Abstract
Gene alteration plays key role in the pathogenesis of intervertebral disc degeneration (IDD). This study aimed to explore the genes related to IDD, and identified potential therapeutic drugs for IDD treatment. Three gene profile datasets of the IDD were used to identified key genes related to IDD pathogenesis. A scRNA-seq dataset of IDD was used to examine the expression of key genes. A diagnostic model for IDD was constructed and validated by external dataset. Functional enrichment analysis was performed for diagnostic model. Connectivity Map (CMap) database was used to identify potential drugs for IDD based on the genes. The clinical samples of discs tissues were applied to test the expression of genes by immunohistochemistry (IHC) staining. Three genes (ELMO1, CKAP4, and SACM1L) were highly expressed in IDD tissues after overlapping DEGs from three gene datasets. The scRNA-seq dataset results revealed that these three genes were high expression in nucleus pulposus cells of severe IDD. A diagnostic model was constructed based on the three genes, and showed high diagnostic value. Functional enrichment analysis revealed the biological function and pathways of model related genes. CMap database and molecular docking revealed that several candidate drug for IDD treatment. The IHC results from clinical disc samples confirmed the elevation of the three genes in IDD tissues. The ELMO1, CKAP4, and SACM1L were critical genes to the pathogenesis of IDD. The diagnostic model using these three genes demonstrated high accuracy for IDD diagnosis, and identified several candidate drugs for IDD treatment.
Similar content being viewed by others
Introduction
Intervertebral disc degeneration (IDD) stands as a principal contributor to low back pain, significantly impairing the quality of life for affected individuals1. The development and progression of IDD are complex, involving an interplay of cellular senescence, inflammatory processes, and mechanical loading2,3. Emerging evidence underscores the pivotal role of genetic elements in IDD etiology. Genetic variations have been shown to critically influence IDD pathophysiology, opening new possibilities for earlier diagnosis and personalized therapeutic strategies4. The advent of next-generation sequencing technologies has facilitated the identification of genetic markers with potential as biomarkers for IDD5,6. These advancements hold promise for enhancing diagnostic precision by enabling the early detection of at-risk individuals prior to the manifestation of severe symptoms. Furthermore, unraveling the genetic underpinnings of IDD is paving the way for more tailored treatment approaches7.
Specific genes implicated in the regulation of the extracellular matrix (ECM), inflammatory responses, and cellular metabolism have been associated with IDD pathology. Polymorphisms within genes such as COL1A1, COL9A2, COL9A38, IL-109, and MMP1210 have been linked to an elevated risk of IDD. For instance, alterations in collagen-coding genes may undermine the structural stability of the intervertebral disc, while dysregulation of pro-inflammatory cytokine genes like IL-1β could exacerbate matrix breakdown and perpetuate chronic inflammation within the disc tissue11. Despite these insights, the precise mechanisms through which genetic modifications contribute to IDD remain partially elucidated. Therefore, integrating multi-omics datasets of IDD to validate gene alterations would help to refine the diagnostic tools and develop targeted therapies for IDD patients. This study aimed to advance our understanding of IDD molecular basis by employing a combination of multi-omics data analysis, bioinformatics techniques, and clinical sample assessments.
In this study, we identified three genes significantly associated with IDD: engulfment and cell motility 1 (ELMO1), cytoskeleton-associated protein 4 (CKAP4), and SAC1-like phosphatidylinositide phosphatase (SACM1L). ELMO1 is a highly conserved cytoplasmic bridging protein implicated in the regulation of cell migration, phagocytosis, and chemotactic signaling through its interaction with DOCK family proteins and activation of small GTPases such as RAC1. Dysregulation of ELMO1 has been linked to inflammatory and fibrotic processes in various tissues, suggesting a potential role in disc inflammation and matrix remodeling12,13. CKAP4 is a type II transmembrane protein predominantly localized to the endoplasmic reticulum and functions as a receptor for Dickkopf-1 (DKK1), an inhibitor of the Wnt/β-catenin pathway14,15,16. SACM1L is an integral ER-resident phosphoinositide phosphatase that regulates phosphatidylinositol-4-phosphate (PI4P) turnover and membrane trafficking17. Perturbations in SACM1L activity can affect ER morphology, autophagy, and cellular stress responses, all processes increasingly recognized as key players in disc cell senescence and degeneration18,19. Together, these genes represent novel molecular candidates whose functional roles in IDD warrant further investigation as potential diagnostic biomarkers or therapeutic targets.
Results
Screening genes associated with IDD pathogenesis
To identify potential disease-related genes, we analyzed three gene expression datasets, including GSE167931, GSE70362, and GSE23130, which comprising a total of 33 normal and 23 IDD samples. DEGs were determined using predefined thresholds (|LogFC|> 0.5 and P value < 0.05), resulting in the identification of 1,283, 2,118, and 1,277 DEGs in each dataset, respectively (Fig. 1A–C; Supplement Table S1). A Venn diagram analysis revealed five overlapping DEGs across all datasets, including ELMO1, ARHGEF11, CKAP4, SCOC-AS1, and SACM1L (Fig. 1D), suggesting their potential involvement in IDD progression. Further investigation of these five genes in the GSE70362 (Fig. 1E) and GSE23130 (Fig. 1F). The results showed that all the five genes increased in IDD tissues compared to controls, but the differences were not statistically significant regarding ELMO1, ARHGEF11, and SCOC-AS1. Given its higher expression in IDD tissues compared with those of ARHGEF11 and SCOC-AS1, ELMO1 was still selected for further analysis along with CKAP4 and SACM1L, which also demonstrated notable expression changes.
Screening genes associated with IDD pathogenesis. (A) Volcano plot of differential expression genes (DEGs) between normal and IDD in GSE167931 dataset; (B) Volcano plot of DEGs between normal and IDD in GSE70362 dataset; (C) Volcano plot of DEGs between normal and IDD in GSE23130 dataset; (D) Venn plot of common DEGs from three datasets; (E) Expression of five common genes in GSE70362 dataset; (F) Expression of five common genes in GSE23130 dataset. Data are presented as means ± SD. Student’s t-test was used to compare two groups. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Analysis the key genes in the scRNA dataset of IDD
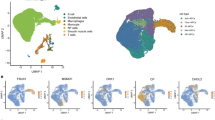
The GSE244889 dataset provided scRNA-seq data from nucleus pulposus (NP) tissues of six mild and seven severe IDD cases. Following quality control and cell type annotation, transcriptomic profiles of 106,937 cells were obtained, representing eight major cell clusters (Fig. 2A,B). The six cell markers plot of NP cells (COL2A1, MT2A, CLU, CDO1, COL3A1, COL1A2) was showed in Fig. 2C. Analysis of ELMO1, CKAP4, and SACM1L across these clusters revealed low expression of ELMO1 and SACM1L in most cell types, whereas CKAP4 was broadly and highly expressed, particularly in NP cells (Fig. 2D). Comparison of gene expression between mild and severe IDD NP cells showed higher expression of all three genes in mild cases; however, only CKAP4 demonstrated statistical significance (Fig. 2E). These findings suggest that CKAP4 may play a more prominent role in early-stage IDD at the single-cell level.
Analysis the key genes in the scRNA dataset of IDD. (A) The distribution of each cluster visualized by UMAP map of GSE244889 scRNA-seq dataset; (B) The distribution of each cell type visualized by UMAP map; (C) The distribution of six cell markers of NP cells; (D) Dot plot of the expression of ELMO1, CKAP4, and SACM1L in each cell type; (E) Comparison of the expression of ELMO1, CKAP4, and SACM1L in nucleus pulposus cells between mild and severe IDD tissues.
Determining the diagnostic value of the genes and constructed the diagnostic model for IDD
Using the GSE70362 dataset (10 IDD and 14 normal samples), we evaluated the diagnostic value of ELMO1, CKAP4, and SACM1L. ROC result indicated moderate to strong diagnostic power for each gene, with AUC values of 0.821 (95%CI 0.595–0.924), 0.779 (95%CI 0.550–0.903), and 0.907 (95%CI 0.684–0.934), respectively (Fig. 3A). When combined into a multivariate model, these three genes achieved an AUC of 0.943 (95%CI 0.733–0.936) (Fig. 3B), demonstrating excellent diagnostic accuracy. Validation of this diagnostic model in the independent GSE23130 dataset yielded an AUC of 0.817 (95%CI 0.529–0.933) (Fig. 3C), supporting the robustness and generalizability of the model.
Determining the diagnostic value of the genes and constructed the diagnostic model for IDD. (A) Diagnostic value of the ELMO1, CKAP4, and SACM1L for IDD patients in GSE70362 dataset; (B, C) Diagnostic value of the combination of the ELMO1, CKAP4, and SACM1L for IDD patients in GSE70362 dataset (B); GSE23130 dataset (C).
Exploring the biological function and pathways of the diagnostic model
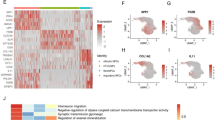
To elucidate the biological roles of the gene-based diagnostic model, functional enrichment analyses were conducted using the GSE70362 dataset. GO analysis revealed significant enrichment in processes such as positive regulation of cell population proliferation, cartilage development, and fibroblast activation (Fig. 4A). KEGG analysis revealed that this model mainly participated in Protein processing in endoplasmic reticulum, p53 signaling pathway, various types of N-glycan biosynthesis (Fig. 4B). GSEA identified pathways related to estrogen response late, cholesterol homeostasis, and reactive oxygen species signaling (Fig. 4C). Additionally, GSVA highlighted the involvement of CHRNA7/E2F signaling, cell cycle G1/S transition, and PDGFR-RAS/ERK pathways (Fig. 4D), suggesting a broad regulatory role of the model in IDD pathophysiology.
Exploring the biological function and pathways of the diagnostic model. (A) Biological process enrichments of the model related genes by GO analysis; (B) KEGG analysis for the model related genes; (C) GSEA of the model related genes by GO analysis; (D) GSVA of the model related genes by GO analysis.
Screening therapeutic drugs and molecular docking of model relate genes
Given the model high diagnostic performance, we explored potential therapeutic interventions by querying the CMap database. Forty-one compounds with potential modulatory effects on the model-related genes were identified, among which BRD-K5229182 exhibited the highest connectivity score, while rolipram showed the lowest (Fig. 5A). Mechanistic classification revealed that most candidates functioned as phosphodiesterase inhibitors (Fig. 5B). Molecular docking simulations were performed between BRD-K5229182, dipyridamole, and vinpocetine and the protein products of ELMO1, CKAP4, and SACM1L. The calculated binding energies were generally below −5 kcal/mol, indicating strong molecular interactions. Three-dimensional interaction maps generated using PyMOL software illustrated specific residue-level binding patterns (Fig. 5C), providing structural evidence for the potential repurposing of these compounds in IDD treatment.
Screening therapeutic drugs and Molecular docking of model relate genes. (A) Candidate drugs scores screened by the CMap database; (B) Candidate drugs classification and number according to the mechanism of action; (C) Molecular docking and visualized for the ELMO1, CKAP4, and SACM1L with BRD-K5229182, dipyridamole, and vinpocetine.
Validation of the model relate genes in clinical samples
Clinical intervertebral disc tissue samples were collected from 6 controls (Pfirrmann grade II–III) and 30 severe IDD (Pfirrmann grade IV–V) patients. MRI assessments using the Pfirrmann classification confirmed the degenerative status of the IDD group (Fig. 6A). Histological examination via H&E staining revealed well-preserved extracellular matrix, abundant NP cells, and organized collagen fibers in non-degenerated discs. In contrast, degenerated tissues exhibited reduced cellularity, fibrosis, disorganized collagen architecture, and matrix loss (Fig. 6B). IHC analysis further confirmed elevated expression levels of ELMO1, CKAP4, and SACM1L in IDD tissues compared to controls (p < 0.05, Fig. 6C). These results corroborate our bioinformatics findings and support the biological relevance of the identified genes in human IDD pathology.
Expression of the model relate genes in clinical samples. (A) Representative MRI images of intervertebral discs of normal subjects and IDD patients. The arrow points to IDD. (B) Representative H&E staining of normal and degenerated human NP tissues. (Scale bar: 100 μm). (C) Comparison of IHC score between normal and IDD NP tissues for ELMO1, CKAP4, and SACM1L. (Scale bar: 100 μm). Data are presented as means ± SD. Student’s t-test was used to compare two groups. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Discussion
In the present study, we identified key genes potentially involved in IDD by integrating and analyzing three independent gene expression datasets. Through comparative analysis of DEGs, we identified three genes (ELMO1, CKAP4, and SACM1L) that were consistently upregulated in IDD tissues compared to normal controls. Further exploration using a scRNA-seq dataset revealed that CKAP4 was notably enriched in NP cells, with significantly higher expression in severe IDD compared to mild cases. In contrast, while ELMO1 and SACM1L also exhibited elevated levels in NP cells from severe IDD samples, these differences did not reach statistical significance. We further evaluated the diagnostic potential of these genes and developed a diagnostic model for IDD. Our results demonstrated that each gene individually showed moderate to high diagnostic accuracy. Validation in an independent dataset confirmed the robustness of the model. Functional enrichment analyses provided insights into the biological processes and signaling pathways associated with the model, including cell proliferation, cartilage development, cholesterol homeostasis, and ERK signaling. Importantly, leveraging the CMap database, we identified several candidate compounds that may modulate the expression of these genes. Molecular docking simulations supported the feasibility of targeting these proteins pharmacologically. Finally, IHC validation in clinical tissue samples confirmed significantly elevated protein expression of all three genes in IDD discs compared to controls, reinforcing their biological relevance in IDD pathogenesis. Collectively, our findings suggest that ELMO1, CKAP4, and SACM1L play pivotal roles in IDD development and that the diagnostic model based on these genes holds strong diagnostic potential. Targeting these molecules with repurposed or novel therapeutics may offer new strategies for managing IDD.
Emerging evidence has highlighted the involvement of genetic and molecular alterations in the pathophysiology of IDD, particularly in relation to ECM dysregulation, inflammatory responses20, and cellular senescence21. Several studies have identified key regulatory genes implicated in IDD progression, such as programmed cell death-related genes like YWHAB, BID, and GSDME22, and signaling mediators including p38/NCoR2, which contributes to vitamin D deficiency-induced ECM degradation23. Additionally, experimental models have shown that FBXW7 can delay IDD by promoting mitophagy and suppressing ferroptosis via the mTOR pathway24. While previous studies have reported associations between ELMO125 and CKAP426 and IDD independently, our work is the first to construct a diagnostic model incorporating these two genes along with SACM1L, demonstrating superior diagnostic performance. Notably, this is the first report implicating SACM1L in IDD, suggesting its novelty in the context of disc degeneration and warranting further investigation.
The identification of ELMO1, CKAP4, and SACM1L as biomarkers for IDD offers promising avenues for early diagnosis and disease monitoring. The high diagnostic accuracy of our multigene model, particularly in cross-dataset validation, underscores its potential applicability in clinical settings. Furthermore, functional annotations suggest that these genes are involved in critical biological processes relevant to disc homeostasis and degeneration, thereby providing mechanistic insights into IDD pathogenesis. The integration of scRNA-seq data allowed us to dissect cell-type-specific expression patterns, revealing CKAP4 as a dominant player in NP cells, especially in advanced stages of IDD. This level of resolution enhances our understanding of the heterogeneity within the disc microenvironment and highlights the importance of cell-specific targeting in future therapeutic strategies. Moreover, the use of the CMap database to identify several small-molecule compounds capable of modulating gene expression represents a novel approach to drug discovery in IDD research. To be noted, little is known of the effect of BRD-K5229182 on diseases. The dipyridamole, is commonly used to reduce risk of a blood clot after valve replacement. Vinpocetine is a synthetic derivative of the vinca alkaloid vincamine, primarily used to enhance memory and protect brain cells, but its efficacy and safety are subjects of ongoing debate. However, no study has reported the effect of these compounds in IDD, therefore, further experiment is need to validate their role in the treatment of IDD.
Currently, ELMO1 has reported to exert its effect via several mechanism. For example, ELMO1 regulates cellular shaping and motility through binding with Dock180 and activation of the Rac1 signaling pathway12. ELMO1 regulates macrophage directed migration and attenuates inflammation via NF-kappaB signaling pathway in primary biliary cholangitis27. ELMO1 ameliorates intestinal epithelial cellular senescence via SIRT1/p65 signaling in inflammatory bowel disease-related fibrosis28. Overexpression of Dock180 and Elmo1 in Melanoma is Associated with Cell Survival and Migration29. Regarding SACM1L, there were studies showed that Sac1 depletion in hepatic cells results in increased levels of intracellular PI4P pools and impaired trafficking of the HBV envelope proteins30. SACM1L negatively regulates basal autophagic degradation in 293A cells31. In addition, SACM1L uses PI4P as its substrate, greatly affects the replication of certain bacteria and viruses in vitro18,19,32.
Compared to previous studies on IDD, this research uniquely combines the identification of key genes with functional analysis and drug discovery, providing a more holistic understanding of IDD pathophysiology. Unlike earlier works that often focused solely on identifying genetic markers or exploring individual signaling pathways33, this study leverages scRNA-seq data to analyze gene expression at the single-cell level, offering deeper insights into cell-type-specific roles of CKAP4, ELMO1, and SACM1L in IDD. Moreover, the application of CMap database to screen for potential therapeutic compounds represents an innovative approach to IDD therapy development. While other studies have made significant contributions to understanding IDD34,35, this research advances the field by proposing a diagnostic model with strong diagnostic potential and suggesting novel therapeutic targets, thereby setting a new benchmark for future investigations in IDD.
However, several limitations should be acknowledged. First, the relatively small number of samples used for DEGs detection and model construction may limit the generalizability of our findings. Second, while we identified multiple candidate drugs, in vivo and in vitro validation of their efficacy and safety is still required. Third, the specific molecular functions of ELMO1 and SACM1L in the context of IDD remain largely unexplored, necessitating further mechanistic studies to elucidate their roles in disc biology and pathology.
Conclusion
The present study identifies ELMO1, CKAP4, and SACM1L associated with the pathogenesis of IDD. We constructed and validated a multigene diagnostic model with high diagnostic accuracy for IDD, offering a potential tool for early detection and prognosis evaluation. Furthermore, we propose several candidate therapeutic agents for IDD treatment.
Materials and methods
Acquisition of IDD gene expression datasets
To investigate gene expression alterations associated with IDD, three publicly available microarray datasets (GSE167931, GSE70362, and GSE23130) were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo). These datasets collectively included a total of 43 intervertebral disc samples, comprising both normal and IDD-affected tissues. The sample composition was as follows: GSE167931 (normal n = 4, IDD n = 5), GSE70362 (normal n = 14, IDD n = 10), and GSE23130 (normal n = 15, IDD n = 8). Raw data were preprocessed using Robust Multi-array Average (RMA) normalization to correct background signals. Differentially expressed genes (DEGs) between control and IDD groups were identified using the “limma” package in R based on the criteria of |LogFC|> 0.05 and P value < 0.05. Visualization of DEGs was performed using the “ggplot2” package for volcano plots and the “ggvenn” package for Venn diagrams.
Single-cell RNA sequencing (scRNA-seq) analysis
The scRNA-seq dataset GSE244889 contained nucleus pulposus tissue profiles from seven patients with mild IDD and six with severe IDD. Raw gene expression matrices were integrated and analyzed using the Seurat package (version 5) following standard quality control procedures. High-quality cells were selected based on the following criteria: mitochondrial gene content < 15%, erythrocyte gene content < 3%, minimum of 500 unique molecular identifiers (UMIs), and detection of more than 200 genes per cell. To minimize batch effects across samples, the “harmony” package was applied. Cell clustering was performed using the “FindClusters” function, and results were visualized via t-distributed stochastic neighbor embedding (tSNE). Cell type annotations were determined by referencing marker genes from the CellMarker 2.0 database (http://bio-bigdata.hrbmu.edu.cn/CellMarker) and the Enrichr platform (https://maayanlab.cloud/Enrichr/).
Development of a diagnostic model
A logistic regression model was constructed based on gene expression profiles from the GSE70362 dataset. This model aimed to predict the likelihood of IDD development. Receiver operating characteristic (ROC) curves and area under the curve (AUC) values were calculated using the “pROC” package in R to assess the model’s diagnostic performance.
Functional enrichment analysis of DEGs
To explore the biological implications of the diagnostic model, functional enrichment analyses were conducted using the GSE70362 dataset. Samples were stratified into high- and low-expression groups based on the median model score. DEGs between these two groups were identified using the “limma” package. Gene Ontology (GO) and KEGG enrichment analysis36,37 was carried out with the “clusterProfiler” package to identify significantly enriched biological processes. Additionally, Gene Set Enrichment Analysis (GSEA) was performed using the c2.cp.v7.2. symbols gene set from the Molecular Signatures Database. Gene Set Variation Analysis (GSVA) was employed to evaluate pathway activity differences between high- and low-expression groups using the h.all. v2024.1.Hs. symbols gene set. Statistical significance was defined as p < 0.05 and false discovery rate (FDR) < 0.20.
Drug repurposing using connectivity map (CMap) database and molecular docking
The top 100 upregulated and downregulated DEGs from the high- versus low-expression comparison in GSE70362 were submitted to the CMap database38 to identify potential therapeutic agents. Compounds showing significant negative connectivity scores (< −0.8, p < 0.05) were prioritized as candidates for drug repurposing. Selected molecules were further evaluated through molecular docking simulations. Protein structures were retrieved from the UniProt database, while compound structures in SDF format were downloaded from PubChem and converted to PDB format using Open Babel. Docking analyses were conducted using AutoDock Vina 1.5.7. Binding affinities were assessed based on predicted binding energies, and 3D interaction visualizations were generated using PyMOL 2.1.0. A binding energy below zero indicates spontaneous binding, with lower values reflecting stronger molecular interactions.
Collection of intervertebral discs samples
The intervertebral discs samples were collected in the Third Affiliated Hospital of Guangxi Medical University from 1st September 2024 to 5th Mach 2025. This study was approved by the Ethics Committee of the Third Affiliated Hospital of Guangxi Medical University. All participants provided written informed consent prior to enrollment. IDD severity was evaluated using magnetic resonance imaging (MRI) according to the Pfirrmann classification. MRI scans were performed using a 3.0-T scanner (General Electric Company, PHILIPS Ingenia Elition). Control samples were collected from spinal trauma patients classified as Pfirrmann grade I–III, while IDD samples were obtained during spinal decompression surgery.
The Pfirrmann grading classification was defined as followed39 grade I: Homogeneous, bright hyperintense signal; normal disc height. grade II: Inhomogeneous, but still hyperintense; clear distinction between nucleus and annulus; normal disc height; a gray horizontal band may be present. grade III: Inhomogeneous, with intermediate gray signal; unclear nucleus-annulus boundary; normal or slightly reduced disc height. grade IV: Inhomogeneous, hypointense dark gray signal; no distinction between nucleus and annulus; slightly to moderately decreased disc height. grade V: Inhomogeneous, hypointense black signal; no nucleus-annulus differentiation; severely collapsed disc space.
Hematoxylin and eosin (H&E) staining
For histological evaluation, intervertebral disc tissues were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at 5 μm thickness. Sections were deparaffinized, rehydrated, and subjected to H&E staining according to standard protocols. Morphological changes were examined under light microscopy.
Immunohistochemical (IHC) staining
IHC was performed on 5 μm tissue sections. After antigen retrieval and blocking, sections were incubated overnight at 4 °C with primary antibodies against primary antibody. The following primary antibodies were used: anti-ELMO1 (Cat No. 26974-1-AP, 1:100; Proteintech, Wuhan, China), anti-CKAP4 (Cat No. 16686-1-AP, 1:100; Proteintech, Wuhan, China), anti-GAPDH (Cat No. 13033-1-AP, 1:80; Proteintech, Wuhan, China). Following PBS washing, sections were treated with horseradish peroxidase (HRP)-conjugated secondary antibody and developed with 3,3’-diaminobenzidine (DAB). Counterstaining was performed with hematoxylin. Image-Pro Plus version 6.0 software (Media Cybernetics Inc.) was used to quantify protein expression levels.
Statistical analysis
All statistical analyses were conducted using R software (version 4.3.0). Continuous variables with normal distribution were compared using two-tailed Student’s t-tests, whereas non-normally distributed data were analyzed using the Wilcoxon rank-sum test. A two-tailed p-value < 0.05 was considered statistically significant.
Data availability
The datasets used and/or analyzed in the study are available from the corresponding author on reasonable request.
References
Lin, C. C. et al. The economic burden of guideline-recommended first line care for acute low back pain. Eur. Spine J. 27, 109–116. https://doi.org/10.1007/s00586-016-4781-0 (2018).
Vergroesen, P. P. et al. Mechanics and biology in intervertebral disc degeneration: a vicious circle. Osteoarthritis Cartilage 23, 1057–1070. https://doi.org/10.1016/j.joca.2015.03.028 (2015).
Wang, Y. et al. Oxidative stress in intervertebral disc degeneration: Molecular mechanisms, pathogenesis and treatment. Cell. Prolif. 56, e13448. https://doi.org/10.1111/cpr.13448 (2023).
Xia, Q. et al. Progress in the study of molecular mechanisms of intervertebral disc degeneration. Biomed. Pharmacother. 174, 116593. https://doi.org/10.1016/j.biopha.2024.116593 (2024).
Astur, N. et al. Next-generation sequencing (NGS) to determine microbiome of herniated intervertebral disc. Spine J. 22, 389–398. https://doi.org/10.1016/j.spinee.2021.09.005 (2022).
Colombini, A. et al. High-throughput gene and protein analysis revealed the response of disc cells to vitamin D, depending on the VDR FokI variants. Int. J. Mol. Sci. 22, 9603. https://doi.org/10.3390/ijms22179603 (2021).
Karchevskaya, A. E., Poluektov, Y. M. & Korolishin, V. A. Understanding intervertebral disc degeneration: background factors and the role of initial injury. Biomedicines 11, 2714. https://doi.org/10.3390/biomedicines11102714 (2023).
Trefilova, V. V. et al. The role of polymorphisms in collagen-encoding genes in intervertebral disc degeneration. Biomolecules 11, 1279. https://doi.org/10.3390/biom11091279 (2021).
Bermudez-Lekerika, P. et al. In-silico proteomic analysis of the role of IL-4 and IL-10 in IVD degeneration: Protein-protein interaction networks for candidate prioritisation. Comput. Struct. Biotechnol. J. 27, 1600–1613. https://doi.org/10.1016/j.csbj.2025.04.015 (2025).
Sun, Y. et al. MMP12-dependent myofibroblast formation contributes to nucleus pulposus fibrosis. JCI Insight 10, e180809. https://doi.org/10.1172/jci.insight.180809 (2025).
Li, H. et al. The mechanisms and functions of IL-1beta in intervertebral disc degeneration. Exp. Gerontol. 177, 112181. https://doi.org/10.1016/j.exger.2023.112181 (2023).
Tocci, S., Ibeawuchi, S. R., Das, S. & Sayed, I. M. Role of ELMO1 in inflammation and cancer-clinical implications. Cell. Oncol. (Dordr) 45, 505–525. https://doi.org/10.1007/s13402-022-00680-x (2022).
Park, Y. L. et al. Engulfment and cell motility 1 (ELMO1) regulates tumor cell behavior and predicts prognosis in colorectal cancer. Anticancer Res. 42, 5343–5355. https://doi.org/10.21873/anticanres.16058 (2022).
Schweizer, A., Rohrer, J., Slot, J. W., Geuze, H. J. & Kornfeld, S. Reassessment of the subcellular localization of p63. J. Cell. Sci. 108(Pt 6), 2477–2485. https://doi.org/10.1242/jcs.108.6.2477 (1995).
Li, S. X., Li, J., Dong, L. W. & Guo, Z. Y. Cytoskeleton-associated protein 4, a promising biomarker for tumor diagnosis and therapy. Front. Mol. Biosci. 7, 552056. https://doi.org/10.3389/fmolb.2020.552056 (2020).
Kikuchi, A., Matsumoto, S. & Sada, R. Dickkopf signaling, beyond Wnt-mediated biology. Semin. Cell Dev. Biol. 125, 55–65. https://doi.org/10.1016/j.semcdb.2021.11.003 (2022).
Bajaj Pahuja, K. et al. Phosphoregulatory protein 14–3–3 facilitates SAC1 transport from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 112, E3199-3206. https://doi.org/10.1073/pnas.1509119112 (2015).
Ishikawa-Sasaki, K., Nagashima, S., Taniguchi, K. & Sasaki, J. Model of OSBP-mediated cholesterol supply to Aichi virus RNA replication sites involving protein-protein interactions among viral proteins, ACBD3, OSBP, VAP-A/B, and SAC1. J. Virol. https://doi.org/10.1128/JVI.01952-17 (2018).
Lee, C. Roles of phosphoinositides and phosphoinositides kinases in hepatitis C virus RNA replication. Arch. Pharm. Res. 35, 1701–1711. https://doi.org/10.1007/s12272-012-1001-2 (2012).
Liu, Y. et al. Tracing the pathways: how inflammatory cytokines and blood metabolites drive intervertebral disc degeneration. Eur. Spine J. 34, 2106–2116. https://doi.org/10.1007/s00586-025-08850-9 (2025).
Li, Z. H. et al. Identification of aging-related biomarkers for intervertebral disc degeneration in whole blood samples based on bioinformatics and machine learning. Front. Immunol. 16, 1565945. https://doi.org/10.3389/fimmu.2025.1565945 (2025).
Zou, M. et al. Bioinformatics analysis reveals hub genes linked to programmed cell death in intervertebral disc degeneration. Appl. Biochem. Biotechnol. https://doi.org/10.1007/s12010-025-05243-y (2025).
Li, X. et al. Vitamin D deficiency promotes intervertebral disc degeneration via p38/NCoR2-mediated extracellular matrix degradation. Eur. J. Nutr. 64, 163. https://doi.org/10.1007/s00394-025-03685-y (2025).
Lu, X. et al. A novel mechanism of FBXW7 in combating intervertebral disc degeneration: Mitigating ferroptosis in nucleus pulposus cells through the regulation of mitophagy. Int. Immunopharmacol. 155, 114668. https://doi.org/10.1016/j.intimp.2025.114668 (2025).
Li, F. et al. LGR6 modulates intervertebral disc degeneration through regulation of macrophage efferocytosis. J. Transl. Med. 23, 475. https://doi.org/10.1186/s12967-025-06427-0 (2025).
Guo, D. et al. SSR1 and CKAP4 as potential biomarkers for intervertebral disc degeneration based on integrated bioinformatics analysis. JOR Spine 7, e1309. https://doi.org/10.1002/jsp2.1309 (2024).
Ma, D. et al. ELMO1 regulates macrophage directed migration and attenuates inflammation via NF-kappaB signaling pathway in primary biliary cholangitis. Dig. Liver Dis. 56, 1897–1905. https://doi.org/10.1016/j.dld.2024.05.012 (2024).
Chen, J. et al. ELMO1 ameliorates intestinal epithelial cellular senescence via SIRT1/p65 signaling in inflammatory bowel disease-related fibrosis. Gastroenterol. Rep. (Oxford) 12, goae045. https://doi.org/10.1093/gastro/goae045 (2024).
Lee, Y. J. et al. Overexpression of Dock180 and Elmo1 in melanoma is associated with cell survival and migration. Ann. Dermatol. 35, 439–450. https://doi.org/10.5021/ad.23.023 (2023).
Popescu, M. A. et al. Sac1 phosphatidylinositol 4-phosphate phosphatase is a novel host cell factor regulating hepatitis B virus particles assembly and release. FEBS J. 289, 7486–7499. https://doi.org/10.1111/febs.16575 (2022).
Nigorikawa, K., Fukushima, Y., Shimada, C., Matsumoto, D. & Nomura, W. CRISPRa analysis of phosphoinositide phosphatases shows that TMEM55A is a positive regulator of autophagy. Biol. Pharm. Bull. 47, 1148–1153. https://doi.org/10.1248/bpb.b23-00865 (2024).
Zheng, J. et al. Lipid phosphatase SAC1 suppresses hepatitis B virus replication through promoting autophagic degradation of virions. Antiviral Res. 213, 105601. https://doi.org/10.1016/j.antiviral.2023.105601 (2023).
Lin, L. et al. Exploring the molecular mechanisms underlying intervertebral disc degeneration by analysing multiple datasets. Sci. Rep. 15, 14748. https://doi.org/10.1038/s41598-025-98070-4 (2025).
Zhang, Y. et al. Identification and validation of circadian rhythm-related genes involved in intervertebral disc degeneration and analysis of immune cell infiltration via machine learning. JOR Spine 8, e70066. https://doi.org/10.1002/jsp2.70066 (2025).
Fan, C. et al. Integrated bulk and single-cell RNA sequencing to identify potential biomarkers in intervertebral disc degeneration. Eur. J. Med. Res. 30, 102. https://doi.org/10.1186/s40001-025-02346-4 (2025).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457-462. https://doi.org/10.1093/nar/gkv1070 (2016).
Lamb, J. et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935. https://doi.org/10.1126/science.1132939 (2006).
Griffith, J. F. et al. Modified Pfirrmann grading system for lumbar intervertebral disc degeneration. Spine (Phila Pa 1976) 32, E708-712. https://doi.org/10.1097/BRS.0b013e31815a59a0 (2007).
Acknowledgements
We thank Dr. Yansheng Liang, Department of Pathology, The Second People’s Hospital of Nanning, for the reviewing of the H&E staining.
Funding
This study was supported by the Guangxi Natural Science Foundation Program (2021GXNSFAA075007), China Youth Entrepreneurship and Employment Foundation (20250702-01 and 20240630-03).
Author information
Authors and Affiliations
Contributions
Study concept and design: LJW; Collection and assembly of data: LTJ, WYS and LBB; Performed the experiment: LTJ, WYS and LBB; Data analysis and interpretation: LTJ, WYS and LBB; Manuscript writing and review: All authors. All authors have read and approved the manuscript in its current state.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approval by the Ethics Committee of the Third Affiliated Hospital of Guangxi Medical University. All the procedures were carried out in accordance with institutional guidelines.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, Tj., Wang, Ys., Lan, Bb. et al. Unveiling key genes for intervertebral disc degeneration prediction and potential drug discovery. Sci Rep 16, 1918 (2026). https://doi.org/10.1038/s41598-025-31675-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-31675-x