels are more variable with higher values for long-reads and hybrid methods. A higher completeness, based on BUSCO score and appropriate CDS number, was measured for short-reads only assemblies.
Assessment of the genome pan-assembly of X. fastidiosa using clustering algorithms based on metrics of the 3 C criterion. Different conditions regarding strategy, assembler and sequencing depth were used to assemble the genome, resulting indifferent sequences that can be compared using a profile based on contiguity, correctness, and completeness (3C). In the panassembly,(A) Hierarchical clustering and (B) Principal Component Analysis (PCA) show that sequencing strategy and algorithm,unlike sequencing depth, impact the assembly when compared to the expected (reference) sequence.
Comparison of key metrics of the 3 C criterion among sequencing strategies for the genome pan-assembly of X. fastidiosa. Violin plots were used to assess distribution of each parameter according to the use of short-reads only, long-reads onlyor hybrid approaches (which included all six algorithms and several levels of sequencing depth). In comparison to the referencesequence, the contiguity (contigs and N50) resulted with best values for long-reads and hybrid strategies, while a more confidentcorrectness (mismatches and indels) was found for short-reads only. Completeness (BUSCO score and CDS) was found to be closeto the reference sequence for short-reads only approaches.
For B. henselae, E. coli, and P. aeruginosa, similar results were obtained during the clustering analysis (Supplementary Figs. 1–3) and median comparison among strategies, shown in Table 2 for each pan-assembly. Interestingly, for all the four models, the number of CDS were higher and BUSCO score were lower under long-reads only approaches, unlike other methods which remained homogeneous and closer to the values of the reference sequence.
In the comparison of assemblers, differences were revealed even using the same sequencing strategy. For X. fastidiosa, results for Megahit and Unicycler (short-reads) were similar among clustering analysis (Fig. 1A, B) and key 3 C metrics (Fig. 3). However, differences in values and dispersion were evidenced using long-reads (Canu vrs Unicycler) or Hybrid (Wengan vrs Unicycler), indicating that the algorithm influences the final assembly. This appreciation was also verified for the other prokaryotic models, as shown in Supplementary Figs. 1–3.
Besides, conditions of those assemblers belonging to the vicinity of the reference sequence were considered as the relevant parameters to obtain the expected sequence. In X. fastidiosa, these conditions were the use of Unicycler as assembler under a hybrid strategy (Fig. 1A,B). This is supported by the high value for the explained variance of 80.7% (PC1 + PC2) in the PCA analysis. The profile of long-reads only approaches (with Canu or Unicycler), as well as a few hybrid cases with Wengan, resulted with the more discordant profiles. Again, proximity to the reference sequence is not associated with the sequencing depth for short- or long-reads used for the assembly (Fig. 1A).
For the other bacterial models (Supplementary Figs. 1–3), hybrid approaches with Unicycler also resulted closer to the reference than other assemblies. Moreover, unlike X. fastidiosa, short-reads approaches tended to present a more different profile of 3 C metrics from reference when compared to the long-reads approaches. In all cases, these patterns were provided with high support for the explained variance of the PCA, with 72.9%, 83.5% and 78% for B. heneselae, E. coli, and P. aeruginosa, respectively.
On the other hand, a comparative genomics approach was implemented to describe the pan-assembly based on gene content. As found in Figs. 4 and 5 for the four models (blue lines: presence of the gene in each assembly), the well-defined clusters suggest that the gene content profile is established -again- by sequencing strategy and assembler, but independent on sequencing depth (after a minimal value). For X. fastidiosa (Fig. 4), gene fragmentations are identified for long-reads only and some hybrids with Wengan (when sequencing depth for short-reads is 12.5X). Gene fragmentations for long-reads, as well as some specific hybrid cases with Wengan, are also present in the other models (Fig. 5). In P. aeruginosa, loss of genes is evidenced when using Wengan even for > 100X depth for short-reads, besides no assemblies were established for lower levels with the same assembler. Unlike this case, using Unicycler in a hybrid mode, sequences were built using at least 25X depth for short-reads.
Comparison of gene content predicted for each sequence of the genome pan-assembly of X. fastidiosa. Based ongene content (presence: blue), a pan-genomic approach was used to assess all assemblies. Expected genes (based on thereference sequence) were found for most cases using short-reads, hybrid approaches with Unicycler and some hybrids with Wengan.Other Wengan-based hybrid assemblies, as well as long-read only cases, resulted in an unexpected profile with a different number ofgenes and apparently several “exclusive” genes. Despite some exceptions, no association between sequencing depth and the genecontent outcome is evidenced.
Comparison of gene content predicted for each sequence of the genome pan-assembly of B. henselae, E. coli , and P. aeruginosa. Based on gene content (presence: blue), a pan-genomic approach was used to assess all assemblies. Expectedgenes (based on the reference sequence) were found for most cases using short-reads, hybrid approaches with Unicycler and somehybrids with Wengan. Other Wengan-based hybrid assemblies, as well as long-read only cases, resulted in an unexpected profilewith a different number of genes and apparently several “exclusive” genes. Despite some exceptions, no association betweensequencing depth and the gene content outcome is evidenced.
A final assessment of the parameters studied here (predictors: strategy, assembler, and sequencing depth) and their effect on the key 3 C metrics (response variable) was done using generalized linear models (GLM) for the pan-assembly of each organism. As summarized in Table 3 for all the four bacteria, significant values (< 0.01) were obtained for the very most cases of the 3 C metrics using strategy and assembler as predictors. Only scarce significant values were evidenced for sequencing depth for short- or long-reads among response conditions.
Finally, the subsequent analysis was conducted to determine the minimal sequencing depth to achieve an assembly with a similar quality to assemblies obtained with depth ≥ 100X data under the same strategy and assembler (Table 4). Using short-reads only approaches, requirements of depth for Unicycler and Megahit were always the same for all the bacterial models (12.5X for E. coli and X. fastidiosa, and 25X for others). A similar situation was observed for long-reads only approaches, in which 25X is enough to assemble the sequence for all bacteria but X. fastidiosa (with 12.5X). In the case of hybrid approaches, Wengan was demonstrated to always need more sequencing depth in comparison to Unicycler, including requirements of > 100X depth for short-reads (B. henselae and P. aeruginosa). In B. henselae, Wengan also demanded 100X depth for long-reads. When sequencing depth ≥ 25X (in some cases even with lower values), hybrid Unicycler was always able to assemble the sequence.
Jointly, whole results of pan-assembly analyses indicate that the assemble sequence and the 3 C metrics are significantly impacted by sequencing strategy and assembler, but not by sequencing depth after a minimal level is considered for the four bacterial models.
Discussion
De novo genome assembly allows for the genome reconstruction of an organism without using a reference sequence31. However, the assembly results depend on various sequencing technologies that generate data with differing fidelity, read lengths, and coverage levels, as well as on performance of a wide variety of assemblers (algorithms)13,32. In this study, we compared pan-assemblies under these conditions and their impact on de novo genome assembly for four bacterial models, using the 3 C criteria -contiguity, correctness, and completeness- for evaluation.
Regarding contiguity, our results indicate that short reads present significant variability in the number of contigs, suggesting high fragmentation in the resulting assemblies and reduced N50 values (low contiguity), in contrast to when long reads are used either alone or in hybrid formats. Thus, the use of long-read technology improves contiguity and potentially leads to better genome reconstruction under this criterion, even allowing for the potential circularization of bacterial genomes, as previously demonstrated22,33. In another benchmarking strategy, the performance of long reads was also outstanding, with a low number of contigs and high N50 values34. However, a detailed inspection of the fragments revealed assembly errors that were not apparent when only contiguity was assessed. These findings have also been highlighted in other comparative studies35,36. This underscores the need for diverse metrics to evaluate assemblies, as we propose with the 3 C criteria14,16.
In the context of correctness or fidelity evaluation, significant differences were reported in the error rates for the four bacterial models. Short reads demonstrated a lower incidence of errors compared to long reads. In other studies, using Illumina and Oxford Nanopore data for various bacterial models, the results support our findings, with a low error rate detected when working with short-read data, including regions containing repeats14,35,36.Thus, in applications where fidelity is a priority, short reads may be preferable to minimize specific errors such as those caused by indels and technical sequencing errors33,37.
Regarding completeness, the best performance was identified in assemblies with short reads and hybrid strategies. This effect was also evident in the pan-assembly analyses based on CDS gene content, with significant reductions mainly in hybrids assemblies using Wengan and low-coverage conditions. Although long reads can span more complex and broader genomic regions, gene prediction is incomplete due to assembly errors, a characteristic of these strategies with lower fidelity38. These findings reflect the capability of short reads to generate assemblies with high completeness, despite inherent limitations such as raw data length and low contiguity33.
For hybrid strategies, the high completeness is associated with the resolution of ambiguities in genomic regions characterized by repetitive or highly complex sequences provided by long reads39,40. In other works, the best results are justified by the resolution achieved with long assembled fragments37,41, although this contrasts with other cases where long reads are associated with lower completeness and gene identification due to frameshifts from sequencing errors and their impact on gene prediction14. This latter effect is also evidenced by the high number of CDS (compared to the reference sequence) found for long reads but not for other strategies.
Due to the lower fidelity of long-read strategies, the need to continue optimizing long-read sequencing techniques and minimizing errors without compromising assembly integrity is emphasized42. Notable advances in this area include the implementation of polishing strategies for assemblies generated from long-read data, which have significantly improved assembly completeness33,43. In parallel, the development of high-fidelity long-read sequencing technologies—such as PacBio HiFi9,38,44 —and optimized ONT platforms incorporating new flow cells and deep learning–based data processing have further enhanced sequencing accuracy and reliability10,11. These two scenarios were not included in this study.
Regarding algorithms, this study observed statistically significant differences in the performance of assemblers for genome reconstruction. Their effect was evident in the size of the contigs produced (N50 and number of contigs), the number of expected genes reconstructed or completeness (BUSCO Score and CDS), fidelity or correctness (indels and mismatches), and the resolution of pan-assemblies based on gene content. Overall, the choice of algorithm has direct implications for the quality and utility of the assembled genome5,45,46. Additionally, as evidenced here, using exactly the same input data within the same assembly strategy, fidelity results increase for certain algorithms47.
Considering all parameters, assemblies using Unicycler performed best in each strategy for the bacterial models studied. As supported by the literature in various benchmarking studies, Unicycler is currently one of the algorithms with the best reported performance for bacterial genome assembly, whether using long reads, short reads, or a hybrid mode14,35,43,48. In studies with only long reads, Unicycler also stood out as the top-performing algorithm35, though in other studies, Canu has been reported as performing better46,47,48. In our study, Canu showed good values in contiguity and moderate accuracy and completeness, but its performance did not surpass that of Unicycler. However, it is worth noting that Canu is optimized for maximizing performance in cluster computing environments40. Regarding the use of Megahit for assemblies with only short reads, it has been reported as a high-performing assembler compared to other strategies48,49,50, even comparable to Unicycler, mainly in terms of completeness and accuracy. A strength of Megahit is its execution time, which is much faster compared to Unicycler. For hybrid assemblies, the other algorithm used was Wengan. In this study, it performed well but depended on high coverage to produce high-quality assemblies, unlike Unicycler in hybrid mode. This observation contrasts with a previous report where Wengan was highlighted for generating quality assemblies even in low-coverage situations25. In other metrics, Wengan has been reported with superior performance compared to other algorithms25,46. However, Wengan showed suboptimal performance in contiguity, with low N50 values in another study38. In summary, Unicycler showed the highest concordance with reference genome data compared to other algorithms, regardless of whether long reads, short reads, or hybrid modes were used. The hybrid mode demonstrated the best overall performance.
Lastly, the final evaluation of the pan-assembly focused on the possible correlation between quality of the assembly and coverage depth. The results suggest that there is no statistical correlation between coverage depth (after a minimum level) and 3 C criteria metrics, with a few exceptions. These results are consistent with several reports in the literature regarding this parameter in genome assembly analyses, showing no association between increased depth (in both long and short reads) and improved assembly quality43,51,52. One study highlighted that values of 40X do not change the results in cases of lower coverage, and ultra-deep sequencing can lead to algorithm saturation and a significant increase in computational resource usage43. It has also been reported that very low coverage levels of 16x are sufficient to assemble a complete genome, but accuracy improves with values around 30X46. In other studies, the minimum coverage level has even been reported as low as 10X51,53. These minimum values are very similar to our results for the four organisms studied. It should be noted that in certain applications, such as genetic variant imputations, there are recommendations for a minimum coverage level for decision-making53,54, but these are not focused on de novo genome assembly. From a practical perspective, this aspect of coverage depth is highly relevant because it means that high-quality assemblies can be generated using moderate coverage levels, without the need for ultra-deep sequencing (> 100X), translating into significantly lower sequencing costs.
In summary, once a minimum data level is reached, coverage depth does not significantly affect assembly quality results across the various characteristics evaluated by the 3 C criteria, for which no statistical correlation was observed. This contrasts with the influence of strategy (technology) type and assembly algorithms on the quality of the resulting assembly. These results are based on genome sequencing, and our previous works on other molecular strategies55,56, will be considered to continue working on these bacterial models at the local biological context.
It should be mentioned that this study has some limitations. This study focused on four bacterial models, each with their own genomic complexity, but it could be extended to other models, including eukaryotic organisms in further analyses. Other methodological strategies, such as genome polishing or high-fidelity long-reads data obtained using HiFi sequencing ot enhanced ONT platforms, were not assessed in this comparison. Additionally, other 3 C criteria metrics, execution time and computational resource usage could be valuable for comparing assembly conditions in other studies.
Conclusions
In this study, the pan-assembly of four bacterial models (B. henselae, E. coli, P. aeruginosa, and X. fastidiosa) was assessed using contiguity, accuracy, and completeness metrics (the 3 C criteria). The benchmarking strategy showed that short-read assemblies presented higher accuracy with fewer errors (high correctness) and a high degree of completeness but lower contiguity due to fragmented assemblies. In contrast, long-read-based strategies showed high contiguity but lower completeness and accuracy. The hybrid strategy yielded the best overall results across all parameters by leveraging the strengths of both types of technology. Regarding assembly algorithms, Unicycler was the top assembler in 3 C metrics, using any of the short-read (compared to Megahit), long-read (compared to Canu), or hybrid strategies (compared to Wengan). Overall, the hybrid approach with Unicycler proved to be the best general approach for genome assembly of the four bacterial models. Finally, regarding coverage depth, increasing depth did not significantly affect assembly quality results if a minimum data level was maintained, indicating that high-quality assemblies can be achieved using moderate coverage levels. In summary, these results of the pan-assembly provide working conditions for de novo genome assembly that can be applied to bacterial models of interest, guiding the selection of optimized experimental and bioinformatics conditions while reducing sequencing costs for generating high-quality sequences.
Data availability
The raw sequencing data (short and long reads), as well as the assembled genomes used in this study, are available in GenBank and SRA databases (NCBI), with the accession number detailed in Table 1.
Change history
15 January 2026
A Correction to this paper has been published: https://doi.org/10.1038/s41598-026-36195-w
References
Simpson, J. T. & Pop, M. The theory and practice of genome sequence assembly. Annu. Rev. Genomics Hum. Genet. 16, 153–172. https://doi.org/10.1146/annurev-genom-090314-050032 (2015).
Segerman, B. The most frequently used sequencing technologies and assembly methods in different time segments of the bacterial surveillance and refseq genome databases. Front. Cell. Infect. Microbiol. 10, 527102. https://doi.org/10.3389/FCIMB.2020.527102/BIBTEX (2020).
Chen, Y., Zhang, Y., Wang, A. Y., Gao, M. & Chong, Z. Accurate long-read de Novo assembly evaluation with inspector. Genome Biol. 22 (1), 1–21. https://doi.org/10.1186/s13059-021-02527-4 (2021).
Miller, J. R., Koren, S. & Sutton, G. Assembly algorithm for Next-Generation sequencing data. Genomics 95 (6), 315–327. https://doi.org/10.1016/j.ygeno.2010.03.001.Assembly (2010).
Dida, F. & Yi, G. Empirical evaluation of methods for de Novo genome assembly. PeerJ Comput. Sci. 7, e636. https://doi.org/10.7717/PEERJ-CS.636 (2021).
Schmeing, S. & Robinson, M. D. Gapless provides combined scaffolding, gap filling, and assembly correction with long reads. Life Sci. Alliance. 6 (7). https://doi.org/10.26508/LSA.202201471 (2023).
Beigel, R., Alon, N., Apaydin, M. S., Fortnow, L. & Kasif, S. An optimal procedure for gap closing in whole genome shotgun sequencing, Proceedings of the Annual International Conference on Computational Molecular Biology, RECOMB, pp. 22–30, https://doi.org/10.1145/369133.369152 (2001).
Amarasinghe, S. L. et al. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 21 (1). https://doi.org/10.1186/S13059-020-1935-5 (2020).
Hon, T. et al. Highly accurate long-read HiFi sequencing data for five complex genomes. Sci. Data. 7 (1). https://doi.org/10.1038/S41597-020-00743-4 (2020).
Delahaye, C. & Nicolas, J. Sequencing DNA with nanopores: troubles and biases. PLoS One. 16 (10), e0257521. https://doi.org/10.1371/JOURNAL.PONE.0257521 (2021).
Lerminiaux, N., Fakharuddin, K., Mulvey, M. R. & Mataseje, L. Do we still need illumina sequencing data? Evaluating Oxford nanopore technologies R10.4.1 flow cells and the rapid v14 library Prep kit for gram negative bacteria whole genome assemblies. Can. J. Microbiol. 70 (5), 178–189. https://doi.org/10.1139/CJM-2023-0175/SUPPL_FILE/CJM-2023-0175SUPPLA.DOCX (2024).
Lantz, H. et al. Ten steps to get started in genome assembly and annotation. F1000Res 7 https://doi.org/10.12688/f1000research.13598.1 (2018).
Bellec, A., Courtial, A., Cauet, S. & Rodde, N. Long read sequencing technology to solve complex genomic regions assembly in plants. J. Next Generation Sequencing Appl. 3 (2). https://doi.org/10.4172/2469-9853.1000128 (2016).
Molina-Mora, J. A., Campos-Sánchez, R., Rodríguez, C., Shi, L. & García, F. High quality 3 C de novo assembly and annotation of a multidrug resistant ST-111 Pseudomonas aeruginosa genome: Benchmark of hybrid and non-hybrid assemblers. Sci. Rep. 10 (1), 1392. https://doi.org/10.1038/s41598-020-58319-6 (2020).
Wang, J. et al. Systematic comparison of the performances of de Novo genome assemblers for Oxford nanopore technology reads from Piroplasm. Front. Cell. Infect. Microbiol. 11, 1. https://doi.org/10.3389/FCIMB.2021.696669/FULL (2021).
Molina-Mora, J. A. & Garcia, F. The 3 C criterion: Contiguity, Completeness and Correctness to assess de novo genome assemblies., BMC Bioinformatics, Bioinformatics: from Algorithms to Applications, vol. 21, no. S20: O7, p. 5,https://doi.org/10.1186/s12859-020-03838-2 (2020).
Zimin, A. V. & Salzberg, S. L. The SAMBA tool uses long reads to improve the contiguity of genome assemblies. PLoS Comput. Biol. 18 (2). https://doi.org/10.1371/JOURNAL.PCBI.1009860 (2022).
Andrews, S. FastQC A Quality Control tool for High Throughput Sequence Data. (Accessed: 10 Apr 2018) https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2018).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30 (15), 2114–2120. https://doi.org/10.1093/bioinformatics/btu170 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. https://doi.org/10.1093/bioinformatics/btp324 (2009).
Okonechnikov, K., Conesa, A. & García-Alcalde, F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data., Bioinformatics, vol. 32, no. 2, pp. 292–4, https://doi.org/10.1093/bioinformatics/btv566 (2016).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13 (6), 1–22. https://doi.org/10.1371/journal.pcbi.1005595 (2017).
Li, D., Liu, C. M., Luo, R., Sadakane, K. & Lam, T. W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31 (10), 1674–1676. https://doi.org/10.1093/bioinformatics/btv033 (2015).
Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation., Genome Res, vol. 27, no. 5, pp. 722–736, (2017). https://doi.org/10.1101/gr.215087.116
Di Genova, A., Buena-Atienza, E., Ossowski, S. & Sagot, M. F. Efficient hybrid de Novo assembly of human genomes with WENGAN. Nat. Biotechnol. 39 (4), 422–430. https://doi.org/10.1038/s41587-020-00747-w (2021).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: quality assessment tool for genome assemblies, Bioinformatics, vol. 29, no. 8, pp. 1072–1075, https://doi.org/10.1093/bioinformatics/btt086 (2013).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 (14), 2068–2069. https://doi.org/10.1093/bioinformatics/btu153 (2014).
Page, A. J. et al. Nov., Roary: rapid large-scale prokaryote pan genome analysis, Bioinformatics, vol. 31, no. 22, pp. 3691–3693,https://doi.org/10.1093/bioinformatics/btv421 (2015).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs, Bioinformatics, vol. 31, no. 19, pp. 3210–3212, Oct. https://doi.org/10.1093/bioinformatics/btv351 (2015).
Nishimura, O., Hara, Y. & Kuraku, S. gVolante for standardizing completeness assessment of genome and transcriptome assemblies. Bioinformatics 33 (22), 3635–3637. https://doi.org/10.1093/bioinformatics/btx445 (2017).
Il Sohn, J. & Nam, J. W. The present and future of de Novo whole-genome assembly. Brief. Bioinform. 19 (1), 23–40. https://doi.org/10.1093/bib/bbw096 (2018).
Ekblom, R. & Wolf, J. B. W. A field guide to whole-genome sequencing, assembly and annotation. Evol. Appl. 7 (9), 1026–1042. https://doi.org/10.1111/eva.12178 (2014).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Completing bacterial genome assemblies with multiplex minion sequencing. Microb. Genom. 3 (10). https://doi.org/10.1099/mgen.0.000132 (2017).
Narzisi, G. & Mishra, B. Comparing de Novo genome assembly: the long and short of it. PLoS One. 6 (4), e19175. https://doi.org/10.1371/JOURNAL.PONE.0019175 (2011).
Breckell, G. L. & Silander, O. K. Do You Want to Build a Genome? Benchmarking Hybrid Bacterial Genome Assembly Methods, bioRxiv, p. 2021.11.07.467652, https://doi.org/10.1101/2021.11.07.467652 (2021).
Chen, Z., Erickson, D. L. & Meng, J. Benchmarking hybrid assembly approaches for genomic analyses of bacterial pathogens using illumina and Oxford nanopore sequencing. BMC Genom. 21 (1), 1–21. https://doi.org/10.1186/S12864-020-07041-8/FIGURES/6 (2020).
Jayakumar, V. & Sakakibara, Y. Comprehensive evaluation of non-hybrid genome assembly tools for third-generation PacBio long-read sequence data. Brief. Bioinform. 20 (3), 866–876. https://doi.org/10.1093/bib/bbx147 (2019).
Espinosa, E. et al. Comparing assembly strategies for third-generation sequencing technologies across different genomes. Genomics 115 (5), 110700. https://doi.org/10.1016/J.YGENO.2023.110700 (2023).
Batty, E. M. et al. Long-read whole genome sequencing and comparative analysis of six strains of the human pathogen Orientia Tsutsugamushi. PLoS Negl. Trop. Dis. 12 (6). https://doi.org/10.1371/journal.pntd.0006566 (2018).
Southwood, D., Rane, R. V., Lee, S. F., Oakeshott, J. G. & Ranganathan, S. Exhaustive benchmarking of de novo assembly methods for eukaryotic genomes, bioRxiv, p. 2023.04.18.537422, https://doi.org/10.1101/2023.04.18.537422 (2023).
Di Genova, A., Buena-Atienza, E., Ossowski, S. & Sagot, M. F. Efficient hybrid de novo assembly of human genomes with WENGAN, Nature Biotechnology 2020 39:4, vol. 39, no. 4, pp. 422–430,https://doi.org/10.1038/s41587-020-00747-w (2020).
Alhakami, H., Mirebrahim, H. & Lonardi, S. A comparative evaluation of genome assembly reconciliation tools. Genome Biol. 18 (1), 1–14. https://doi.org/10.1186/s13059-017-1213-3 (2017).
Zhang, X. et al. Benchmarking of long-read sequencing, assemblers and Polishers for yeast genome. Brief. Bioinform. 23 (3), 1–13. https://doi.org/10.1093/BIB/BBAC146 (2022).
Wenger, A. M. et al. Aug., Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome, Nature Biotechnology 2019 37:10, vol. 37, no. 10, pp. 1155–1162, https://doi.org/10.1038/s41587-019-0217-9 (2019).
Harvey, W. T. et al. Whole-genome long-read sequencing downsampling and its effect on variant calling precision and recall. BioRxiv https://doi.org/10.1101/2023.05.04.539448 (2023).
Gavrielatos, M., Kyriakidis, K., Spandidos, D. A. & Michalopoulos, I. Benchmarking of next and third generation sequencing technologies and their associated algorithms for de novo genome assembly, Mol Med Rep, vol. 23, no. 4, Apr. https://doi.org/10.3892/MMR.2021.11890 (2021).
Latorre-Pérez, A., Villalba-Bermell, P., Pascual, J. & Vilanova, C. Assembly methods for nanopore-based metagenomic sequencing: a comparative study, Scientific Reports 2020 10:1, vol. 10, no. 1, pp. 1–14, Aug.https://doi.org/10.1038/s41598-020-70491-3 (2020).
Zhang, Z., Yang, C., Veldsman, W. P., Fang, X. & Zhang, L. Benchmarking genome assembly methods on metagenomic sequencing data, Brief Bioinform, vol. 24, no. 2, pp. 1–17, https://doi.org/10.1093/BIB/BBAD087 (2023).
Fuentes-Trillo, A. et al. Benchmarking different approaches for Norovirus genome assembly in metagenome samples. BMC Genom. 22 (1), 1–12. https://doi.org/10.1186/S12864-021-08067-2/FIGURES/2 (2021).
Meyer, F. et al. Apr., Critical Assessment of Metagenome Interpretation: the second round of challenges, Nature Methods 2022 19:4, vol. 19, no. 4, pp. 429–440, https://doi.org/10.1038/s41592-022-01431-4 (2022).
Song, G. et al. Aug., Integrative Meta-Assembly Pipeline (IMAP): Chromosome-level genome assembler combining multiple de novo assemblies, PLoS One, vol. 14, no. 8, p. e0221858, https://doi.org/10.1371/JOURNAL.PONE.0221858 (2019).
Babarinde, I. A. & Hutchins, A. P. The effects of sequencing depth on the assembly of coding and noncoding transcripts in the human genome. BMC Genom. 23 (1), 1–14. https://doi.org/10.1186/S12864-022-08717-Z/FIGURES/5 (2022).
Jiang, Y., Jiang, Y., Wang, S., Zhang, Q. & Ding, X. Optimal sequencing depth design for whole genome re-sequencing in pigs. BMC Bioinform. 20 (1), 1–12. https://doi.org/10.1186/S12859-019-3164-Z/FIGURES/7 (2019).
Homburger, J. R. et al. Low coverage whole genome sequencing enables accurate assessment of common variants and calculation of genome-wide polygenic scores, Genome Med, vol. 11, no. 1, pp. 1–12, Nov.https://doi.org/10.1186/S13073-019-0682-2/FIGURES/5 (2019).
Molina-Mora, J. A. et al. Assessment of Mathematical Approaches for the Estimation and Comparison of Efficiency in qPCR Assays for a Prokaryotic Model, DNA, vol. 4, no. 3, pp. 189–200, https://doi.org/10.3390/DNA4030012 (2024).
Molina-Mora, J. A. & García, F. Molecular determinants of antibiotic resistance in the Costa Rican Pseudomonas aeruginosa AG1 by a Multi-omics approach: A review of 10 years of study. Phenomics 1, p. (3). https://doi.org/10.1007/s43657-021-00016-z (2021).
Funding
This work was funded by projects “C1163 pro-NGS 2.0: Protocolos operativos estandarizados de análisis de datos moleculares obtenidos por NGS o afines y de algoritmos de inteligencia artificial en modelos biológicos”, Vicerrectoría de Investigación, Universidad de Costa Rica (period 2021–2023) and “C4604 iPAT: Plataforma genómica, bioinformática y de inteligencia artificial para la vigilancia de patógenos”, Vicerrectoría de Investigación, Universidad de Costa Rica (period 2024–2026).
Author information
Authors and Affiliations
Contributions
J.A.M.M. participated in the conception and design of the study. H.R.M., V.M.L. and J.A.M.M. performed experimental assays and data analysis. J.A.M.M. drafted the manuscript. All authors were involved in its revision and the final approbation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error, where the scientific name ‘Bartonella henselae’ was incorrectly given as ‘Brucella henselae’. Modifications have been made to the Abstract and the Introduction. Full information regarding the corrections made can be found in the correction for this Article.
Supplementary Information
Below is the link to the electronic supplementary material.
41598_2025_26847_MOESM2_ESM.png
Supplementary Figure 1 Assessment of the genome pan-assembly of B. henselae using clustering analysis and distribution of keymetrics of the 3C criterion among assemblers. Principal Component Analysis (PCA, left) was used to compare the profile defined bymetrics of the 3C criterion, as well as the distribution of six key metrics were represented using violin plots (right).
41598_2025_26847_MOESM3_ESM.png
Supplementary Figure 2 Assessment of the genome pan-assembly of E. coli using clustering analysis and distribution of key metricsof the 3C criterion among assemblers. Principal Component Analysis (PCA, left) was used to compare the profile defined by metricsof the 3C criterion, as well as the distribution of six key metrics were represented using violin plots (right).
41598_2025_26847_MOESM4_ESM.png
Supplementary Figure 3 Assessment of the genome pan-assembly of P. aeruginosa using clustering analysis and distribution of keymetrics of the 3C criterion among assemblers. Principal Component Analysis (PCA, left) was used to compare the profile defined bymetrics of the 3C criterion, as well as the distribution of six key metrics were represented using violin plots (right).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rojas-Miranda, H., Madrigal-Ly, V. & Molina-Mora, J.A. Benchmarking genome assemblers for four bacterial models based on contiguity, correctness, and completeness. Sci Rep 15, 42858 (2025). https://doi.org/10.1038/s41598-025-26847-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-26847-8