Abstract
Critical for cellular maintenance and repair, autophagy recycles damaged or redundant cellular components. Dysregulated autophagy is associated with progressive cellular injury in various liver pathologies. However, the role of autophagy in intestinal failure-associated liver disease has not previously been investigated. Using a preclinical murine model of short bowel syndrome, we explored the role of autophagy impairment at early and late time points. Regardless of sex, small bowel resected (SBR) mice demonstrated significant liver injury compared to sham controls, exhibiting elevated serum transaminases, altered liver histology, and increased mRNA expression of endoplasmic reticulum stress marker (Ddit3) and oxidative stress marker (Nqo1). Autophagy impairment was supported by increased p62 mRNA and protein expression in SBR liver and then confirmed with qualitative autophagy flux assay which suggested alteration of autophagy induction in SBR liver. Single nuclear RNA sequencing identified a cluster of hepatocytes in the SBR liver with increased p62 expression. These hepatocytes showed enrichment of genes related to the PI3K-Akt and ErbB pathways both which converge on mTORC1. Immunohistochemical distribution of pS6 confirmed increased mTORC1 activation in the SBR hepatocytes. Overall autophagy dysregulation is correlated with mTORC1 activation in SBR mice and is associated with greater liver injury.
Similar content being viewed by others
Introduction
Short gut syndrome is a highly comorbid condition resulting from extensive small bowel resection (SBR). A complication of short gut syndrome is intestinal failure associated liver disease (IFALD). While previously thought to be a direct consequence of prolonged parenteral nutrition, recent evidence has shown that IFALD has a multifactorial etiology that includes bowel resection, shifting microbiome, perturbed bile acid metabolism and disrupted enterohepatic circulation1. As such, we have developed a parenteral nutrition-independent model of IFALD using bowel resection alone. Multiple investigations have been published using this model2,3,4,5. Recently, we described the presence of endoplasmic reticulum (ER) stress and subsequent aberrant unfolded protein response in the liver after SBR in mice6. Pursuant to this finding, we sought to understand the role of autophagy in processing these misfolded proteins.
Autophagy is a conserved, lysosomal-dependent catabolic cellular pathway by which abnormal cytoplasmic components (including damaged organelles, lipid droplets, and protein aggregates) are degraded, and their constituents recycled. The mechanisms regulating cellular autophagy involve multiple intersecting pathways7,8,9. The formation of autophagosomes requires autophagy-related genes (ATGs) that produce six functional protein groups, including the ATG1 (Unc-51 like kinase in mammals) kinase complex, the ATG9 vesicle, the ATG14-containing PtdIns 3-kinase complex, the ATG2–ATG18 complex, the ATG12 conjugation system, and the ATG8 conjugation system10. Activated by stressors such as starvation, hypoxia, oxidative stress, protein aggregation or ER stress, autophagy begins by stimulation of the Unc-51 like kinase-1 complex. This activated complex phosphorylates the PI3KC3 complex 1 to then build an omegasome at the ER membrane producing phosphatidylinositol-3-phosphate (PI3P). PI3P recruits additional ATGs including mammalian ATG8 homologs referred to as microtubule-associated protein 1 light chain 3 (LC3). This recruitment results in conjugation of LC3 to phosphatidylethanolamine, which creates the characteristic phospholipid of autophagosomes. Ubiquinated-p62 cargo then binds the LC3. The autophagosomal membrane is elongated and sealed using cellular donated membranes resulting in a mature autophagosome. A lysosome fuses with the autophagosome and the acidic hydrolases degrades the cargo. Disruption of this process can result in buildup of various damaged proteins and organelles and may result in cellular injury. Specific to our present study, dysregulated autophagy has been associated with a variety of liver pathologies9.
An important step in the autophagy mechanism is the tagging of cargo destined for recycling. One of these tagging proteins is p62 (Sqtsm1 gene). Considered to be a selective autophagy receptor, one of its core functions is to bind cargo destined for autophagy11,12. In conditions of impaired autophagy, p62 levels accumulate. Thus, researchers have used this phenomenon as a surrogate marker for impaired autophagy12. Static observations of autophagic vesicles or machinery are often misleading and do not provide a nuanced understanding of all cellular processes. Because of this, investigations on autophagy require direct observation and/or measurement of autophagy flux using LC3 or p62 as a surrogate13.
In this study we demonstrate that autophagy is impaired in hepatocytes following SBR using both surrogate and direct observations. We further use single nuclear RNA sequencing to garner more nuanced insights into the potential mechanisms underlying autophagy impairment.
Results
SBR results in characteristic body morphological changes and liver injury
As validated in previous studies6,14, SBR mice demonstrated characteristic initial weight loss after resection with a nadir between 2–3 weeks followed by gradual weight gain over the subsequent 8 weeks (Figure S1a). Compared to female SBR mice, male SBR mice had decreased postoperative weight gain over time. Analysis of body composition at 10 weeks revealed the SBR cohorts had a reduced percentage of fat mass and greater lean body mass compared to sham mice (Figure S1b). Furthermore, SBR mice exhibited evidence of intestinal adaptation with increased villus height compared to the same mouse’s intraoperative small bowel sample taken at the time of SBR surgery (Figure S1c-e). Adaptation was evident at both 2 and 10 weeks after resection.
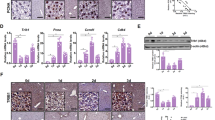
Liver injury was evaluated by measurement of serum transaminases, qualitative histology, Sirius red staining for collagen deposition, and rt-qPCR for markers of ER and oxidative stress. At both 2 and 10 weeks, the SBR mice exhibited elevated transaminases compared to sham-operated controls (Fig. 1a-b). The magnitude of serum transaminases decreased over the study course in SBR mice. Male SBR mice had an early increase in AST/ALT compared to female SBR mice. Dungubat et. al. reported a similar difference in male and female mice in a non-alcoholic fatty liver disease murine model. Male mice exhibited higher transaminases compared to female mice at earlier time points but normalized at later time points15. In our study, after 2 weeks, both male and female SBR mice had increased liver injury documented by uptrending Nonalcoholic Steatohepatitis Clinical Research Network (NAS CRN) composite scores (Fig. 1d) of H&E liver sections (Fig. 1c) that became more apparent by week 10. Notably the composition of the NAS CRN composite scores differed between sexes, with steatosis a more dominant feature in the female cohort (Figure S2a). The degree of lobular inflammation appeared to increase overtime in both sexes, while hepatocyte ballooning was present early in females and gradually appeared in males (Figure S2b-c). Despite the differences in steatohepatitis features, all SBR mice, regardless of sex, exhibited similar degrees of liver fibrosis at 10 weeks (Fig. 1c and e, S2d). As shown in our previous study, damage inducible transcript 3 (Ddit3), an ER stress marker, was increased in SBR livers at both time points regardless of sex (Figure S2e-f). Additionally, the relative increase in mRNA expression of the antioxidant NAD(P)H quinone dehydrogenase 1 (Nqo1) indicated that SBR livers exhibit increased and persistent oxidative stress.
SBR induced liver injury at 2 and 10 weeks after surgery. a-b. Serum alanine transaminase (ALT) and aspartate transaminase (AST) were increased in SBR mice compared to sham mice at 2 weeks (a) and 10 weeks (b) (2 week—female sham n = 7, female SBR n = 8, male sham n = 11, male SBR n = 6; 10 week—female sham n = 6, female SBR n = 7, male sham n = 7, male SBR n = 11) c. Representative histological images and Sirius red staining of male liver from the 2- and 10-week cohorts taken at 20X objective. SBR liver demonstrated progressive liver injury over time with steatosis (yellow circle), hepatocyte ballooning (blue arrow), periportal inflammation (red arrow) and liver fibrosis (black arrow) at 10 weeks (female liver histology was similar and is reported in Supplementary Fig. 2d). d. NAS CRN composite scores for each week across sexes showed SBR has increased markers for steatohepatitis compared to sham. Female SBR mice appeared to have a more pronounced steatohepatitis phenotype than males. Individual scores for steatosis, lobular inflammation and hepatocyte ballooning are in Supplementary Fig. 2a-c (note only a subset of samples were submitted for histology scoring, n = 5 were not scored). e. NAS CRN fibrosis scores at 10 weeks demonstrated SBR livers have increased fibrosis compared to shams but equal fibrotic signatures between the sexes (only a subset of samples underwent Sirius Red staining, n = 5 were not stained). For panels a-b, statistical analysis completed with ANOVA and Tukey Correction. Due to the ordinal nature, a nonparametric (Kruskal–Wallis) ANOVA with Dunn correction was completed at each time point for panels d-e. Bar height represents mean; error bars represent SD. *P < 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
SBR-associated liver injury is characterized by dysregulated autophagy
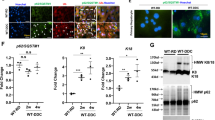
To determine the role of autophagy in the SBR induced liver injury model, we assayed the surrogate marker p62 using immunohistochemistry (Fig. 2a). At both 2 and 10 weeks after surgery, p62 was detected mostly in the hepatocytes of SBR liver, while sham liver showed minimal staining in both male and female mice. The increase in p62 mRNA and protein expression was directly confirmed with western blot (females Fig. 2b; males Fig. 2c, unedited western blots Figure S3a-d) and rt-qPCR (Fig. 2d). Taken together, this relative buildup of p62 in SBR liver indicated autophagy alteration.
p62 (a marker for autophagy) was increased in SBR liver. a. Representative immunohistochemistry slides of p62 in male livers at both 2 and 10 weeks post resection showed increased p62 protein expression in SBR liver (female images are not shown but have similar phenotype). Black arrows represent areas of p62 deposition. b-c. Representative western blots of sham and SBR liver showed increasing protein expression at both 2- and 10-weeks post resection regardless of sex. Both p62 and GAPDH were run on the same gel. Rows were cut and arranged for easier analysis. Unedited blots are in Supplementary Fig. 3a-d. d. rt-qPCR at both 2 and 10 weeks showed elevated p62 mRNA expression in the SBR mice irrespective of sex (2 week—female sham n = 7, female SBR n = 8, male sham n = 10, male SBR n = 5; 10 week—female sham n = 6, female SBR n = 7, male sham n = 7, male SBR n = 8) For panels b, c and d, statistical analysis was completed with unpaired t-test and ANOVA with Tukey correction as appropriate. Bar height represents mean; error bars represent SD. *P < 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
We employed a qualitative autophagy assay to confirm impaired autophagy in hepatocytes16. Hepatocytes were isolated from a subset of 2-week male SBR and sham mice. Viable hepatocytes were cultured and infected with GFP-labelled lipated light chain 3 (GFP-LC3II). LC3II is a product of the autophagy cascade and a necessary component of the autophagophore development. Autophagy was then induced via starvation. Hepatocytes were also co-stained with Tetramethylrhodamine methyl ester (TMRM), a mitochondrial fluorescent dye that is membrane potential dependent. After the stimulus of starvation, sham hepatocytes mobilized their lipid stores through lipophagy as expected (Fig. 3a). GFP-LC3II vesicles were noted to form around lipid droplets within the sham hepatocyte (white asterisk). In contrast, the SBR hepatocyte showed no mobilization of lipid stores; GFP-LC3II was localized to the ER with no surrounding lipid droplet staining observed (Fig. 3b). These observations indicated that autophagy was inhibited at an early point of regulation in the liver after SBR. Additionally, co-staining with TMRM revealed that sham mitochondria tend to have a high intensity of mitochondrial staining (Fig. 3c). In contrast, the SBR hepatocytes demonstrated a relatively low intensity of mitochondrial staining (Fig. 3d). This suggested SBR mitochondria have an altered membrane potential, most likely from mitochondrial membrane uncoupling. In the sham hepatocyte, we observed evidence of ongoing mitophagy with GFP-LC3II vesicles encircling these dysfunctional mitochondria (Fig. 3e, white ovals). Overall, both surrogate (p62) and qualitative visualization of autophagy alteration revealed that sham hepatocytes had intact autophagy (lipophagy and mitophagy), while SBR hepatocytes had impaired autophagy despite the presence of damaged mitochondria.
Confocal analysis of autophagy flux in isolated hepatocytes at 2 weeks post resection. Isolated hepatocytes from sham-operated (n = 2) and SBR mice (n = 2) were subjected to culture with infection of GFP-LC3II (green) and staining of mitochondria with TMRM, a membrane potential dependent fluorescent dye (red). Autophagy was induced via starvation of the cultured hepatocytes. a. Representative image of a sham operated hepatocyte showing successful initiation of lipophagy as evidenced by the GFP-LC3II-positive vesicle formation around a lipid droplet (example shown by white asterisk). b Conversely, a representative image of SBR hepatocytes demonstrates no evidence of lipophagy. c. Another representative image of the sham operated hepatocytes demonstrating brightly stained TMRM mitochondria suggestive of normal resting membrane potential. This panel also contains blue and purple boxes highlighting areas of increase magnification. d. Another representative image of SBR hepatocytes showing decreased TMRM staining suggesting a disrupted membrane potential. e. Increased magnification of 2 instances of visualized mitophagy in the sham operated mice with GFP-LC3II-positive vesicle formation surrounding mitochondria (white ovals). All images were taken in live hepatocytes with the Zeiss 880 confocal microscope. Panels a-d are taken at same magnification; images in panel e are 12X magnified from panel c.
Single nuclear RNA sequencing reveals a potential mechanism related to mTORC1 activation in hepatocytes after SBR
Single nuclear RNA sequencing was performed at the 2 week postoperative timepoint in male livers. Annotation of recovered cells demonstrated populations of hepatocyte, stellate, cholangiocyte, liver sinusoidal, and immune cells. Notably a cluster of hepatocytes (cluster 5 hepatocytes) were present more prominently in SBR compared to sham operated livers (Fig. 4a). We previously detailed these hepatocytes as marked by Annexin A2 and postulated they contributed to liver injury at two weeks17. Interestingly, these hepatocytes also display the highest relative mRNA expression of p62 (Sqtsm1 gene) (Fig. 4b). By comparing the differentially expressed genes of cluster 5 hepatocytes, we found substantial differential expression between the sham and SBR conditions (Fig. 4c). Kyoto Encyclopedia of Genes and Genomes (KEGG) overrepresentation analysis showed cluster 5 hepatocytes had enrichment of genes related to the ErbB signaling and PI3K-Akt pathways, as well as gene sets related to cancer (Fig. 4d). KEGG gene set enrichment analysis (GSEA) further demonstrates that these unique hepatocytes had enrichment of genes related to the ER protein processing, ubiquitin mediated proteolysis, and ribosomal biogenesis with suppression of hepatic metabolism gene sets such as fatty acid degradation and amino acid metabolism (Fig. 4e).
Single nuclear RNA sequencing revealed a hepatocyte cluster with increased p62 (Sqtsm1) expression in SBR liver. a. UMAP plot of recovered nuclei in both Sham (n = 3) and SBR (n = 2) conditions. The hepatocytes labeled “Cluster 5 Hepatocytes” was representative of the SBR condition. b. A feature plot and dot plot of p62 (Sqtsm1) expression demonstrated these Cluster 5 hepatocytes have increased mRNA expression of p62 compared to other cell types. c. Volcano plot of the cluster 5 hepatocytes in the SBR and sham conditions showed large differential gene expression. d. Kyoto Encyclopedia of Genes and Genomes (KEGG) overrepresentation analysis of the differentially expressed genes in the SBR cluster 5 hepatocytes relative to the sham cluster 5 hepatocytes showed upregulation of genes related to the PI3K-Akt and ErbB pathways. e. KEGG gene set enrichment analysis showed the predicted genes sets that are activated in the cluster 5 hepatocytes relative to the sham cluster 5 hepatocytes. Abbreviations: liver sinusoidal endothelial cell (LSEC), conventional dendritic cell (cDC), plasmacytoid dendritic cell (pDC), macrovascular endothelial cell (MaVEC).
Considering these enriched pathways in the cluster 5 hepatocytes, we noted that ErbB signaling pathways and PI3K-Akt can both converge on the mTOR canonical pathway. The mTOR protein kinase has 2 different functional complexes: mTORC1 and mTORC2. mTORC2 promotes cellular proliferation and survival while mTORC1 promotes cell growth, metabolism, and ribosomal translation. mTORC1 activation is a known inhibitor of the formation of the Unc-51 like kinase complex, a necessary complex for autophagy induction. Since our flux assay suggested early inhibition of the autophagy flux, we hypothesized that mTOR is activated in our model and results in the SBR associated autophagy inhibition. Further evidence indicative of mTOR activation was the increased ribosomal biogenesis gene set observed in the GSEA analysis. To confirm activated mTORC1, we stained liver with phosphorylated ribosomal protein S6 (p-S6), a marker of mTOR activation. At both 2 and 10 weeks, SBR liver had increased p-S6 activation compared to sham livers on immunohistochemistry (IHC) (Fig. 5a-b). This staining suggests our model activates mTORC1 in the liver. Future work is necessary to determine the precise cause of mTORC1 activation as well as the contribution of this mTORC1 mediated inhibition of autophagy in SBR induced liver injury.
mTORC1 was activated in the SBR hepatocytes. a. Representative female liver immunohistochemistry samples of pS6, a downstream effector of mTORC1 activation (male images not shown but have similar phenotype). Black arrows designate areas of pS6 deposition. b. Quantification of pS6 in a 4 × field using QuPath software showed both male and female SBR livers had increased pS6 staining at both two and ten weeks, implying persistent mTORC1 activation ((2 week—female sham n = 3, female SBR n = 3, male sham n = 6, male SBR n = 6; 10 week—female sham n = 3, female SBR n = 3, male sham n = 2, male SBR n = 2)). Statistical analysis was performed for panel b with an unpaired t-test. Bar height represents mean; error bars represent SD. *P < 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Discussion
In this study, we found dysregulated autophagy appears to be present within a SBR-associated liver injury model. p62 was a consistent marker for liver injury and suggestive of dysregulated autophagy. Qualitative autophagy assay provided additional evidence that autophagy may be dysregulated and likely inhibited at an early point in the autophagy cascade. We further hypothesize that mTOR activation was a possible mechanism for autophagy inhibition.
Selective autophagy has been shown to be dysfunctional in other liver injury models9. In experimental metabolic-associated syndrome liver disease (MASLD) animal-models, observations of failed autophagosome-lysosome fusion, decreased transcription of lysosomal hydrolases, and suppressed biogenesis of lysosomes portend a late phase dysregulation of the autophagy flux18. Zeng et. al. were able to recover autophagy flux using a biodegradable acid-activated acidifying nanoparticle that selectively enhanced lysosomal function and thus reduced hepatic steatosis in a murine model of NAFLD19. In the hepatocellular carcinoma (HCC) model, autophagy flux dysregulation was likely secondary to aberrant signaling pathways such as the Sqstm1-Nrf2 and mTORC1 pathways9. However, to our knowledge, autophagy impairment has not been investigated in the setting of IFALD. While exact mechanisms contributing to dysfunctional autophagy remain unknown, several key observations provide hypotheses for further examination.
Previously, we have found that surgical resection alters intestinal lymphatic structures and transcriptional profile of the remaining ileum3,4. These alterations may affect dietary lipid absorption possibly shunting lipids through the portal system. Additionally, we have previously demonstrated that SBR leads to increased gut permeability and portal serum endotoxin that correlates with the degree of resection20. Finally in this study, we have demonstrated persistent endoplasmic reticulum (ER) stress, reactive oxidative stress (increased Nqo1 expression), and mitochondrial injury in the SBR hepatocytes6. Taken together, we hypothesize that disrupted lipid metabolism and gut microbiome changes after small bowel resection may be drivers of liver injury. In fact, studies have shown that an overload of saturated free fatty acids increased the ER stress in hepatocytes21 and that ER stress itself resulted in misfolded proteins, ultimately stimulating the autophagy response22. Additionally, excessive fatty acids can overload the beta-oxidation flux in mitochondria resulting in excessive mitochondrial reactive oxygen species, uncoupling, and ultimate permeability23. Endotoxins may also cause direct liver injury by inducing an inflammatory cascade and oxidative stress within hepatocytes24. In a normal state, these processes should stimulate selective autophagy and mitophagy. However, our qualitative autophagy assay suggested autophagy impairment at the level of induction in SBR mice. Single nuclear transcriptomics and IHC analyses suggest the PI3K-Akt-mTORC pathway is a possible contributor to this inhibition.
The canonical mTOR pathway is involved in the proliferation, survival, growth, and metabolism of cells as well as ribosomal translation and is highly regulated by the PI3K-Akt pathway25. While the exact mechanism is unclear, both MASLD and HCC liver injury models have demonstrated evidence of mTOR activation26,27. Additionally, multiple studies have demonstrated hepato-protective effects of mTOR inhibitors in both NAFLD and HCC pre-clinical models28. Furthermore, clinical trials of PI3K-Akt-mTOR inhibitors in HCC have shown some efficacy towards tumor regression and life prolongation29. Despite these observations, the exact etiology of PI3K-Akt-mTOR activation in these pre-clinical models remains unclear and appears to be multifactorial involving lipid metabolism, intestinal microbiota, and oxidative stress26. A recent study by Bu et. al. found that palmitic acid can cause palmitation of Akt resulting in activation of Akt and its downstream effectors30. They theorized this palmitation may be one of the inciting events in the NAFLD disease progression. While these pathways have not been detailed in the IFALD literature, it is possible that enteral feeding post bowel resection may result in a similar overload of free fatty acids in the portal system that would deliver similar liver insults. Subsequently, this hypothesis could be extended to parenteral nutrition liver injury models whereby a high volume of fatty acids reaches the liver bypassing entero-lymphatic processing. Overall, more studies are needed to elucidate the exact mechanism of PI3K-Akt-mTOR activation in IFALD.
We acknowledge this study contains certain limitations. First our autophagy assay was qualitative in nature; while we believe this visual evidence is convincing, we acknowledge a quantitative assay could have been beneficial. Additionally, the mechanistic insights are speculative in nature. Pharmacological rescue with a mTOR inhibitor is part of our future work. Another limitation is that the autophagy assay and single nuclear sequencing were performed only on a subset of male mice. Interestingly, we found that female mice seemed to have increased hepatic steatosis compared to males. While we plan to investigate the sex-differences in future work, we found that surrogate markers for autophagy (p62) and mTOR activation (pS6) to correlate between sexes. Finally, this study assumed that autophagy dysregulation is a main driver of liver injury in this model. While we did not explore the relative impact of defective autophagy in our model, we argue the defective autophagy observed in other liver injury models implies the presence of dysfunctional autophagy in our model ultimately contributes towards liver injury. Future experiments would be needed to ascertain if recovery of autophagy can mitigate liver injury after SBR.
Conclusion
This study highlights the dysregulated autophagy response in IFALD. Bioinformatic analyses suggest the PI3K-Akt-mTORC pathway may be involved. Future work includes pharmacological interventions to block this axis and possibly restore autophagy.
Methods
Animal study
All animal studies were approved by Washington University in St. Louis IACUC (#23–0421). All experiments were performed in accordance with relevant named guidelines and regulations including the ARRIVE guidelines. 8-week-old male (n = 50) and female (n = 40) C57BL/6 J mice (purchased from The Jackson Laboratory; Bar Harbor, Maine) received sham or 75% proximal SBR (extra male mice were needed for autophagy flux assay). A full description of the operations has been described in prior papers6,14. Briefly, 8-week old mice underwent operations. The sham operation involved a transection of the small bowel 6 cm proximal to the cecum with primary end-to-end anastomosis with 9–0 nylon sutures placed in an interrupted fashion. For the 75% SBR operations, the small bowel was transected at 2 points: 1-2 cm distal to the ligament of Treitz and 6 cm proximal to the cecum. The intervening mesentery was tied off and the intervening small bowel was resected. An end-to-end primary anastomosis was completed with 9–0 nylon sutures placed in an interrupted fashion. Postoperative care included no food for 24 h and 1 week of incubator housing. All mice were fed a liquid enteral diet (LD101 #0,007,551, Test Diet; Richmond, Indiana) ad lib. The mice were then randomized into 2- and 10-week cohorts. Body composition (lean and fat mass) at the 10-week time point was captured using MRI (Echo Medical Systems; Houston, Texas). After survival, final cohort numbers were n = 7, 9, 11, 6 for Sham F, SBR F, Sham M, and SBR M at 2 weeks and n = 6, 7, 7, and 11 for Sham F, SBR F, Sham M, and SBR M at 10 weeks.
At the time of harvest, peripheral blood was collected, centrifuged and stored at –80 °C. Serum transaminases (aspartate aminotransferase [AST]) and alanine aminotransferase [ALT]) were obtained using the AMS Diagnostics Liasys 330 Chemistry Analyzer (AMS Diagnostics; Weston, Florida). The left anterior lobe of each liver was sectioned and stored in the following manner: formalin for histological analysis, flash frozen for western blot, RNeasy lysis buffer (Qiagen; Germantown, Maryland) then frozen for rt-qPCR measurements, and RNALater (Thermo Fisher Scientific Baltics UAB; Vilnius, Lithuania) then frozen for single nuclear isolation. Intestinal samples were taken from the proximal end of the remaining bowel distal to the anastomosis. All formalin fixed samples were embedded in paraffin. Liver and intestinal tissue was stained with hematoxylin and eosin. Liver samples were stained with Sirius Red to measure fibrotic liver changes. Representative histological images were captured using the Nikon Eclipse 80i microscope with the Nikon DS-Ri2 camera attachment under a 20 × objective (Nikon Instruments, Inc.; Melville, New York). NIS-Elements Advanced Research software version 4.51 was used for image acquisition (Nikon Instruments, Inc.). Intestinal adaptation was calculated by comparing intraoperative and harvest villus height and crypt depth measurements under a 10X objective (NIS elements AR 4). At least 20 measurements per villus and crypt were taken from quality sections.
Histopathology analysis
Blinded to treatment groups and sex, a board-certified pathologist was given liver H&E and Sirius Red stains and scored each slide according to the Nonalcoholic Steatohepatitis Clinical Research Network (NAS CRN) scoring system31. Each H&E slide was scored according to the NAS CRN composite score which is composed of the percent steatosis on low-medium powered field, number of lobular inflammation foci per 200 × field and qualitative hepatocyte ballooning assessment (none, few, many). Scores of 0–2 are consistent with no steatohepatitis, 3–5 are indeterminant, and greater than 5 are diagnostic of steatohepatitis. Sirius Red slides were graded according to the NAS CRN guidelines: score 0 is no fibrosis, score 1 is perisinusoidal or periportal fibrosis (mild), score 2 is perisinusoidal and portal/periportal fibrosis (moderate), score 3 is bridging fibrosis (severe) and score 4 is cirrhosis.
RT-qPCR
Liver RNA was extracted using the RNeasy Mini Kit (Qiagen; Germantown, Maryland). RNA concentrations confirmed via the NanoDrop Spectrophotometer (ND-1000, Nanodrop Technologies; Wilmington, Deleware). p62, Ddit3, Nqo1 and Gapdh primers were obtained from Applied Biosystems (Life Technologies; Carlsbad, California). qPCR was performed using the TaqMan RNA to C(T) 1-Step Kit on Quantstudio 3 Real Time PCR system (Applied Biosystems; Waltham, Massachusetts). Reactions were normalized to sample RNA concentrations, run in duplicate, and reported as a relative quantification to the endogenous control Gapdh.
Western
Liver protein lysate was obtained via mechanical homogenization (ServiceBio; WuHan, People’s Republic of China) and protein concentration was determined using RC DC kit (BioRad; Hercules, California). Antibodies from Cell Signaling Technologies (Danvers, Massachusetts) – p62 (#23214) and GAPDH (#5174) were used to determine protein expression. Protein detection and quantification was performed using Image Lab Software (version 6.1, Bio-Rad, https://www.bio-rad.com/en-us/product/image-lab-software?ID=KRE6P5E8Z).
IHC
Formalin-fixed tissues embedded in paraffin were sectioned and deparaffinized using xylene and serial dilutions of ethanol. Endogenous peroxides were blocked with hydrogen peroxide. To retrieve the antigen, Diva Decloaking solution (BioCARE Medical; Pacheco, California) was added to the slides and incubated in a pressure cooker at 120 °C. Slides were blocked with Avidin-pink and biotin-blue (BioCARE Medical) then incubated with primary antibody overnight at 4 °C. The slides were visualized using biotinylated goat anti-rabbit IgG, followed by streptavidin–horseradish peroxidase, diaminobenzidine and hematoxylin counterstaining. The antibodies used in this study were p-S6 (#5364) for mTOR activation and p62 (#23214) for autophagy surrogate. Both antibodies were procured from Cell Signaling Technologies (Danvers, MA).
Images were captured using the Nikon Eclipse 80i microscope with the Nikon DS-Ri2 camera attachment under a 4 × objective (Nikon Instruments Inc.). The images were saved using the NIS-Elements Advanced Research software version 4.51 (Nikon Instruments Inc.). These images were uploaded into QuPath (version 0.5.1, https://qupath.readthedocs.io/en/0.6/). They were viewed under brightfield H-DAB setting. Any artifacts or shadowing were highlighted and deselected for analysis. A DAB pixel thresholder was created under the 3.061 resolution, 1.0 smoothing factor and threshold of 0.30. The area of quantification was divided by the total selected area to obtain the percent of staining area.
Single nuclear preparation and bioinformatic analyses
Single nuclear suspensions were produced in 3 sham and 3 SBR samples according to the protocol outlined by Nadelmann et. al.32 Samples were submitted to the Gene Techology Access Center at Washington University in St. Louis. cDNA was prepared after the GEM generation and barcoding, followed by the GEM-RT reaction and bead cleanup steps. Purified cDNA was amplified for 11–13 cycles before being cleaned up using SPRIselect beads. Samples were then run on a Bioanalyzer to determine the cDNA concentration. GEX libraries were prepared as recommended by the 10 × Genomics Chromium Single Cell 3’ Reagent Kits User Guide (v3.1 Chemistry Dual Index) with appropriate modifications to the PCR cycles based on the calculated cDNA concentration. For sample preparation on the 10 × Genomics platform, the Chromium Next GEM Single Cell 3’ Kit v3.1, 16 rxns (PN-1000268), Chromium Next GEM Chip G Single Cell Kit, 48 rxns (PN-1000120), and Dual Index Kit TT Set A, 96 rxns (PN-1000215) were used. The concentration of each library was accurately determined through qPCR utilizing the KAPA library Quantification Kit according to the manufacturer’s protocol (KAPA Biosystems/Roche) to produce cluster counts appropriate for the Illumina NovaSeqX instrument. Normalized libraries were sequenced on a NovaSeqXPlus Flow Cell using the 151 × 10x10 × 151 sequencing recipe according to manufacturer protocol. Basecalls and demultiplexing were performed with Illumina’s DRAGEN and BCLconvert.v4.2.4 software. Read 1 was trimmed to the 10 × Genomics recommendation of 28 bp. A median sequencing depth of 50,000 reads/cell was targeted for each Gene Expression Library. The fastq files were then aligned and quantitated with 10x’s CellRanger v7.2.0 software against 10x’s standard refdata-gex-mm10-2020-A reference according to manufacturer protocol.
Using RStudiov2023.09, output files were converted into Seurat.v5 objects using Read10X and CreateSeuratObject, with parameters including 200 features per cellular barcode and features detected in at least 3 cellular barcodes. Each object was processed using thresholds of total number of unique transcripts detected within a nucleus (nCount_RNA) > 500, genes detected in each nucleus > 250, mitochondrial percent < 1%, and ratio of log10 number of genes per unique molecular indicator > 0.8. One SBR sample demonstrated poor sequencing depth and was removed from analysis.
Object counts were normalized, scaled, and variable features were detected. Doublets were identified with DoubletFinder and removed using a multiplet rate threshold calculated from the number of sequenced nuclei33. Genes were annotated with predicted cell cycle associations34. Stratifying by cell cycle phases demonstrated little variability. To regress out mitochondrial genes, interquartile ranges were quantified for percent of mitochondrial genes and stratified. A portion of variability could be explained by medium/high (50–75%) and high (75–100%), so variability was removed with the SCTransform. Using the SCT assay, the RunPCA and RunUMAP functions were subsequently performed.
Due to variability between operative groups, we proceeded with Harmony integration35. Harmony algorithm was applied using the IntegrateLayers function with HarmonyIntegration method accounting for sample, surgical procedure and percent mitochondrial genes. Our data converged after 4 iterations of the harmony algorithm. Using FindNeighbors, the k nearest neighbors were found and a shared nearest neighbor (SNN) graph was constructed on the integrated Seurat object. FindClusters optimized the modularity function of the SNN to identify clusters according to specified resolutions. The RNA assay count data was then re-normalized and scaled after integration.
PrepSCTFindMarkers was run on the SCT assay and FindAllMarkers was employed to find differentially expressed genes (DEGs) for each of the clusters. Clusters were subsequently annotated using both automated (SingleR36, SC-type37) and manual (Cell Marker 2.038, Annotation of Cell Types39) annotation. Hepatic lobule zones were annotated by using the methods of recent work from Hu et al.,40 MacParland et al.,41 Halpern et al.,42,43 and Paris et al.44 Cells were labeled and count matrices exported for each cell type. Differential gene expression was found using the DESeq2 package45. Gene Ontology Biological Processes and KEGG databases were used for overrepresentation (ORA) and gene set enrichment analysis (GSEA) of differentially regulated genes using the clusterProfiler package46.
Hepatocyte isolation and qualitative autophagy assay
Hepatocyte isolation was performed according to the detailed protocol outlined by Biel et. al.16 Two sham and 2 SBR mice underwent hepatocyte isolation. Mice were anesthetized with ketamine, and the IVC was cannulated and secured with 3–0 silk. Perfusion buffers and digestion buffers containing collagenase IV were administered to perform enzymatic digestion separating the liver cells. The liver was then harvested and placed on a cold petri dish. A plating medium was added, and homogenization was performed by use of pipette. Viable hepatocytes were plated on collagen coated plates. After washing non-viable cells, a maintenance medium was added.
Autophagic flux was qualitatively measured by starving the cultured hepatocytes for 2 h. They were infected with adenovirus-GFP-LC3 overnight. Tetramethylrhodamine methyl ester (TMRM) was administered for 30 min. Fluorescence images were obtained using the Zeiss 880 confocal microscope (Zeiss; Oberkochen, Germany).
Statistics
Bar graphs of serum transaminases, reverse transcription quantitative polymerase chain reaction, western blot, and IHC quantification were analyzed using GraphPad Prism software (version 10.2.3, La Jolla, California, https://www.graphpad.com/features). Data were summarized with mean and standard deviation. For parametric data, unpaired t-test and one-way ANOVA with multiple comparison were conducted for 2 group and multigroup comparisons respectively. Tukey HSD corrections were conducted on p-values for multiple comparisons. Nonparametric ANOVA (Kruskal–Wallis) with Dunn correction was used for ordinal data. A p-value less than 0.05 was considered significant.
Data availability
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository, GSE293958. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE293958. Data can be provided upon request made to corresponding author, Jun Guo (jguo22@wustl.edu).
Abbreviations
- ATG:
-
Autophagy related gene
- Ddit3:
-
DNA damage inducible transcript 3
- EGF:
-
Epidermal growth factor
- ER:
-
Endoplasmic reticulum
- GFP -LC3II:
-
Green fluorescent protein tagged lipated light chain 3
- GSEA:
-
Gene set enrichment analysis
- IFALD:
-
Intestinal failure associated liver disease
- IHC:
-
Immunohistochemistry
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- LC3:
-
Microtubule associated protein 1 light chain 3
- mTOR:
-
Mechanistic target of rapamycin
- mTORC1:
-
Mechanistic target of rapamycin complex 1
- PI3k-AKT:
-
Phosphoinositide 3-kinase protein kinase B
- PI3kc3:
-
Phosphoinositide 3-kinase catalytic subunit type 3
- PI3P:
-
Phosphoinositide 3-phosphate
- pS6:
-
Ribosomal subunit S6
- rt-qPCR:
-
Reverse transcription quantitative polymerase chain reaction
- SBR:
-
Small bowel resection
- SGS:
-
Short gut syndrome
- SQSTM1/P62:
-
Sequestosome-1
- TMRM:
-
Tetramethylrhodamine methyl ester
- ULK1:
-
Unc-51-like autophagy-activating kinase 1
References
Zafirovska, M., Zafirovski, A. & Rotovnik Kozjek, N. Current insights regarding intestinal failure-associated liver disease (IFALD): A narrative review. Nutrients 15, 3169 (2023).
Onufer, E. J. et al. Effects of high-fat diet on liver injury after small bowel resection. J. Pediatr. Surg. 55, 1099–1106 (2020).
Onufer, E. J. et al. Lymphatic network remodeling after small bowel resection. J. Pediatr. Surg. 54, 1239–1244 (2019).
Onufer, E. J. et al. Lipid absorption and overall intestinal lymphatic transport are impaired following partial small bowel resection in mice. Sci. Rep. 12, 11527 (2022).
Courtney, C. M. et al. Small bowel resection increases paracellular gut barrier permeability via alterations of tight junction complexes mediated by intestinal TLR4. J. Surg. Res. 258, 73–81 (2021).
Steinberger, A. E. et al. A novel maladaptive unfolded protein response as a mechanism for small bowel resection-induced liver injury. Am. J. Physiol. Gastrointest. Liver Physiol. 323, G165–G176 (2022).
Dikic, I. & Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364 (2018).
Liu, S., Yao, S., Yang, H., Liu, S. & Wang, Y. Autophagy: Regulator of cell death. Cell Death Dis. 14, 648 (2023).
Wu, Y. et al. Molecular mechanisms of autophagy and implications in liver diseases. Liver Res. 7, 56–70 (2023).
Al-Kuraishy, H. M. et al. Defective autophagy and autophagy activators in myasthenia gravis: A rare entity and unusual scenario. Autophagy 20, 1473–1482 (2024).
Kumar, A. V., Mills, J. & Lapierre, L. R. Selective autophagy receptor p62/SQSTM1, a pivotal player in stress and aging. Front. Cell Dev. Biol. 10, 793328 (2022).
Tan, C. T., Soh, N. J. H., Chang, H.-C. & Yu, V. C. p62/SQSTM1 in liver diseases: The usual suspect with multifarious identities. FEBS J. 290, 892–912 (2023).
Yoshii, S. R. & Mizushima, N. Monitoring and measuring autophagy. Int. J. Mol. Sci. 18, 1865 (2017).
Phelps, H. M. et al. Intestinal knockout of peroxisome proliferator-activated receptor-alpha affects structural adaptation but not liver injury following massive enterectomy. J. Pediatr. Surg. 58, 1170–1177 (2023).
Dungubat, E. et al. Age-dependent sex difference of non-alcoholic fatty liver disease in TSOD and db/db mice. PLoS ONE 17, e0278580 (2022).
Biel, T. G. et al. Sirtuin 1 suppresses mitochondrial dysfunction of ischemic mouse livers in a mitofusin 2-dependent manner. Cell Death Differ. 23, 279–290 (2016).
Park, A. et al. Single nuclear RNA sequencing reveals unique hepatocyte population in small bowel resection-induced liver injury. Cell Mol. Gastroenterol. Hepatol. 19, 101609 (2025).
Jin, S. et al. Mechanisms and therapeutic implications of selective autophagy in nonalcoholic fatty liver disease. J. Adv. Res. https://doi.org/10.1016/j.jare.2024.01.027 (2024).
Zeng, J. et al. Restoration of lysosomal acidification rescues autophagy and metabolic dysfunction in non-alcoholic fatty liver disease. Nat. Commun. 14, 2573 (2023).
Han, Y.-H. et al. Enterically derived high-density lipoprotein restrains liver injury via the portal vein. Science 373, eabe6729 (2021).
Han, J. & Kaufman, R. J. The role of ER stress in lipid metabolism and lipotoxicity. J. Lipid Res. 57, 1329 (2016).
Kwon, J., Kim, J. & Kim, K. I. Crosstalk between endoplasmic reticulum stress response and autophagy in human diseases. Anim. Cells Syst. 27, 29 (2023).
Rial, E. et al. Lipotoxicity, fatty acid uncoupling and mitochondrial carrier function. Biochimica et Biophysica Acta (BBA) Bioenergetics 1797, 800–806 (2010).
Nakadate, K. et al. Advances in understanding lipopolysaccharide-mediated hepatitis: Mechanisms and pathological features. Curr. Issues Mol. Biol. 47, 79 (2025).
Panwar, V. et al. Multifaceted role of mTOR (mammalian target of rapamycin) signaling pathway in human health and disease. Sig. Transduct. Target Ther. 8, 1–25 (2023).
Feng, J. et al. mTOR: A potential new target in nonalcoholic fatty liver disease. Int. J. Mol. Sci. 23, 9196 (2022).
Ferrín, G., Guerrero, M., Amado, V., Rodríguez-Perálvarez, M. & De la Mata, M. Activation of mTOR signaling pathway in hepatocellular carcinoma. Int. J. Mol. Sci. 21, 1266 (2020).
Sharma, N., Singh, L., Sharma, A., Kumar, A. & Mahajan, D. NAFLD-associated hepatocellular carcinoma (HCC) – A compelling case for repositioning of existing mTORc1 inhibitors. Pharmacol. Res. 208, 107375 (2024).
Sun, E. J., Wankell, M., Palamuthusingam, P., McFarlane, C. & Hebbard, L. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Biomedicines 9, 1639 (2021).
Bu, L. et al. High-fat diet promotes liver tumorigenesis via palmitoylation and activation of AKT. Gut 73, 1156–1168 (2024).
Kleiner, D. E. et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41, 1313–1321 (2005).
Nadelmann, E. R. et al. Isolation of nuclei from mammalian cells and tissues for single-nucleus molecular profiling. Curr. Protoc. 1, e132 (2021).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cells 8, 329–337 (2019).
tinyatlas/cell_cycle/Mus_musculus.csv at master hbc/tinyatlas. GitHub https://github.com/hbc/tinyatlas/blob/master/cell_cycle/Mus_musculus.csv.
Korsunsky, I. et al. Fast, sensitive, and accurate integration of single cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019).
Ianevski, A., Giri, A. K. & Aittokallio, T. Fully-automated and ultra-fast cell-type identification using specific marker combinations from single-cell transcriptomic data. Nat. Commun. 13, 1246 (2022).
Hu, C. et al. Cell Marker 2.0: An updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 51, D870–D876 (2023).
Quan, F. et al. Annotation of cell types (ACT): A convenient web server for cell type annotation. Genome Med. 15, 91 (2023).
Hu, Y. et al. Unveiling the power of microenvironment in liver regeneration: An in-depth overview. Front. Genet. 14, 1332190 (2023).
MacParland, S. A. et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 9, 4383 (2018).
Halpern, K. B. et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017).
Halpern, K. B. et al. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat. Biotechnol. 36, 962–970 (2018).
Paris, J. & Henderson, N. C. Liver zonation, revisited. Hepatology 76, 1219–1230 (2022).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Xu, S. et al. Using clusterProfiler to characterize multiomics data. Nat. Protoc. https://doi.org/10.1038/s41596-024-01020-z (2024).
Acknowledgements
We thank the Genome Technology Access Center at the McDonnell Genome Institute at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH. Additionally, we would also like to thank Chunxiao Shi and Cristi King for their dedication and service to our lab. Finally, we would like to thank the Washington University in St. Louis Division of Surgical Sciences Editorial Services for manuscript review.
Funding
National Institutes of Health (RO1DK079879), Mid-America Transplant Foundation (07201903), Foundation for Barnes-Jewish Hospital (4776, 5153), the Washington University Department of Surgery Pilot Grant, National Institutes of Health R01DK128169, National Institutes of Health R01DK119147, Pediatric Gastro T32 grant (#DK077653) and NIH Washington University DDRCC Grant No. NIDDK P30 DK052574, National Institutes of Health DK119437, HL 151328.
Author information
Authors and Affiliations
Contributions
J.G. and B.W.W conceived the study. D.M.A., A.P., K.S., N.P., E.M., D.M.A, J.S.K, X.L. and J.G. contributed to the acquisition and analysis of data with additional interpretation and dialogue from B.W.W., N.O.D. and D.R. D.M.A., J.G., and B.W.W wrote the manuscript with edits from all authors. B.W.W. and J.G. supervised experiments and obtained compliance approvals.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Alligood, D.M., Park, A., Swanson, K. et al. Liver injury in a preclinical model of short bowel syndrome is associated with impaired autophagy. Sci Rep 16, 714 (2026). https://doi.org/10.1038/s41598-025-30243-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-30243-7