Abstract
The hepato-canalicular adenosine triphosphate (ATP)-binding cassette transporter ABCB4/MDR3 is responsible of the secretion of phosphatidylcholine (PC) into bile. Variations in ABCB4 gene induce a spectrum of cholestatic liver diseases, the most severe form is progressive familial intrahepatic cholestasis type 3 (PFIC3). The purpose of our study was to investigate the impact and potential rescue of four ABCB4 missense variants identified in patients, two of which (T437I and T1077M) affect homologous amino acids in the Walker A motifs in the two nucleotide-binding sites NBS2 and NBS1, respectively and the other two (S242R and S346I) affect residues of the transmembrane helices 4 and 6, respectively. The functional role of the four amino acids was assessed by analysis of three-dimensional (3D) structures and molecular dynamics (MD) simulations in a lipid bilayer. For functional validation, the mutants were reproduced in a plasmid encoding the human ABCB4 protein. The localization, the processing and the PC secretion activity of the mutants were studied after transfection in cell models. As the wild-type ABCB4, all four mutants expressed and trafficked efficiently to the canalicular membrane of HepG2 cells, but were dramatically impaired in PC secretion activity. This behavior can be explained by the critical positions of the amino acids in the NBSs and in a region participating in the substrate transport. The Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) potentiators ivacaftor and Small Binder of CFTR 219 (SBC219) significantly rescued the function defect of the mutants with differential sensitivity. Our results demonstrate the importance of the four mutated residues for ABCB4 function, which may explain the pathogenic phenotype. They provide an experimental evidence that targeted pharmacotherapy for genetic diseases caused by ABCB4 deficiency is likely to be mutation-specific.
Similar content being viewed by others
Introduction
The ABCB4 gene encodes the ABCB4 protein (also known as multidrug resistance 3 MDR3 protein), which is an adenosine tri-phosphate (ATP)-binding cassette (ABC) transporter expressed at the canalicular membrane of hepatocytes where it flops the membrane phospholipid, phosphatidylcholine (PC) from the inner to the outer leaflet of the canalicular membrane1,2. PC is crucial to ensure solubilization of cholesterol into mixed micelles and to prevent bile acids cytotoxicity and the formation of gallstones3. Failure to flop enough PC results in cytotoxic bile, leading to inflammation and cholestasis4. At present, more than 500 variations of the ABCB4-encoding gene have been reported, they cause a spectrum of rare liver and biliary diseases, among which the progressive familial intrahepatic cholestasis type 3 (PFIC3) is the most severe one. PFIC3 is a rare autosomal recessive disease affecting patients who are generally homozygous or compound heterozygous, it occurs early in childhood and is characterized by the early onset of persistent cholestasis that rapidly evolves to cirrhosis, most often requiring liver transplantation5,6,7. PFIC3 has also been identified as cryptogenic liver cirrhosis cause in adults8. Heterozygous mutations of ABCB4 can cause less severe cholestatic diseases such as low-phospholipid-associated cholelithiasis syndrome (LPAC)9 and intrahepatic cholestasis of pregnancy (ICP)10. These cholestasis of the adult generally improve under treatment with ursodeoxycholic acid (UDCA), a bile acid with low hydrophobicity which decreases the toxicity of bile acids11.
We conducted a comprehensive analysis of more than sixty ABCB4 variants12,13,14,15,16,17, our unpublished data) and introduced the first functional classification of these variations based on their impact on the production (class I), traffic (class II), secretion activity (class III) or membrane stability of the protein (class IV)13. This classification is imperative for precision drug development, including repositioning of clinically approved molecules. Regarding the repositioning strategy, we have shown that ivacaftor, a highly effective potentiator clinically approved to treat cystic fibrosis patients carrying mutations affecting cystic fibrosis transmembrane conductance regulator (CFTR) channel gating, was able to rescue significantly some class III ABCB4 mutants that are localized in the conserved and functionally critical motifs of ABC transporters, but not those localized within the transmembrane helices14,15. Therefore, a major challenge is to find pharmacological treatment for class III ABCB4 mutants not potentiated by ivacaftor.
ABCB4 has two membrane-spanning domains (MSDs), each composed of six transmembrane helices involved in PC translocation, and two nucleotide-binding domains (NBDs) that bind and hydrolyze ATP to provide the energy required for PC transport18. The NBDs of ABCB4 form two consensus ATP-binding sites (NBS1 and NBS2) that contain several conserved motifs involved in ATP binding and hydrolysis, such as the Walker A and Walker B motifs, the A-, D-, H-, and Q-loops and the ABC signature. The cryogenic electron microscopy (cryo-EM) studies of Locher and coworkers provided high-resolution three-dimensional (3D) structures of ABCB419,20, which allows a deep understanding of the impact of ABCB4 variants at the molecular and cellular level on clinical phenotypes and to search for rational alternative therapy.
In the current study, we combined 3D structure analysis and experimental approaches to characterize four ABCB4 variants, two of which affect homologous amino acids in the Walker A motifs in the two ATP-binding sites (T437I, NBS2 and T1077M, NBS1) and the other two affect amino acids of the transmembrane (TM) helices (S242R, TM4 and S346I, TM6). We found that all four mutants expressed and trafficked efficiently to the canalicular membrane of HepG 2 cells but were dramatically impaired in PC secretion activity. As part of potential repositioning and therapeutic strategies, we tested the effect of ivacaftor and another CFTR potentiator (Small Binder of CFTR, SBC 219) that have been shown to act in a synergic way with ivacaftor on class II and III CFTR mutants21. We showed that ivacaftor, but not SBC219, rescues PC secretion activity of T437I and T1077M mutants. Interestingly, SBC219 but not ivacaftor, enhances the PC secretion activity of S242R and S346I mutants. Our findings highlight a specificity of CFTR potentiators toward ABCB4 mutations depending on their location.
Results
ABCB4 variations and patients’ data analyses
The main characteristics of the patients are shown in Table 1. The S242R and T437I variations were identified with a heterozygous status in patients with LPAC syndrome (this study). The S346I and T1077M variations were identified with homozygous status in patients with PFIC37,22,23.
3D structure analysis of ABCB4 variants and prediction of their impact
The locations of the four variations studied on the human ABCB4 3D structures are shown in Fig. 1. To assess the conformational dynamics of WT transporter in a lipid bilayer, we performed MD simulations starting from four different conformational states: ATP-bound, collapsed (PDB 6S7P19), apo inward-facing (7NIU20), inhibited inward-facing (7NIW, complex with PC and posaconazole20, and occluded (7NIV, complex with PC20, . Root Mean Square Deviation (RMSD) profiles for the protein backbone (Cα atoms) during these simulations are presented in Figure S1. Among the different conformations, the ATP-bound, collapsed state exhibited the most stable behavior throughout the simulations, as indicated by lower RMSD fluctuations compared to the other states.
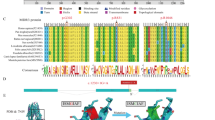
T437I and T1077M variations are located at homologous positions of the Walker A of NBD1 and NBD2, respectively. Analysis of the ATP-bound 3D structure (pdb 6S7P19, indicated that the T437 and T1077 hydroxyl groups make H-bonds with oxygens atoms of ATP a-phosphate (Fig. 2A, B). Furthermore, hydroxyl groups of both threonine residues are also hydrogen-bonded to the side chain oxygen atoms of the conserved tyrosine residues (Y403 and Y1043) of the A-loop (adenine staking), which may stabilize them (Figure S2). Substitution of these residues is thus expected to prevent or at least disturb ATP-binding and thereby ATP-induced NBD dimerization, resulting in ABCB4 dysfunction.
Positions of the homologous amino acids Thr437 (T437) and Thr1077 (T1077) in the human ABCB4 ATP-binding sites. The experimental 3D structure of the human ABCB4 in the ATP-bound, collapsed conformation (PDB: 6S7P) was considered for highlighting the amino acids of the conserved motifs of NBD1 (blue) and NBD2 (orange) involved in the formation of the ATP-binding site 2 (A) and ATP-binding site 1 (B). Amino acids Thr437 (T437) and Thr1077 (T1077) whose mutations were analyzed in this study are colored green and the other ones black. T437 belongs to NBD1, it interacts with the ATP a-phosphate oxygen and the tyrosine (Y403) of the A-loop of NBD1. T1077 belongs to NBD2, it interacts with the ATP a-phosphate oxygen and the tyrosine (Y1043) of the A-loop of NBD2.
S242R and S346I are located in transmembrane helices TM4 and TM6, respectively, which have been described to be important to the binding and the transport of PC by ABCB419,20. The role of these two serine residues is to be investigated in the context of the four conformations observed for ABCB4 in cryo-EM studies (Fig. 3A). The “apo inward-facing” state features a side opening to the inner leaflet of the bilayer (note that CFTR also has a lateral opening involving TM4 and TM6 (TM4-TM6 portal), but at the level of intracellular loops (ICLs))24, while the “occluded” state refers to kinked conformations of TM4 and TM10 helices, generating a Y-shaped occluded cavity at the center of the transporter. TM4’s conformational dynamics is therefore essential for (i) generating the opening to the lipid bilayer and (ii) forming this “occluded” conformation.
Positions of amino acids Ser242 (S242) and Ser346 (S346) in the human ABCB4 membrane-spanning domains. (A) General view of the experimental 3D structures of human ABCB4 membrane-spanning domains in the ATP-bound, collapsed (pdb 6S7P19, , the apo inward-facing (pdb 7NIU20, , the inhibited inward-facing (pdb 7NIW, complex with PC and posaconazole20, and the occluded (pdb 7NIV, complex with PC20 conformations. Amino acids Ser242 (S242) and Ser346 (S346) whose mutations were analyzed in this study are colored green. The three aromatic amino acids (W234, F345, H989) participating in the substrate binding pocket are depicted in orange. DLP stands for 1,2-Dilinoleoyl-Sn-Glycero-3-Phosphocholine. (B) Enlarged view of TM4 (in magenta) and TM6 (in blue) at the level of amino acid S242 in the ATP-bound, collapsed (pdb 6s7p19, and occluded (pdb 7NIV, complex with PC20 conformations. (C) Contact established between S242 and the headgroup of a POPS molecule in the MD simulation of the apo inward-facing conformation (second replica, also see Table S2) (D) Enlarged view of TM4 (in magenta) and TM6 (in blue) at the level of amino acid S346 in the four conformations depicted in panel A. At right are shown the details of the H-bonds that S346 forms with I342 (red dashed lines). In the red box is shown the tight contact formed between TM4 (A231-A232) and TM6 (G348) in the occluded conformation.
On the one hand, S242 (TM4) plays a critical role in the formation (and stabilization) of the different conformational states (Fig. 3B). It “locks” the ATP-bound, collapsed state by forming a bidentate hydrogen bond (H-bond) with the oxygens of the side and main chains of N359 (TM6), within a network incorporating at a greater distance K293 (TM5), which is salt-bridged to TM4 E245 (Fig. 3B-left, and Figure S2-red barplots). Remarkably, S242 also plays a critical role in ensuring the “kink” essential for the formation of the PC-bound “occluded” conformation (Fig. 3B-right), involving H-bonds of the oxygen and nitrogen main chain atoms of S242 to the carbonyl oxygen atom of S239 (i-i + 3 H-bond), modifying the standard i-i + 4 network observed in the collapsed, apo and apo-inhibited forms (corresponding to unbent helices). Interestingly, MD simulations revealed the potential for additional interactions. In particular, contacts were observed between S242 and residues Q823 and Q825 in TM9, as well as with D355 in TM6 and F993 in TM12 (Figure S2), suggesting a broader network of interactions that may contribute to the stabilization of the kinked helical conformation in the occluded state.
In the inward-facing conformations, represented by the apo inward state and the inhibited state, S242 also appears to play a role in lipid interaction and possibly lipid transport. In these conformations, MD simulations revealed that S242 can form transient contacts with membrane lipids headgroups, suggesting its involvement in facilitating phospholipid access to the transporter cavity (Table S2). Notably, these inward states exhibited the highest RMSD fluctuations among all simulated conditions (Figure S1), highlighting their structural flexibility. This dynamic behavior, especially the varying degrees of inward opening at the TM4–TM6 portal, may be crucial for lipid accommodation, entry and subsequent transport. Specifically, we observed S242 participating in the initial entrance of a palmitoyl-oleoyl phosphatidylserine (POPS) molecule into the transporter in one MD replica of the apo inward simulation (Table S2 and Fig. 3C). Additionally, in simulations of the inhibited state, already containing a bound palmitoyl-oleoyl phosphatidylcholine (POPC) and an inhibitor, S242 displayed potential interactions with another POPC molecule approaching the cavity (Table S2).
On the other hand, S346 (TM6) is located in the vicinity of the substrate binding pocket, which includes the aromatic amino acids H989, W234 and F345 (shown in orange in Fig. 3C, with the substrate (phosphatidylcholine) shown in yellow), with possible direct contacts with H989. Interestingly, and as proposed by the Locher group19,20, W234 (TM4) appears to play multiple roles depending on the conformational state of the transporter. In the apo inward-facing state, it is positioned near the lipid bilayer, likely facilitating phospholipid recruitment and initial PC entry. In the inhibited and occluded states, W234 relocates to the central cavity, where it forms a strong cation–π interaction with the headgroup of PC, stabilizing the substrate. In the ATP-bound, collapsed state, W234 tilts outward again toward the membrane, coinciding with the loss of the PC-binding pocket. Altogether, these observations suggest that W234 contributes not only to substrate binding but also to the conformational transitions by adopting distinct orientations across the transport cycle.
It would appear from the analysis shown in Fig. 3D (bottom views) that the conformational differences observed for helix TM6 between the ATP-bound collapsed/occluded and apo inward-facing/inhibited inward-facing states correlate with the presence/absence of a bidentate H bond between the carbonyl oxygen atom of I342 and the nitrogen atom (main chain) and hydroxyl group (side chain) of S346.
In the apo inward-facing/inhibited inward-facing states, the S346 hydroxyl groups are not involved in H-bonds. However, MD simulations of these states revealed side-chain contacts with T201 and, in one replica, H989. Sporadic interactions with Q197 were also observed. Both T201 and Q197 are also found in contact with S346 in the occluded and ATP-bound, collapsed states, with the most stable and frequent interactions observed in this last conformation (Figure S2). In the inhibited state, S346 additionally forms transient contacts with lipid tails and, in some cases, cholesterol molecules, suggesting a possible additional role in stabilizing the membrane environment (Figure S2).
Worth noting is the close proximity of TM6 (G348) and TM4 (A231-A232) in the occluded state (Fig. 3D-red box), further emphasizing the critical position of the S346 neighbor in these conformational transitions.
In conclusion, S242 (TM4) and S346 (TM6) play key roles in ABCB4 function through their ability to form state-specific hydrogen bonds and contacts during the transport cycle. S346, located near the substrate binding site, engages with similar residues across different conformations, but with varying stability depending on the state. In particular, in the inward-facing conformations - especially the inhibited state - S346 also forms transient contacts with lipid tails, suggesting an additional role in membrane sensing or local stabilization. S242 displays a broader range of state-dependent contacts, reflecting its dynamic role across the transport cycle, and participates in lipid headgroup interactions in inward-facing states, potentially facilitating substrate entry. Mutations at these positions, like S242R and S346I, are thus expected to disrupt substrate binding/uptake or membrane interactions, leading to ABCB4 dysfunction.
Expression and maturation of ABCB4 mutants
The expression and the maturation of the mutants were assessed by Western blot analyses and compared to that of ABCB4-wt. Representative blots are shown in Fig. 4A (full immunoblots are shown in Figure S3), ABCB4-wt migrated essentially with an apparent molecular weight of 160 kDa. A very faint band migrating at 140 kDa was also detected, as previously reported14. Although the expression level of the T437I and S346I mutants is lower than that of the S242R and T1077M mutants, the four mutants displayed the same electrophoretic pattern, they were found predominantly under the slow-migrating 160 kDa form (Fig. 4A). Quantification of replicate data sets showed that the mutants were similar to the wt protein and migrated predominantly as a mature form (Fig. 4B). From these results, we concluded that none of the four mutations impaired the intracellular processing of ABCB4, although we cannot formally exclude the possibility of an altered post-translational processing or stability defect for The T437I and S346I mutants.
(A) Representative Western blot analysis of wt and mutants ABCB4. The expression and the processing of ABCB4-wt and the mutants was examined by Western analysis of whole-cell lysates from stably transfected HEK293 cells. ABCB4 expression was detected following SDS-PAGE and immunoblotting with the P3-II-26 antibody. Tubulin was used as a loading control. Molecular masses are indicated on the right (in kDa). Presented data were cropped from full immunoblots shown in supplementary Figure S3. (B) Densitometric quantification of protein amount was performed with iBright Analysis Software. The mature and immature bands were separately quantified on gels, and their relative amounts were calculated. Results are means (± SD) of at least three independent experiments.
Subcellular localization of ABCB4 mutants in HepG2 and HEK293 cells
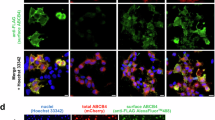
The mutated complementary DNAs reproducing the four ABCB4 variations were transfected in polarized HepG2 cells and in nonpolarized epithelial human embryonic kidney 293 (HEK293) cells. HepG2 cells derived from a human hepatocarcinoma form neo-bile canaliculi in culture and allow localization studies, whereas HEK293 cells are suitable for studies of PC secretion activity. No endogenous ABCB4 was detected in either cell line in our experimental conditions (data not shown). The canalicular/plasma membrane localization of the S346I mutant was previously reported in HepG2 and HEK293 cells13. Forty-eight hours after transfection of HepG2 cells, immunofluorescence showed that ABCB4-wt was exclusively detected at the canalicular membrane where it co-localizes with Multidrug Resistance associated Protein 2 (MRP2) used as a canalicular marker (Fig. 5A), as previously shown12,14,15. In HEK293 cells, subcellular localization of the mutants was studied after stable transfection and selection of stable cell populations. Confocal microscopy showed that as in HepG2 cells, the four mutants were detected exclusively at the plasma membrane (Fig. 5B). These observations indicate that the S242R, S346I, T437I and T1077M mutants did not impair the intracellular trafficking and the plasma membrane localisation of ABCB4.
Subcellular localization of wt and mutants ABCB4 in HepG2 and HEK293 cells by immunofluorescence and confocal microscopy. (A) HepG2 cells were transiently transfected with ABCB4-wt or mutants. Forty-eight hours later, cells were fixed and permeabilized, and processed for immunofluorescence using the anti-ABCB4 (P3-II-26) and anti-MRP2 (M2-I-4) monoclonal antibodies, followed by goat anti-IgG2b Alexa Fluor 488- and goat anti-IgG1 594-conjugated secondary antibodies, and visualized by confocal microscopy. yellow denotes colocalization of ABCB4-wt and all mutants with endogenously expressed MRP2 used as a canalicular marker in merged images. Nuclei are stained in blue with Draq5. Transfected cells are indicated by dashed lines. Bile canaliculi are indicated by asterisks. Bars: 10 μm. (B) Localization of ABCB4-wt or the mutants in stably transfected HEK293 cells was assessed by indirect immunofluorescence using anti-ABCB4 antibodies as in (A). Bars: 10 μm.
PC secretion activity of ABCB4 mutants and effect of CFTR potentiators
3D structure analyses of S242R, S346I revealed that these two serine residues play a critical role in the stabilization of the different conformations adopted by TM4 and TM6 allowing PC binding and translocation, their substitution is expected to destabilize the PC-binding pocket, resulting in a loss of function of ABCB4. Regarding T437I and T1077M mutants, as they are predicted that their PC secretion activity would be altered, a possibility further supported by their normal intracellular processing and canalicular localization. The PC secretion activity of ABCB4 mutants was measured in transiently transfected HEK293 cells and compared to that of ABCB4-wt. The amount of PC released over 24 h was calculated after normalization to the level of the mature ABCB4 protein expressed in the corresponding cell culture well. We found that PC secretion activity of the mutants located in the transmembrane helices S242R and S346I represented only 21% and 14%, respectively and that of the mutants located at equivalent positions of the Walker A of NBD1 and NBD2 T437I and T1077M represented only 16% and 7%, respectively (Fig. 6A, B). These results confirmed that, as predicted by structural analyses, disease-causing mutations in the transmembrane helices and in the conserved motifs impaired the PC secretion activity of ABCB4, highlighting their critical role in ABCB4 function. We next investigated the effects of the CFTR potentiators ivacaftor (VX-770) and SBC219 on the four ABCB4 mutants. To quantify the efficacy with which the CFTR potentiators restore the secretion activity of the mutants, we measured the PC secretion activity of ABCB4-wt and mutants in transiently transfected HEK293 cells treated with 10 µmol/L ivacaftor or 0,1 µmol/L of SBC219 for 24 h. As shown in Fig. 6A, treatment with VX-770 allowed a significant increase of PC secretion activity of T437I and T1077M mutants but not that of S242R and S346I mutants. Conversely, SBC219 treatment allowed a significant increase of PC secretion activity of S242R and S346I but not that of T437I and T1077M (Fig. 6B). Finally, as for wt CFTR, a potentiation of the function of the wt ABCB4 was observed after treatment with SBC219 (Fig. 6B). Altogether, these results indicate that depending on their location (highly conserved NBDs motifs or transmembrane helices), ABCB4 mutants display differential sensitivity to CFTR potentiators.
PC secretion activity of ABCB4-wt and the mutants and response to potentiators. HEK293 cells were transiently transfected with plasmids encoding ABCB4-wt or the indicated mutants, and PC secretion was measured after 24 h in the absence (-) or presence (+) of 10 µmol/L of ivacaftor (A) and 0,1 µmol/L of SBC219 (B). PC secretion was normalized to expression level of the mature form of the respective protein (ABCB4-wt or mutants) and expressed as a percentage of the secretion activity of ABCB4-wt. Results are means (± SD) of at least four independent experiments performed in triplicate. *p < 0.05; ** p < 0.01;***p < 0.001, **** p < 0.0001.
Discussion
In the present study, we show that four disease-causing missense variations in ABCB4, two of which affect homologous residues in the Walker A motifs in the two ATP-binding sites (T437I, NBS2 and T1077M, NBS1) and the other two affect residues of transmembrane helices (S242R, TM4 and S346I, TM6), lead to a loss of ABCB4 function, which can differentially rescued by two distinct CFTR potentiators, ivacaftor and SBC219.
The four mutations studied were identified in patients. Regarding the mutations that affect homologous residues in the Walker A motifs in the two NBDs, the T437I variation was identified in heterozygous patient with LPAC (no.3, Table 1; this study). The T1077M variation was identified in a homozygous 2,5-years-old Yemeni patient with PFIC3 (no.4, Table 1). The patient was not improved by medical therapy and required liver transplantation six months after diagnosis22. A percutaneous liver biopsy revealed established cirrhosis with bile ductular proliferation. In the patient’s liver, immunohistochemistry showed a canalicular staining for ABCB422. This suggested that in the liver, in vivo, the mutation did not affect the targeting of the protein to the canalicular membrane of hepatocytes. In agreement with this observation, we found that ABCB4-T1077M was exclusively detected at the canalicular membrane of HepG2 cells and the plasma membrane of HEK293 cells. For mutations in the transmembrane helices, the S242R variation was identified in heterozygous patient with LPAC (no.1, Table 1; this study). The S346I variation was identified in a homozygous patient with PFIC3, whose case was previously reported (no.2, Table 1)7. In the patient’s liver, immunohistochemistry showed a faint canalicular staining for ABCB4, and the level of phospholipids in bile was 1% (jacquemin et al., 2001). These results support the current observation that the S346I mutant was exclusively detected at the canalicular membrane of HepG2 cells but was dramatically impaired in PC secretion activity.
T437I/T1077M mutations are located in the NBD1:NBD2 homologous positions within the highly conserved Walker A motifs of ABC transporters, which are involved in binding of ATP (they form the phosphate binding loop)18. Indeed, 3D structure analysis indicated that residues T437 and T1077 interact on the one hand with the ATP a-phosphate of the nucleotide-binding sites 2 and 1, respectively and, on the other hand, with the aromatic residues Y403/Y1043 of the A-loops (which stacks the ATP adenine). Their substitution may thus lead to disturb ATP binding via two pathways, affecting adenine and phosphates, resulting in ABCB4 dysfunction. Consistent with this analysis, T437 and T1077 are highly conserved among ABCB4 from different species (supplementary Fig S4). In agreement with the 3D structure analysis, we found that the T1077M mutant was normally processed and targeted to the canalicular membrane of transfected HepG2 cells, whereas its PC secretion activity was dramatically decreased. Despite being predominantly expressed in a mature form and exclusively localized at the canalicular membrane of transfected HepG2 cells, the T437I mutants showed a decreased expression level compared to the wild-type, which may reflect a defect in post-translational processing or a defect in its membrane stability that remain to be determined. Nevertheless, it also exhibited a PC secretion defect, consistent with the in-silico predictions. Likewise, the homologous variation in ABCB11 (T463I) also showed a functional defect25,26. S242R and S346I mutants are located in the TM4 and TM6, respectively. 3D structures of ABCB4 have been solved in multiple conformational states19,20, which were all considered for analysis and molecular dynamics simulation in a lipidic environment (detailed earlier in the results section), highlighting the importance of residues S242 and S346 in the dynamics of the transporter and in the conformational transitions allowing PC entry and binding to the substrate binding pocket. In particular, both amino acids form state-specific hydrogen bonds and contacts during the transport cycle. These transient contacts are established not only with other amino acids, but also with lipids (lipid tails in the case of S346, located near the substrate binding site, lipid headgroups in the case of S242, located at the level of the transporter entrance), suggesting a role in conformational transition, as well as in membrane sensing, local stabilization and substrate uptake. In line with these observations, we found that, although correctly targeted to the canalicular membrane of transfected HepG2 cells, S242R and S346I displayed a major activity defect. As for the T437I mutant, we cannot rule out the possibility that, in addition to the functional defect, the S346I mutant, whose expression level is lower than that of S242R and wt-ABCB4, may also display a processing or membrane stability defect that remain to be determined. According to our functional classification, and as the vast majority of ABCB4 studied variants, the four variants studied herein belong to class III variations13. A major challenge is to find ways of restoring function to these mutants. The clinical success of CFTR-targeting drugs has paved the way for therapeutics options targeting other ABC transporters whose mutations are at the root of rare genetic diseases27. Indeed, we and others previously provided experimental evidence for the successful preclinical use of the clinically approved CFTR potentiator ivacaftor in restoring the PC secretion activity of class III ABCB4 mutants14,15, the bile acid transport function of class III ABCB11 mutants26 and the lipid transport function of class III ABCA3 mutants28. As ivacaftor was initially identified to specifically potentiate CFTR class III gating mutations, it is not surprising that its beneficial effect was exclusive on class III mutants affecting the conserved ATP binding sites of human ABCB4, ABCB11 and ABCA3 that show significant similarity to those of human CFTR. It is thus tempting to speculate that ivacaftor might rescue the function of mutants located in the ATP binding sites of all ABC transporters. However, these speculations remain to be validated experimentally. In line with this observation, we found that ivacaftor was ineffective on S242R and S346I mutants located in TM4 and TM6, respectively, suggesting again selectivity of ivacaftor towards mutants located in the highly conserved ATP binding sites. So far, no specific therapeutic tool has been proposed to rescue the function of ABCB4 class III mutations located in transmembrane helices. In line with our strategy of drug repositioning, we were interested in promising new CFTR potentiators, including the small binder of CFTR SBC21921. SBC219 has been shown to act as a potentiator in a synergic way with ivacaftor on several CFTR mutants21. Interestingly, we showed that SBC219 specifically rescued the function of class III ABCB4 mutants located in transmembrane helices, but was ineffective on those located in the conserved ATP binding sites, suggesting that mutants of nucleotide binding sites and those of membrane-spanning domains display distinct pharmacological profiles. Furthermore, contrary to what has been shown for CFTR potentiation, we did not observe a synergic effect when SBC219 was combined with ivacaftor (our unpublished results). Of particular interest will be to identify the binding sites of ivacaftor and SBC219 in human ABCB4, which may help understanding their mechanisms of action. Of note here is that the ivacaftor-binding site identified in human CFTR from cryo-EM studies, located in a region highly specific to the anion channel and including an unusual conformation TM829, is not present in ABCB4.
In conclusion, the results obtained in the present in vitro study enable the further characterization at the structural and cellular level of the impact of four ABCB4 mutants. Furthermore, the results allow us to refine our previous functional classification and propose an advanced mechanistic classification of functional ABCB4 mutations, mutants still expressed at the canalicular membrane of hepatocytes but with deficient activity, distinguishing (i) defects caused by impaired ATP-binding or hydrolysis which are rescued by the clinically approved CFTR potentiator, ivacaftor; (ii) defects linked to disruptions of conformational transitions associated with PC transport that can be corrected by the CFTR potentiator SBC219. Finally, our results provide a proof of concept and a first step for the development of targeted pharmacotherapy for severe cholestatic liver diseases caused by ABCB4 mutations, for which liver transplantation remains the only effective therapy.
Materials and methods
Patients’ data analyses
Two previously reported13,22,23 PFIC3 and two LPAC patients were included in the present study (Table 1). ABCB4 gene analysis was performed in patients referred to the Reference Center for Inflammatory Biliary Diseases (Hôpital Saint-Antoine, Paris, France), as previously reported30. Clinical phenotypes of the patients were classified according to current spectrum of liver diseases related to ABCB4 gene variations. This study was conducted in accordance with the declaration of Helsinki. The protocol was approved by the local ethical committee. Informed consent was obtained from all participants.
DNA constructs and mutagenesis
The construction of the human wild-type (wt) ABCB4, isoform A (NM_000443.3), in the pcDNA3 vector was reported13. Site-directed mutagenesis was performed using the QuikChange II XL mutagenesis kit from Agilent Technologies (Massy, France). DNA primers used for ABCB4 mutagenesis were from Invitrogen-Life Technologies and are listed in supplementary Table S1. All constructs were verified by automated sequencing.
Antibodies and reagents
Mouse monoclonal anti-ABCB4 (P3-II-26) and anti-MRP2 (multidrug resistance-associated protein 2; M2-I-4) antibodies were purchased from Alexis Biochemicals (San Diego, CA. Goat anti-immunoglobulin G (IgG) 2b Alexa Fluor 488 and goat anti-IgG1 Alexa Fluor 594 secondary antibodies, peroxidase-conjugated secondary antibodies and culture media were from Invitrogen-Life Technologies (Saint-Aubin, France)/GE Healthcare. Ivacaftor (VX-770) was from Selleckchem (Munich, DE, USA). SBC219 was synthesized by J.L Decout (University Grenoble Alpes, France). Sodium taurocholate (NaTC), fatty-acid-free bovine serum albumin (BSA) was from Sigma-Aldrich (Lyon, France).
Cell culture and transfection
Human Embryonic Kidney HEK-293 (ATCC®-CRL-1573™) cells and Human hepatocellular carcinoma HepG2 (ATCC®- HB-8065™) cells were obtained from ATCC (Manassas, VA). HepG2 cells and human embryonic kidney (HEK) 293 were grown at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM) with 10% heat-inactivated fetal bovine serum, 1% penicillin/streptomycin and 1% Sodium pyruvate. Under these conditions HepG2 cells form canaliculi and show no endogenous ABCB4 expression. ABCB4-wt and mutant encoding vectors were transiently transfected in HepG2 and HEK293 cells using jetPRIME® transfection reagent and Turbofect, respectively (Ozyme, France and Thermo-Fisher Scientific, Villebon-sur-Yvette, France), following the manufacturer’s instructions. Transient transfection was used for PC secretion assays, as the protein expression levels were compatible with our measurement method. Stable transfection was used for Western blot and immunofluorescence to ensure homogenous expression levels and to avoid signal oversaturation due to overexpression observed with transient transfection. In order to establish cells that stably express the different constructs, 1 million cells are transfected with 2.5 µg of plasmid DNA using Turbofect (ratio 1/2, plasmid/Turbofect). Twenty-four hours post-transfection, the cells are trypsinized and selected for 3 weeks with 400 µg/ml of G418. The total populations are maintained with 100 µg/ml of G418.
Western blot and Immunofluorescence
Transfected cells were lysed on ice for 10 min in 20 mM Tris HCl, 150 mM NaCl, 1 mM ethylenediaminetetra-acetic acid, pH 7.4, containing 1% (wt/vol) Triton X-100 in the presence of a protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland). Lysates were centrifuged at 11,000 rpm for 10 min to remove insoluble materials. Equal amounts of protein were directly processed for sodium dodecyl sulfate polyacrylamide gel electrophoresis on 7,5% polyacrylamide gels. Immunoblotting was performed using the mouse monoclonal anti-P3-II-26 or anti-tubulin antibodies followed by horseradish peroxidase–conjugated mouse-specific secondary antibody. Development of peroxidase activity was performed with the ECL Plus detection kit. Images were obtained using the iBright reader and analysed with the iBright Analysis software.
For immunofluorescence, HepG2 and HEK293 cells were grown and transfected on glass coverslips. For the detection of ABCB4, cells were fixed with methanol for 30 s at − 20 °C, and processed for immunolabeling with the mouse monoclonal anti-ABCB4 P3-II-26 and the mouse monoclonal anti-MRP2 (M2-I-4) as described31. Nuclei were stained with Dapi. Confocal imaging was acquired with Olympus FV3000 microscope with a 20x immersion objective, 1.4 zoom. Digital images were processed with Image J software (NIH, MA).
Treatment and PC secretion measurement
HEK293 cells were seeded on poly-L-lysine precoated six-well plates at a density of 1.3 × 106 cells/well. At least four hours after seeding, cells were transiently transfected with 1 µg of ABCB4-encoding plasmids. Twenty-four hours post-transfection, cells were washed twice with Hank’s balance salt solution, and then the medium was replaced by phenol red-free Dulbecco’s Modified Eagle’s Medium containing 0.5 mmol/L of NaTC and 0.02% fatty-acid-free BSA in the presence or absence of 10 µmol/L of ivacaftor or 0,1 µmol/L /L of SBC219. Media were collected 24 h later. Collected media were centrifugated at 6 000 rpm for 7 min at 4 °C to remove aggregates and cell debris. Total lipid content was extracted from supernatants by chloroform/methanol/water partition, as previously described32. After collection and evaporation of the organic phase, lipid samples were resuspended in 110 µl of phosphate-buffered saline containing 0.1% Triton X-100 (%w/v), incubated for 1 h at 60 °C, sonicated for 10 min and centrifugated 5 min at 1500 rpm. Measurement of PC content was performed with a fluoro-enzymatic methods based on the measurement of choline released after phospholipase D treatment as described16. Choline contents measured were normalized to the expression levels of mature form of ABCB4, which were quantified from immunoblots of the associated cell lysates. PC secretion was quantified using the following formula:
nPC = [(FPPLD+-FPLD+) – (FPPLD−-FPLD−)] / D.
PC secretions of ABCB4-wt and ABCB4-mutant were quantified using the fluorescent intensities measured for the tested condition in the absence (FPPLD−) or presence (FPPLD+) of phospholipase D, and the background fluorescent intensities in the absence (FPLD−) or presence (FPLD+) of phospholipase D. PC secretions were then normalized to the mature protein level (D) of their respective mature form and expressed as percentage of the wild-type.
3D structure analysis and molecular dynamics simulations
Experimental 3D structures of human ABCB4 were extracted from the Protein Data Bank (PDB; https://www.rcsb.org/) and visualized using Chimera33.
Four cryo-EM structures were used as starting points for molecular dynamics (MD) simulations: the ATP-bound, collapsed state (PDB ID: 6S7P)19, the apo inward-facing conformation (7NIU)20, the inhibited inward-facing conformation in complex with phosphatidylcholine (PC) and posaconazole (7NIW)20 and the occluded state bound to PC (7NIV)19. Each protein model was embedded in a mixed lipid bilayer composed of POPC, palmitoyl-oleoyl phosphatidylethanolamine (POPE), POPS, DSM and cholesterol in a 2:1:1:1:1 ratio. The position of the membrane bilayer was estimated using the PPM server34. All systems were solvated using a solution of 150mM NaCl. The protein, lipids and ions were modeled using the CHARMM36m force field35 and TIP3P model was employed for water. The simulations were performed using different versions of Gromacs (2019, 2020 and 2023)36 depending on the specific computational platform used. Standard input files generated by CHARMM-GUI were used for system preparation, including energy minimization and equilibration. During these initial phases, harmonic positional restraints were initially applied to the heavy atoms of the protein and the lipid headgroups, and gradually released over 1.2 ns.
Production MD simulations were then conducted in the NPT ensemble, without any restraints, at temperature of 310 K and a pressure of 1 bar, respectively, using the Nose–Hoover thermostat37 and Parrinello–Rahman barostat38. Periodic boundary conditions were applied, and long-range electrostatic interactions were treated using the particle mesh Ewald algorithm39, with a switching function employed between 10 and 12 Å for non-bonded interactions. Bond lengths involving hydrogen atoms were constrained using LINCS40. The integration timestep was set at 2 fs, and the total trajectory length was ~ 1000 ns.
For the trajectory analysis of the simulations, Root Mean Square Deviation (RMSD) calculation were performed using MDAnalysis in python341, considering only the Cα atoms of the protein. To assess specific interactions, contact analysis were performed using the VLDM (Voronoi Laguerre Delaunay for Macromolecules) program42,43. For each amino acid of interest, we quantified the frequency of contacts with neighboring residues and surrounding lipids. Contact occurrences were plotted using custom in-house R scripts.
Statistical analysis
Data are shown as means ± SD. Graphics were performed using Prism version 8.4.3 (GraphPad software, La Jolla, CA, USA). Statistical significance among means was determined using the Student t test. A P value < 0.05 was considered significant.
Data availability
The data that support the findings of this study are available from the corresponding author (T.A.S) upon reasonable request.
References
Ruetz, S. & Gros, P. Phosphatidylcholine translocase: a physiological role for the mdr2 gene. Cell 77, 1071–1081 (1994).
van Helvoort, A. et al. MDR1 P-glycoprotein is a lipid translocase of broad specificity, while MDR3 P-glycoprotein specifically translocates phosphatidylcholine. Cell 87, 507–517 (1996).
Boyer, J. L. Bile formation and secretion. Compr. Physiol. 3, 1035–1078 (2013).
Reichert, M. C. & Lammert, F. ABCB4 gene aberrations in human liver disease: an evolving spectrum. Semin Liver Dis. 38, 299–307 (2018).
Stättermayer, A. F., Halilbasic, E., Wrba, F., Ferenci, P. & Trauner, M. Variants in ABCB4 (MDR3) across the spectrum of cholestatic liver diseases in adults. J. Hepatol. 73, 651–663 (2020).
Sticova, E. & Jirsa, M. ABCB4 disease: many faces of one gene deficiency. Ann. Hepatol. 19, 126–133 (2020).
Jacquemin, E. et al. The wide spectrum of multidrug resistance 3 deficiency: from neonatal cholestasis to cirrhosis of adulthood. Gastroenterology 120, 1448–1458 (2001).
Nalbantoglu, I. & Jain, D. Cryptogenic cirrhosis: old and new perspectives in the era of molecular and genomic medicine. Semin Diagn. Pathol. 36, 389–394 (2019).
Rosmorduc, O. et al. ABCB4 gene mutation-associated cholelithiasis in adults. Gastroenterology 125, 452–459 (2003).
Floreani, A. et al. Hepatobiliary phospholipid transporter ABCB4, MDR3 gene variants in a large cohort of Italian women with intrahepatic cholestasis of pregnancy. Dig. Liver Dis. Off J. Ital. Soc. Gastroenterol. Ital. Assoc. Study Liver. 40, 366–370 (2008).
Poupon, R. Ursodeoxycholic acid and bile-acid mimetics as therapeutic agents for cholestatic liver diseases: an overview of their mechanisms of action. Clin. Res. Hepatol. Gastroenterol. 36 (Suppl 1), S3–12 (2012).
Delaunay, J. L. et al. A missense mutation in ABCB4 gene involved in progressive familial intrahepatic cholestasis type 3 leads to a folding defect that can be rescued by low temperature. Hepatol. Baltim. Md. 49, 1218–1227 (2009).
Delaunay, J. L. et al. A functional classification of ABCB4 variations causing progressive familial intrahepatic cholestasis type 3. Hepatol. Baltim. Md. 63, 1620–1631 (2016).
Delaunay, J. L. et al. Functional defect of variants in the adenosine triphosphate-binding sites of ABCB4 and their rescue by the cystic fibrosis transmembrane conductance regulator potentiator, Ivacaftor (VX-770). Hepatol. Baltim. Md. 65, 560–570 (2017).
Delaunay, J. L. et al. Ivacaftor-Mediated potentiation of ABCB4 missense mutations affecting critical motifs of the NBDs: repositioning perspectives for hepatobiliary diseases. Int. J. Mol. Sci. 24, 1236 (2023).
Gautherot, J. et al. Phosphorylation of ABCB4 impacts its function: insights from disease-causing mutations. Hepatol. Baltim. Md. 60, 610–621 (2014).
Khabou, B. et al. Comparison of in Silico prediction and experimental assessment of ABCB4 variants identified in patients with biliary diseases. Int. J. Biochem. Cell. Biol. 89, 101–109 (2017).
Kroll, T., Prescher, M., Smits, S. H. J. & Schmitt, L. Structure and function of hepatobiliary ATP binding cassette transporters. Chem. Rev. 121, 5240–5288 (2021).
Olsen, J. A., Alam, A., Kowal, J., Stieger, B. & Locher, K. P. Structure of the human lipid exporter ABCB4 in a lipid environment. Nat. Struct. Mol. Biol. 27, 62–70 (2020).
Nosol, K. et al. Structures of ABCB4 provide insight into phosphatidylcholine translocation. Proc. Natl. Acad. Sci. U. S. A. 118, e2106702118 (2021).
Froux, L. et al. Targeting different binding sites in the CFTR structures allows to synergistically potentiate channel activity. Eur. J. Med. Chem. 190, 112116 (2020).
Shankar, S. et al. A new variant of an old itch: novel missense variant in ABCB4 presenting with intractable pruritus. J. Clin. Exp. Hepatol. 12, 701–704 (2022).
Chen, R. et al. Clinical and genetic characterization of pediatric patients with progressive familial intrahepatic cholestasis type 3 (PFIC3): identification of 14 novel ABCB4 variants and review of the literatures. Orphanet J. Rare Dis. 17, 445 (2022).
Mornon, J. P., Hoffmann, B., Jonic, S., Lehn, P. & Callebaut, I. Full-open and closed CFTR channels, with lateral tunnels from the cytoplasm and an alternative position of the F508 region, as revealed by molecular dynamics. Cell. Mol. Life Sci. CMLS. 72, 1377–1403 (2015).
Liu, H. et al. Structural basis of bile salt extrusion and small-molecule Inhibition in human BSEP. Nat. Commun. 14, 7296 (2023).
Mareux, E. et al. Functional rescue of an ABCB11 mutant by ivacaftor: A new targeted pharmacotherapy approach in bile salt export pump deficiency. Liver Int. Off J. Int. Assoc. Study Liver. 40, 1917–1925 (2020).
Moore, J. M., Bell, E. L., Hughes, R. O. & Garfield, A. S. ABC transporters: human disease and pharmacotherapeutic potential. Trends Mol. Med. 29, 152–172 (2023).
Kinting, S. et al. Potentiation of ABCA3 lipid transport function by Ivacaftor and genistein. J. Cell. Mol. Med. 23, 5225–5234 (2019).
Liu, F. et al. Structural identification of a hotspot on CFTR for potentiation. Science 364, 1184–1188 (2019).
Ziol, M. et al. ABCB4 heterozygous gene mutations associated with fibrosing cholestatic liver disease in adults. Gastroenterology 135, 131–141 (2008).
van Til, N. P. et al. Alteration of viral lipid composition by expression of the phospholipid floppase ABCB4 reduces HIV vector infectivity. Retrovirology 5, 14 (2008).
Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Lomize, A. L., Todd, S. C. & Pogozheva, I. D. Spatial arrangement of proteins in planar and curved membranes by PPM 3.0. Protein Sci. Publ Protein Soc. 31, 209–220 (2022).
Huang, J. & MacKerell, A. D. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013).
Abraham, M. J. et al. High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2. GROMACS, 19–25 (2015).
Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 81, 511–519 (1984).
Parrinello, M. & Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981).
Darden, T., York, D. & Pedersen, L. Particle mesh ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Hess, B. P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4, 116–122 (2008).
Gowers, R. J. et al. MDAnalysis: A Python package for the rapid analysis of molecular dynamics simulations. scipy (2016). https://doi.org/10.25080/Majora-629e541a-00e
Elbahnsi, A., Retureau, R., Baaden, M., Hartmann, B. & Oguey, C. Holding the nucleosome together: A quantitative description of the DNA-Histone interface in solution. J. Chem. Theory Comput. 14, 1045–1058 (2018).
Esque, J., Oguey, C. & de Brevern, A. G. A novel evaluation of residue and protein volumes by means of Laguerre tessellation. J. Chem. Inf. Model. 50, 947–960 (2010).
Acknowledgements
We are grateful to Romain Morichon for confocal microscopy imaging which was performed with financial support from “ITMO Cancer of Aviesan within the framework of the 2021-2030 Cancer Control Strategy, on funds administered by Inserm”. We thank Dr. Véronique Barbu for providing patients’ data, Anne-Marie Durand-Schneider and Chantal Housset for fruitful discussions and support during this study, and Yves Chrétien for his help with Adobe softwares. MD simulations were performed using High-Performance Computing (HPC) resources from Grand Equipement National de Calcul Intensif (GENCI)/Centre Informatique National de l’Enseignement Supérieur (CINES) (Grant 2023-A0150314556 on Adastra MI250 machine). C.M. was supported by the « Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation ». T.A.S. was supported by grants from the Fondation pour la Recherche Médicale (FRM-EQU-2020-03010517), the association Mucoviscidose-ABCF2 and FILFOIE (Filière de santé des maladies rares du foie, Paris, France).
Author information
Authors and Affiliations
Contributions
C.M., J.L.D. (Jean-Louis Delaunay) and T.A.S. designed the study. C.M., J.L.D. (Jean-Louis Delaunay), A.E., I.C. and T.A.S. performed the experiments. C.M., J.L.D. (Jean-Louis Delaunay), A.E., A.S., S.L. (Sylvie Lagaye), P.C., S.L. (Sara Lemoinne), C.C., B.B., N.C., J.G., J.L.D. (Jean-Luc Décout), I.C., and T.A.S. analysed the data and provided intellectual contribution. T.A.S., A.E. and I.C. wrote the manuscript, which was reviewed and approved by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Madry, C., Elbahnsi, A., Delaunay, JL. et al. ABCB4 disease-causing variants S242R, S346I, T437I and T1077M significantly impair its function and display differential sensitivity to potentiators. Sci Rep 15, 44544 (2025). https://doi.org/10.1038/s41598-025-28407-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-28407-6