Abstract
Advances in computer science and medicine have led to the emergence of artificial intelligence as a key tool in the medical and scientific fields. Its application in the diagnosis and treatment of diseases, such as cancer, has proven to be fundamental in improving early detection and saving lives. This article presents a proposal based on Deep Learning to develop a model capable of detecting melanomas in the skin from clinical images. The aim is to provide doctors with a tool to support early identification of this type of cancer, considering additional factors such as sun exposure and the patient’s skin tone. To optimize diagnostic accuracy and prevent data silos, a collaborative learning technique called Federated Learning is implemented. The FL framework employs a weighted averaging algorithm for model aggregation, allowing locally trained models to contribute to a continuously improving global model without sharing patient data. Experiments show that the proposed federated model achieved an accuracy of 89.1% and a ROC AUC of 0.9251, demonstrating performance comparable to centralized training while preserving privacy. In addition, a web application is presented to manage and process the information efficiently, making it easier for doctors to consult and analyze the results.
Similar content being viewed by others
Introduction
Melanoma remains one of the deadliest forms of skin cancer, responsible for the majority of skin cancer–related deaths despite accounting for less than 5% of total cases. According to the latest GLOBOCAN 2022 report, more than 331,722 new melanoma cases and over 60,000 deaths, reflecting a continued upward trend in incidence across all age groups1. Early detection, therefore, remains critical for improving prognosis, as survival rates exceed 95% when the disease is identified at an early stage.
Among the most pressing challenges in medical imaging data healthcare is the early detection of cancer. Identifying cancer in its initial stages is crucial for improving treatment outcomes and patient survival rates. Over the last decade, the integration of Artificial Intelligence (AI) and Deep Learning (DL) techniques into medical imaging has enabled unprecedented diagnostic capabilities, achieving expert-level accuracy in dermatology2,3,4 through the eHealth paradigm. The eHealth paradigm has redefined healthcare by integrating advanced digital technologies into medical practice, improving traditional processes and enabling unprecedented possibilities for diagnosing, treating, and monitoring diseases5,6. This approach enhances clinical decision-making that may be overlooked in traditional analysis, improving both sensitivity and diagnostic consistency.
A significant challenge in developing AI-driven diagnostic tools is the need for large and diverse datasets to train models effectively. In medical applications, acquiring such datasets is often constrained by the sensitive nature of patient data and the relatively infrequent occurrence of certain conditions7. Moreover, traditional centralized training strategies face significant challenges in medical contexts. The sensitive nature of patient data, strict privacy regulations, and institutional heterogeneity limit the feasibility of pooling datasets into a single repository7. Moreover, centralized models often suffer from reduced generalization when applied to unseen populations, as data biases–such as overrepresentation of fair-skinned patients can propagate into model predictions, compromising fairness and reliability.
To overcome these limitations, Federated Learning (FL) has emerged as a promising paradigm that enables collaborative model development without centralizing data8,9. This approach ensures user privacy while enabling the collaborative training of a global model across multiple institutions without the need to share raw data. The FL architecture allows the model to learn from a diverse set of distributed datasets, thereby improving its robustness, accuracy, and applicability across various healthcare environments. By periodically aggregating updates from locally trained models, the global model is continuously refined and redistributed, maintaining high performance while preserving the privacy and security of patient information.
Therefore, in this work, we propose a FL framework for melanoma detection that combines a convolutional neural network (CNN) with a distributed training protocol to collaboratively build a high-performing and privacy-preserving diagnostic model. The system is complemented by a user-friendly web application and a secure API that facilitate model interaction, clinical consultation, and seamless integration into existing healthcare workflows.
While a few previous works have explored FL in dermatological imaging, most have remained limited to proof-of-concept simulations or small-scale experiments without addressing deployment, usability, or model interpretability10,11,12. In contrast, this work advances the state of the art by integrating a complete FL-based melanoma detection pipeline with a clinical web interface and API, enabling both collaborative training and real-world usability. Furthermore, our study conducts a detailed comparative assessment with respect to a centralised reference, quantifying improvements in different metrics, such as accuracy, sensitivity, or specificity.
In summary, this paper’s contributions include the following:
-
Federated Melanoma Detection Framework: Development of a DL model integrated within an FL architecture that enables decentralized training across institutions while preserving patient privacy.
-
Performance and Privacy Evaluation: Comprehensive assessment of the federated model against a centralized baseline, demonstrating comparable accuracy and higher sensitivity in early melanoma detection.
-
User-Friendly Platform: A web application and API designed to ensure the solution is accessible, practical, and seamlessly integrated into the workflows of healthcare professionals.
The rest of the document is organized as follows. The next section reviews recent related works on the use of AI on dermatology, as well as FL in eHealth. The Methods section presents the design and implementation in detail. The results are shown in the Experimental results section. Then, the Discussion section discusses the results and limitations detected. Finally, conclusions and future work are drawn in the last section.
Literature review
The rapid integration of AI into medical imaging has catalyzed major advances in diagnostic accuracy, particularly in dermatology, where automated systems support clinicians in early melanoma detection. However, despite notable progress in DL and FL, existing research remains fragmented across methodological, clinical, and privacy dimensions. This section reviews the most relevant literature related to (i) DL methods applied to dermatological image analysis and melanoma classification, and (ii) recent FL-based frameworks in medical imaging and skin cancer detection. A critical analysis is provided to highlight the current limitations–such as data imbalance, model interpretability, and lack of large-scale validation–and to identify the research gap addressed by the proposed federated melanoma detection framework.
Deep learning in dermatology
The success of AI and Deep Learning DL in medical imaging has been widely demonstrated across several diagnostic domains. Recent works have applied DL to lung nodule localization in CT scans13, cataract and glaucoma detection using transfer learning with MobileNet architectures14, and brain tumor identification through ensemble learning on MRI images15. These examples highlight the versatility and robustness of DL models in diverse clinical imaging contexts.
In dermatology, DL methods have achieved substantial progress in the automatic detection and classification of skin lesions. Esteva et al.2 pioneered the use of convolutional neural networks (CNNs) for large-scale skin lesion classification, achieving dermatologist-level accuracy. Haenssle et al.3 further validated CNN-based models in clinical settings, demonstrating performance comparable to expert dermatologists. More recent approaches4,16 have refined these architectures by utilising transfer learning, adaptive feature extraction, and improved data augmentation strategies, resulting in enhanced generalisation to diverse lesion types.
Despite these advances, centralized DL models depend heavily on large, balanced, and diverse datasets. Public repositories such as ISIC17 and HAM10000 Footnote 1 remain dominated by lighter skin types, which can introduce biases and reduce model performance in underrepresented populations. Moreover, centralized data aggregation poses ethical and regulatory challenges under frameworks such as GDPR and HIPAA, motivating the exploration of privacy-preserving solutions such as FL.
Federated learning in dermatology and medical imaging
Federated Learning has emerged as a promising approach to mitigate these issues by enabling decentralized model training across multiple institutions without sharing raw data8,9. In the FL framework, clients (hospitals or clinics) train local models on their private datasets and only exchange model parameters with a coordinating server for aggregation. This setup preserves privacy while allowing the model to benefit from heterogeneous and distributed data sources.
In the dermatology domain, several studies have explored FL for skin cancer detection, though most remain at a proof-of-concept stage. For instance, Ain et al.18 proposed a privacy-aware FL model for skin lesion classification, achieving comparable accuracy to centralized training while maintaining data confidentiality. Similarly, Yaqoob et al.19 conducted a systematic review on FL in skin cancer detection, highlighting advances in privacy-preserving approaches but also the lack of validation on large-scale, multi-institutional datasets. More recently, Yaqoob et al.20 presented an asynchronous weighted FL framework that improves communication efficiency and model convergence in heterogeneous dermatological datasets.
Beyond dermatology, numerous FL applications have been reported in radiology, pathology, and cardiology, demonstrating comparable or even superior performance to centralized models while complying with data privacy regulations10,21. Privacy-preserving techniques such as differential privacy, secure aggregation, and homomorphic encryption have been integrated into FL frameworks to further protect model updates during communication. However, these methods often increase computational overhead and may affect convergence speed, which limits their applicability in real-time clinical environments.
Challenges and research gap
Table 1 summarizes the most relevant studies discussed in this section, highlighting their methodologies, datasets, privacy mechanisms, and key advantages and limitations.
Although previous studies have provided valuable insights into the potential of Federated Learning (FL) for dermatology, they still present important limitations. Most implementations remain limited to small-scale or simulated environments with homogeneous data sources, lacking evaluation on real, heterogeneous clinical datasets. In addition, prior works rarely address practical aspects such as model deployment, usability, or integration into healthcare workflows, which are essential for clinical adoption. Few studies perform a detailed comparison between federated and centralized learning to quantify the trade-offs in diagnostic performance. Moreover, interpretability and explainability–key factors for medical decision support–are often underexplored, limiting their reliability in real-world practice.
In contrast, the present work introduces a comprehensive FL framework for melanoma detection that integrates weighted model aggregation (FedAvg), privacy-preserving communication, and an intuitive clinical web interface to facilitate real-world usability. The proposed system not only demonstrates technical feasibility but also delivers a rigorous comparative evaluation against a centralized baseline, evidencing improvements in sensitivity, specificity, and overall robustness. By bridging the gap between experimental FL research and clinically deployable AI tools, this study contributes a practical and scalable solution toward privacy-aware melanoma diagnosis.
Methods
This section outlines the framework adopted in this work to develop an AI-based system for melanoma detection. It begins with a detailed description of the data acquisition process, including the source and classification of dermatological images. Subsequently, it addresses the preprocessing steps required to prepare the dataset for training, followed by the architecture and configuration of the DL model employed. The training methodology, including optimization strategies and evaluation metrics, is also presented. Finally, the integration of the trained model into a complete system–comprising an API and a web interface–is described, emphasizing its deployment and interaction within an FL environment.
Data acquisition
The first step in this research was to acquire a high-quality dataset of skin lesion images suitable for training and validating the DL model. This task posed significant challenges due to the difficulty of locating large and well-annotated datasets of skin images, a prerequisite for developing robust diagnostic tools.
The dataset utilized for this work was sourced from the ISIC Challenge (International Skin Imaging Collaboration), an annual competition designed to foster advancements in dermatological imaging and diagnosis17. These datasets are carefully curated and validated by experts, ensuring their reliability for scientific purposes.
The dataset included a detailed Excel file linking each image to its diagnostic category, facilitating a direct association between the visual data and its corresponding clinical information. The data set classifies the images into six diagnostic categories, summarized as follows.
-
Melanoma (MEL): A malignant cancer originating in melanocytes. Typically dark brown or black, though some melanomas may appear pink, pale, or white.
-
Nevus (NV): Commonly known as moles or freckles, these are benign proliferations of melanocytes.
-
Basal Cell Carcinoma (BCC): A type of skin cancer originating in basal cells, often presenting as a transparent bump on sun-exposed skin.
-
Actinic Keratosis (AKIEC): A rough, scaly patch resulting from long-term sun exposure, often a precursor to more serious conditions.
-
Benign Keratosis (BKL): A non-cancerous skin growth that increases with age, often brown or waxy in appearance.
-
Dermatofibroma (DF): A common, benign skin tumor that typically appears as a small, firm nodule on the lower extremities.
-
Vascular Lesions (VASC): Damage or anomalies in blood vessels, often caused by aging, trauma, or other health conditions.
For the purposes of this approach, images and metadata were utilized to develop a binary classification system distinguishing between malignant and non-malignant lesions. The malignant group encompassed the categories MEL, BCC, AKIEC, and DF, while the non-malignant group included NV, BKL, and VASC. This approach was chosen to simplify the model’s output, focusing on providing an initial assessment to guide medical professionals, who would then conduct further diagnostic tests as needed.
Data processing
To construct a baseline model for melanoma detection, it is essential to preprocess the dataset by aligning images with their respective labels. This step ensures that the model can be trained effectively using labelled data. This section describes the methodology employed to prepare the dataset, including data extraction, label assignment, and data partitioning for training and validation.
Data labelling and preparation
The dataset from the ISIC includes metadata in .csv format, which links each image to its corresponding diagnosis. Using Python’s Pandas library, the relevant columns were extracted, as outlined in the data acquisition process. To create binary labels indicating malignancy, a new column was added to the dataset. For each image, a value of 1 was assigned if any of the following columns–MEL, BCC, AKIEC, or DF–had a value of 1, indicating malignancy. Otherwise, a value of 0 was assigned, classifying the image as non-malignant (categories NV, BKL, and VASC). This binary labelling simplified the classification task to focus on distinguishing between malignant and non-malignant lesions.
Data partitioning
Once the labelled data was prepared, it was divided into two subsets: a training set and a validation set. Typically, and in this case, 80% of the data is allocated to training and the remaining 20% to validation. The training set is used to adjust the model’s parameters, such as weights in the neural network, enabling the model to learn patterns and features relevant to melanoma detection. The validation set, on the other hand, serves to evaluate the model’s performance on unseen data and monitor its ability to generalize beyond the training data.
Image preprocessing
In parallel with the label preparation, the corresponding images were retrieved and processed. Images were resized to ensure a consistent input size for the DL model, a necessary step given the variability in image dimensions. Additionally, the images were standardized to a uniform format and structure to facilitate efficient processing and compatibility with the model architecture. The images were then split into training and validation subsets, mirroring the distribution of the labels. This ensured that the training and validation datasets were paired consistently, maintaining the integrity of the labelled data throughout the preprocessing pipeline.
Model definition
For this purpose, we employ a Convolutional Neural Network (CNN) as the core architecture for melanoma detection. CNNs are ideal for image-based tasks due to their ability to automatically extract and analyze hierarchical features, from simple patterns like edges to complex structures. Their effectiveness in capturing subtle visual cues makes them particularly suitable for medical imaging, where distinguishing between malignant and non-malignant lesions often depends on nuanced differences. The following sections outline the design and configuration of the CNN model.
CNN model definition
To design the Convolutional Neural Network (CNN) for melanoma detection, a Sequential model was selected as the foundational structure. The Sequential model is the most basic and straightforward approach for creating DL models, particularly CNN. Listing 1 shows the architecture of the CNN model:
For a better understanding of the structure of the model, Table 2 shows a summary of the model structure grouped by stages, which is explained in detail below:
Input and feature extraction layers. These layers are responsible for extracting significant features from the input images, such as edges, textures, and patterns. They include convolutional layers to detect local features, normalization layers to stabilize training, and pooling layers to reduce spatial dimensions.
-
Conv2D: Applies convolutional filters to detect local features in the input images with a size of 450x600px. Each filter focuses on specific patterns such as edges or textures.
-
BatchNormalization: Normalizes the outputs of the convolutional layers, improving training stability and speed by reducing internal covariate shift.
-
MaxPooling2D: Reduces the spatial dimensions (width and height) of feature maps, retaining the most important information while reducing computational complexity.
In the model, these layers progressively increase the number of filters, starting with smaller numbers (32) to capture basic features and incrementally increasing to 256 filters for more complex patterns.
Global pooling and dense layers. Once the features are extracted, the model applies a global average pooling operation to reduce each feature map to a single value, followed by dense layers that act as the classifier. These layers learn high-level patterns from the pooled features.
-
GlobalAveragePooling2D: Aggregates each feature map into a single scalar by computing the average, reducing dimensionality and preventing overfitting while preserving global information.
-
Dense: Fully connected layers that learn complex patterns from the pooled features. They use the ReLU activation function to introduce non-linearity, enabling the model to capture intricate relationships in the data.
In this model, the dense layers start with 128 neurons in the first layer and reduce to 64 neurons in the subsequent layer, simplifying the extracted features for the final classification.
Output and regularization layers. These layers are crucial for producing the final output of the model while mitigating overfitting during training. Regularization techniques like dropout are applied to improve the model’s generalization capabilities.
-
Dropout: Deactivates a random fraction (50%) of neurons during training, preventing the model from overfitting and relying too heavily on specific neurons.
-
Dense: The final dense layer uses a sigmoid activation function to output a single probability value, indicating the likelihood of the input being malignant (1) or non-malignant (0).
Neuron distribution. The neuron distribution is designed to capture features effectively, starting with fewer neurons in early layers for basic patterns and increasing to more complex ones. Dense layers reduce and simplify features, while dropout prevents overfitting, ensuring robust classification.
-
Feature Extraction Layers: The number of neurons (filters) increases progressively from 32 to 256 to capture both basic and complex features of the images. The earlier layers detect simple elements like edges, while later layers extract higher-level patterns.
-
Dense Layers: The number of neurons starts high (128) to retain as much information as possible from the feature maps and decreases (64) to simplify the feature representation before classification.
-
Regularization and Output: Dropout layers deactivate 50% of neurons during training to prevent overfitting, while the final dense layer outputs a probability for binary classification.
Model training
The training process of the CNN model involved several steps to ensure efficient resource utilization and achieve optimal performance. This section details the techniques, configurations, and methodologies applied before, during, and after the training phase to enhance the model’s efficiency and effectiveness.
Firstly, the image data was normalized by dividing each pixel value by 255.0 to scale the range to [0, 1]. This normalization enhances model stability and accelerates convergence. Additionally, data augmentation was applied using the ImageDataGenerator shown in Listing 2 to improve generalization by introducing variations in the input images (e.g., rotations, zooms, flips). This step ensures the model adapts to different cases and scenarios, leading to better performance on unseen data.
After that, the compilation step is essential to prepare the CNN model for training, as it configures the model’s behaviour in terms of optimization, loss calculation, and performance evaluation. Listing 3 shows the components selected to align with the binary classification task and the characteristics of the dataset:
-
optimizer: The optimizer adjusts the model’s weights based on the calculated loss during training. For this model, the Adam optimizer was chosen due to its adaptability to large-scale problems and its effectiveness in handling noisy datasets. A learning rate of \(1\times 10^{-4}\) was explicitly set to ensure stable and gradual convergence. Adam combines the benefits of both the RMSprop and Stochastic Gradient Descent (SGD) optimizers, making it well-suited for this task.
-
loss: The loss function evaluates how well the model’s predictions match the expected values. The Binary Crossentropy loss function was selected because it is specifically designed for binary classification tasks where the target labels are either 0 or 1. This function calculates the difference between the predicted probabilities and the true binary labels, providing a reliable metric for guiding the optimizer.
-
metrics: Metrics provide insights into the model’s performance during training and validation. In addition to accuracy, which measures the proportion of correct predictions, the model also tracks precision, recall, and the area under the ROC curve (AUC). These metrics are particularly important in medical contexts, as they offer a more detailed evaluation of the model’s ability to distinguish between malignant and non-malignant cases.
Next, to optimize the training process and ensure efficient resource usage, two key techniques were employed as shown in Listing 4. These methods dynamically adjust the training process, preventing overfitting and unnecessary resource consumption while maximizing model performance.
-
Early stopping: Early Stopping monitors the validation AUC (val_auc) during training and halts the process if there is no improvement after a specified number of epochs (patience). This technique minimizes overfitting and reduces training time. Three key parameters define Early Stopping:
-
monitor: The metric chosen to monitor was val_auc, as it reflects the model’s ability to distinguish between classes, which is crucial in binary medical classification tasks.
-
mode: Set to ”max” to indicate that training should continue as long as the AUC improves.
-
patience: Set to 7 epochs, meaning the training will stop if there is no improvement in validation AUC for seven consecutive epochs. This value allows the model enough time to overcome small fluctuations while avoiding excessive training.
-
restore_best_weights: Set to True, ensuring that the final model uses the weights from the epoch with the best validation performance.
-
-
ReduceLROnPlateau: This technique adjusts the learning rate dynamically when the validation AUC stagnates. It includes:
-
monitor: As with Early Stopping, val_auc was chosen to guide the adjustments based on model discrimination capability.
-
mode: Also set to ”max”, ensuring learning rate reductions only occur when AUC plateaus.
-
patience: Set to 3 epochs, allowing a quicker response to performance stagnation.
-
factor: Set to 0.3, meaning the learning rate is reduced by 70% when triggered, allowing more gradual fine-tuning.
-
min_lr: Set to \(1\times 10^{-6}\) to prevent the learning rate from becoming too small and halting learning.
-
Model evaluation
After training the model, the next critical step is to evaluate its performance and analyze the results. For this work, several evaluation methods were employed to assess how effectively the trained model performs on unseen data. These methods are detailed below:
-
Predictions using model.predict(validation_data): The function model.predict was used to make predictions on the validation dataset. By iterating through the predictions and their corresponding actual labels, a visual comparison was conducted to evaluate how closely the model’s predictions matched the true labels.
-
Model evaluation using model.evaluate(validation_data): The model.evaluate function computes the model’s performance metrics on the validation data. For this binary classification problem, metrics such as validation loss and accuracy were returned. Validation loss serves as a measure of how well the model generalizes, while accuracy quantifies the proportion of correct predictions.
-
Calculating accuracy percentage: The model’s accuracy was further analyzed using a relative threshold. For this, predictions within a defined margin of the actual labels (e.g., 10% deviation) were identified, and the percentage of predictions meeting this criterion was calculated. This method offers additional insight into the model’s reliability and practical applicability.
-
Saving the trained model: To ensure the model can be reused without retraining, the trained network was exported and saved as a file. This allows the model to be tested with new datasets or integrated into applications such as an API for real-time melanoma detection. Saving the model also facilitates reproducibility and future improvements.
-
Visualization of training and validation loss: Using Matplotlib library22, the training and validation loss were plotted across all epochs to provide a visual representation of the learning process. These plots help identify trends, such as overfitting or underfitting, and demonstrate the model’s convergence during training.
Federated model
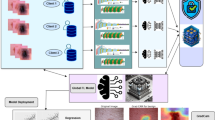
A weighted federated averaging strategy (FedAvg) was employed to synthesize a single global network from locally fine-tuned replicas trained at each institution. Figure 1 illustrates the overall workflow, which comprises three main stages: data acquisition, local model training, and model aggregation. In this setup, medical centers train models on their private data and share only the resulting model parameters, thereby preserving data confidentiality throughout the process.
A weighted federated averaging strategy (FedAvg) was employed to synthesize a single global network from locally fine-tuned replicas trained at each institution. Figure 1 illustrates the overall workflow, which comprises three main stages: data acquisition, local model training, and model aggregation. In this setup, medical centers train models on their private data and share only the resulting model parameters, thereby preserving data confidentiality throughout the process.
Let \(\varvec{w}_k \in \mathbb {R}^P\) denote the parameter vector (all trainable weights and biases, layer-wise concatenated) learned at client k using \(n_k\) local samples, and let \(N=\sum _k n_k\) be the total number of samples across all clients. The server-side aggregation computed the global model as a layer-wise convex combination
which was then redistributed as the starting point for subsequent rounds or used as the final model at the end of the federation. This rule approximate empirical risk minimization under heterogeneous sizes while preserving privacy, since no raw data were exchanged, only model parameters.
Data acquisition. Each participating institution collected dermoscopic images and associated metadata from its local repository while maintaining data within its secure environment. Before training, datasets were anonymized to remove any identifiable patient information, and standardized preprocessing pipelines were applied to harmonize image resolution and color balance across sites.
Local model training. Each participating medical center fine-tuned an identical CNN architecture to ensure shape compatibility during aggregation (see the local training blocks in Fig. 1). All models were initialized from a common checkpoint, freezing the earlier convolutional layers while adapting the final dense layers to local data characteristics. Data augmentation and class weighting were applied to mitigate class imbalance. Training was monitored with early stopping based on validation AUC and a learning-rate reduction on plateau, both of which improved robustness and minimized unnecessary local epochs before aggregation.
Model aggregation. As depicted in the rightmost section of Fig. 1, at the end of each federated round, the server collected model weights and corresponding sample counts \(n_k\) from all clients through an encrypted communication protocol. A layer-wise weighted sum of parameters was computed and normalized by N to produce the global model \(\varvec{w}^{(t+1)}\), following the FedAvg rule in Eq. 1. The aggregated weights were then compiled and persisted for evaluation and redistribution. Operationally, the API endpoint /federatedLearning executed this pipeline, automatically scanning client checkpoints, performing weighted averaging, and saving the updated global parameters.
This aggregation step consolidated heterogeneous knowledge learned across institutions, each with distinct data distributions and sizes, into a single global model while preserving patient privacy. Weighting by \(n_k\) ensured that clients with larger datasets exerted a proportionally greater influence on the consensus parameters, yielding a balanced operating point suitable for clinical screening scenarios where sensitivity and specificity must be considered jointly.
Listing 5 presented the final model aggregation algorithm (averageWeightsModel), which loaded all models’ checkpoints, inferred the per-client sample counts \(n_k\) from local label logs, computed the sample-size–weighted, layer-wise average (FedAvg), persisted checkpoints/average/average.weights.h5, and then invoked create_average_model.
Listing 6 detailed the materialization step (create_average_model), which reconstructed the architecture, loaded the averaged weights, compiled the network (Adam \(10^{-4}\), binary cross-entropy; accuracy/precision/recall/AUC), and saved the deployable model at models/average/melanoma_average.h5.
API
The API is a fundamental component of the melanoma detection system, serving as the interface between the trained model, the web application, and the FL framework. Its primary purpose is to enable seamless integration of key functionalities, including image predictions, local model retraining, FL coordination, and data management. By supporting these operations, the API ensures the system’s scalability, usability, and adaptability to clinical environments, making it suitable for real-world healthcare applications.
The API is essential for implementing the FL architecture, which allows distributed model training while preserving data privacy. Through this architecture, the API facilitates the aggregation of model weights from multiple nodes, enabling the creation of a robust global model without the need for centralized data storage. This is particularly important in healthcare, where privacy and compliance are critical.
API core functions
-
Image processing: Uploaded images are preprocessed to ensure compatibility with the trained model. This includes decoding images to RGB format, resizing them to the required dimensions, and normalizing pixel values. These steps ensure the accuracy and consistency of predictions, aligning with the preprocessing pipeline used during model training.
-
Label management: Prediction results are stored in a text file for future use, enabling traceability and supporting local retraining. If the file does not exist, it is created, and the prediction label is appended.
-
Base model generation: This function initializes the base model architecture and loads the corresponding weights. It is used when clinicians upload weights for local retraining or when FL updates the global model.
-
Loading models and weights: Upon user login, the system loads the clinician’s base model and applies the latest weights available for that user. This ensures the predictions and retraining processes are up-to-date with the most recent data.
-
Training data preparation: This function divides the dataset into training and validation subsets (e.g., 80% training, 20% validation). It is used during local model retraining to prepare data for the model.
-
Weight aggregation for FL: For FL, this function collects the weights from all participating nodes, computes the average, and generates a new global model with the aggregated weights.
-
Performance visualization: After training or retraining, accuracy and loss graphs are generated using the model’s performance metrics. These visualizations provide clinicians with insights into the training process and model improvements.
-
Report generation: When a prediction is made, this function creates a detailed report, including the prediction results, user information, and other relevant details. Reports are stored in a structured format, facilitating easy retrieval.
Table 3 summarizes the API endpoints implemented in the proposed system, detailing their functionalities, and roles within the federated learning workflow.
Web
The web application, developed using Angular, plays a pivotal role in ensuring accessibility and usability of the melanoma detection platform for healthcare professionals. Its primary objective is to provide clinicians with a secure, intuitive, and efficient interface for interacting with the system’s core functionalities, thus facilitating its integration into routine clinical workflows.
From the authentication process onward, the platform guarantees that only authorized users can access patient data and diagnostic tools, thereby upholding privacy and regulatory compliance. The application enables clinicians to easily upload dermatological images, which are then preprocessed and analyzed by the AI model through seamless communication with the backend API. Upon completion of the analysis, the application displays the diagnostic prediction and related metrics in a clear and interpretable format, supporting informed decision-making.
Additionally, the web interface grants access to historical records and detailed diagnostic reports generated by the system. This feature allows clinicians to review previous cases, monitor patient evolution over time, and compare diagnostic outcomes, all within a unified environment.
Login screen: user authentication
Upon accessing the platform at the /login route, users are presented with a minimalist and intuitive login interface designed to guarantee secure access. When credentials are submitted, the application sends a POST request to the backend API’s /login endpoint. Upon successful verification, the API issues an access token that secures subsequent interactions with the platform. This authentication mechanism is required before accessing any clinical features of the system.
Main dashboard: report generation ( /home )
After successful authentication, users are redirected to the main dashboard, accessible at the /home route. This screen serves as the starting point for generating new diagnostic reports and managing clinical interactions with the system.
The interface includes the following elements:
-
Patient selector: Allows users to choose a patient from a predefined list. The application retrieves patient data by querying the backend API’s /patients endpoint.
-
Affected area: A text field where users specify the anatomical area under examination.
-
Sun exposure selector: A dropdown menu for indicating the patient’s degree of sun exposure, supporting contextual risk assessment.
-
File upload: Users can select and upload a dermatological image, which is processed as part of the diagnostic workflow.
-
Generate Report button: Initiates the analysis process. When clicked, the application sends a request–including the selected patient, affected area, sun exposure, and image file–to the backend API’s /predict endpoint. The API processes the image, applies the AI model, and returns a diagnostic prediction.
In addition to the report generation form, the top navigation bar provides access to further functionality:
-
Home: Returns the user to the dashboard for generating new reports.
-
Reports: Displays a list of diagnostic reports previously generated by users in the same medical center. This section interacts with the API’s /informs endpoint to retrieve the data.
-
Account: Opens the user profile section, where personal information is displayed and additional actions such as model retraining and FL can be performed.
-
Logout: Logs the user out, invalidates the session, and redirects back to the login screen (/login).
This layout allows clinicians not only to generate new diagnostic reports but also to easily review past reports, manage their accounts, and securely log out from the system. The integration with the corresponding API endpoints ensures smooth communication between the web application and backend services.
Reports overview ( /informs )
After accessing the Reports section, users are presented with an overview page (see Fig. 2) that displays all available diagnostic reports for the currently authenticated medical center.
The layout consists of two main sections:
-
Report list (left panel): Shows a scrollable list of reports, each presented as a card. Each card contains the patient’s name, the affected area, the test date, and a thumbnail of the original image. The currently selected report is visually highlighted for clarity.
-
Report details (right panel): Displays detailed information about the selected case, including a larger view of the associated dermatological image, patient name, area affected, age, sex, sun exposure, test date, and the malignancy probability estimated by the AI model.
Users can quickly browse through different cases by clicking on report cards, which updates the detail view on the right. All report data are retrieved from the backend through the /informs/user_id endpoint, while images are loaded dynamically based on the URLs provided in each report.
Account configuration ( /account )
The /account page provides users with access to their personal and professional details, as well as configuration options relevant to the diagnostic workflow. The main components include:
-
Professional registration number: Displays the clinician’s unique identifier within the platform.
-
Full name: Shows the name of the logged-in user.
-
Sun exposure factor: Editable fields for both High” and Half” sun exposure levels. These parameters allow each medical center to set custom multipliers that adjust the AI model’s melanoma risk assessment according to local clinical criteria or environmental conditions. By modifying these values, clinicians can tailor the model’s predictions to better reflect the realities of their specific patient population.
-
Save settings: Saves any changes made to the sun exposure factors or user details by issuing a PUT request to the /users/settings/user_id endpoint.
-
Improve local model: Initiates retraining of the AI model using recent clinical cases from the center via a POST request to the /retrain_model/user_id endpoint.
-
Apply FL: Submits the updated local model for federated averaging by calling the /federatedLearning endpoint, supporting collaborative model improvement across all participating centers.
This settings page empowers medical centers to adjust model parameters, customize sun exposure risk multipliers, and actively participate in the continuous improvement of the global diagnostic model through FL.
Data and code availability
The source code utilized and analyzed in our research is publicly accessible at the DOI 10.5281/zenodo.15805545, or as a direct link in reference23. The leveraged dataset from ISIC is specifically ISIC Challenge Datasets 2018, which is also available atFootnote 2. The dataset was acquired as-is, leveraging the existing annotations from the dataset’s metadata. The dataset’s quality was analyzed considering the balance among the MEL and No-MEL classes, and the system was designed with this imbalance in consideration.
Experimental results
This section presents the results obtained from a series of experiments designed to evaluate the performance of the proposed melanoma detection system. The goal is to compare the effectiveness of a centralized learning approach versus a simulated FL strategy, both aimed at distinguishing melanoma from non-melanoma skin lesions. The analysis includes details on training configuration, evaluation metrics, and visualizations such as loss curves and confusion matrices to provide a comprehensive assessment of each model’s behaviour. The results are first structured with the description of the evaluation setup, followed by detailed results for the centralized and federated scenarios.
Evaluation setup
To evaluate the effectiveness of the proposed melanoma detection system, two different training scenarios were tested: a Centralized Learning Strategy (CLS) and a FL Simulation (FLS). In both cases, the task was formulated as a binary classification problem, distinguishing between melanoma (MEL) and non-melanoma (No-MEL) lesions.
In the CLS setup, the model was trained using a dataset of 10,015 labelled dermatological images. In contrast, the FLS setup involved a base model trained with 4,000 images, followed by three independent user-specific training phases using datasets of 2,000, 2,000, and 2,015 images, respectively. Each training phase represented a distinct medical center (as the workflow shown in Fig. 1), contributing to the global model without sharing raw data. All federated partitions were derived from the same original dataset of 10,015 images using a Python script that ensured non-overlapping subsets and preserved class distribution across clients.
To ensure fair and unbiased evaluation, all trained models were tested on an independent dataset composed of 202 labelled images, which was not used during any phase of training or validation.
All training and evaluation experiments were conducted on a personal laptop equipped with an AMD Ryzen 7 5800H processor, 16 GB of RAM, and an NVIDIA RTX 3060 GPU, which provided sufficient computational capacity to execute the models efficiently while simulating a realistic clinical research setting.
The objective of this evaluation is to rigorously assess the performance of the proposed melanoma detection system under both the CLS and FLS strategies. The evaluation process is structured in two phases. During the training phase, the goal is to monitor the model’s learning behaviour and internal performance using metrics such as accuracy, precision, recall, F1-score, and ROC AUC (Area Under the Receiver Operating Characteristic Curve), along with the analysis of loss curves to detect overfitting or convergence issues. In the subsequent evaluation phase, the trained models are tested on the independent test set to measure their generalization capability. This phase includes the computation of the same classification metrics and the generation of a confusion matrix to visualize the distribution of correct and incorrect predictions, providing insight into sensitivity and specificity in detecting melanoma.
CLS results
The centralized model was trained on a dermatoscopic dataset with a marked class imbalance: 8,902 No-MEL cases and 1,113 MEL cases. To address this, class weights were computed to compensate for the disparity, and real-time data augmentation was applied using random transformations–such as rotation, translation, shear, zoom, and horizontal flipping–during training. The full configuration of the data generators is provided in the Model training subsection. Although the number of images remained constant, these transformations effectively increased the diversity of the training data across epochs.
The training process achieved a final accuracy of 0.7193, with a precision of 0.2603, recall of 0.8292, and ROC AUC of 0.8429. These results indicate that the model was optimized for sensitivity, effectively detecting most melanoma cases despite a higher false positive rate. The optimal classification threshold, obtained through F1-score maximization, was set at 0.697 to favour recall.
Figure 4ashows the loss curves during training. The consistent downward trend in both training and validation loss suggests stable convergence and absence of overfitting.
Evaluation on the independent test set yielded an overall accuracy of 0.9119 and a ROC AUC of 0.9251. For the melanoma class, the model achieved a precision of 0.5833, recall of 0.6667, and an F1-score of 0.6222. For the non-melanoma class, the recall was 0.9419, indicating strong performance on negatives while maintaining a sensitivity-oriented behaviour suitable for clinical screening. It is worth noting that, in this context, recall for the melanoma class corresponds to sensitivity, whereas recall for the non-melanoma class corresponds to specificity.
Figure 3 presents the confusion matrix. The model correctly identified 162 of the 172 non-melanoma cases, with only 10 false positives. For melanoma, 14 out of 21 cases were correctly classified, resulting in 7 false negatives. This distribution confirms the model’s ability to detect the majority of melanoma cases while maintaining strong performance on the negative class.
FLS results
The federated training process was structured as an initial model followed by three independent training phases, each simulating a local update from a distinct clinical node. Upon completion of all phases, model weights were aggregated to update the global model without exchanging raw patient data.
The results reported below correspond to the base model and subsequent iterations within this federated workflow, all evaluated on the same independent test set used in the centralized setup.
FLS - base model
The base model in the federated setup was trained on a subset consisting of 3,524 non-melanoma (No-MEL) cases and 476 melanoma (MEL) cases. As with the centralized scenario, techniques such as data augmentation and class weighting were applied to mitigate the class imbalance and support learning from minority cases.
After training, the model reached an accuracy of 0.7237, with a precision of 0.2811, a recall of 0.8478, and AUC of 0.8544. These results demonstrate a clear focus on recall, consistent with the system’s intended role in medical triage. The classification threshold that maximized the F1-score was determined to be 0.637.
While the recall is notably strong, it is important to consider that the model was trained on fewer and less diverse samples than the centralized counterpart. This may partially explain the higher sensitivity, possibly at the cost of generalization. The reduced variety in the training set could make the model more responsive to patterns present in the test set, inflating its apparent performance.
Figure 4bdisplays the loss curves over epochs. The behaviour suggests stable convergence, with validation loss closely tracking the training loss, and no signs of overfitting.
Evaluation on the independent test set yielded an overall accuracy of 0.8705 and a ROC AUC of 0.9297. For the melanoma class, the model achieved a precision of 0.4474, a recall (sensitivity) of 0.8095, and an F1-score of 0.5763. For the non-melanoma class, the recall (specificity) was 0.9012. These metrics reinforce the model’s high sensitivity, despite a moderate rate of false positives.
FLS – medical center 1
Following the first user-specific training phase using 2,000 images, the model reached an accuracy of 0.7125 and a ROC AUC of 0.8459. The recall remained high at 0.8313, while precision decreased to 0.2421, highlighting a trade-off favouring sensitivity. The optimal classification threshold was set at 0.696. As shown in Fig. 5a, both training and validation loss continued to decline steadily, suggesting that the model effectively integrated new data without signs of overfitting.
FLS – medical center 2
The second user-specific training phase used 2,000 images from a distinct medical center. The model achieved an accuracy of 0.6862, precision of 0.2286, recall of 0.8081, and ROC AUC of 0.8216. Despite the lower accuracy, the recall remained consistent, while loss curves (Fig. 5b) showed continued, stable convergence.
FLS – medical center 3
The third user-specific training phase incorporated 2,015 images from another medical center. The model obtained an accuracy of 0.7246, precision of 0.2505, recall of 0.8012, and ROC AUC of 0.8342. The learning curves (Fig. 5c) confirmed sustained training progression with no overfitting across the individual updates.
FLS – federated model
After completing the three user-specific training phases, a final global model was obtained by applying weighted federated averaging. In this approach, each medical center’s model contributed to the global update proportionally to the number of training samples used. This ensured that more representative nodes had a greater influence on the final weights while preserving fairness across participants.
The aggregation process involved summing the weights of each layer across all models, scaled by their sample counts, and then normalizing by the total number of samples. This yielded a new set of parameters representing a consensus across all local models.
The resulting federated model was then evaluated using the independent test set of 202 images. To ensure comparability with previous training phases, the classification threshold was fixed at 0.637, the value that maximized the F1-score in the federated base model.
The final model achieved an accuracy of 0.8601 and a ROC AUC of 0.9321. For the melanoma class, the model obtained a precision of 0.4210 and a recall (sensitivity) of 0.7619. For the non-melanoma class, the recall (specificity) was 0.8720, indicating a balanced operating point that improves precision over earlier phases while maintaining strong detection performance.
Figure 6 presents the confusion matrix. The model correctly classified 150 of 172 non-melanoma cases and 16 of 21 melanoma cases. The 22 false positives and 5 false negatives reflect a solid trade-off between specificity (0.8720) and sensitivity (0.7619), suitable for clinical scenarios where early detection is critical.
Discussion
To assess the effectiveness of the FL approach, the federated model obtained through aggregation was compared against the centralized model trained on the full dataset. Both were evaluated using the same independent test set of 202 labelled images, and all metrics were calculated using their respective optimal F1-score thresholds (0.697 for the centralized model, 0.637 for the federated one).
Performance comparison
Table 4 summarizes the key evaluation metrics for both models. The performance comparison between the centralized (CLS) and federated (FLS) models highlights key differences in their evaluation metrics. The CLS achieved a higher accuracy of 0.9119 and a higher precision for melanoma cases (MEL) of 0.5833, suggesting fewer false positives. In contrast, the FLS reached a slightly lower accuracy of 0.8601 and a precision of 0.4210.
However, the federated approach outperformed the centralized model in terms of sensitivity, achieving 0.7619 compared to 0.6667, meaning it correctly identified a larger proportion of actual melanoma cases. Regarding specificity, the CLS reached 0.9419 while the FLS obtained 0.8720, indicating that the centralized model was more effective at correctly classifying non-melanoma samples.
Moreover, the FLS also obtained a higher ROC AUC of 0.9321 compared to 0.9251 for the CLS, indicating a superior overall discrimination ability. These results suggest that while the centralized model offers better precision and specificity, the federated model may be better suited for early detection scenarios where higher sensitivity to melanoma cases is critical. To serve as a comparison point,24 is compared with sensitivity, specificity, and accuracy, which are reported in detail in Table 4. Overall, except for sensitivity, the results are similar. Interestingly, sensitivity is the least relevant metric of the three for this use case, as it is a first screening method: false positives will be detected eventually by more sensitive screening methods, and the key is to correctly detect true negatives. These results are reinforced by the p-value analysis, performed at a 99% significance. The p-values, also reported in Table 4, would show that the differences for each type of result (true positives, true negatives, false positives, and false negatives) are statistically significant, were the p-value be below 0.01. As all p-values are multiple orders of magnitude below 0.01, the difference between CLS and FLS is considered significant.
Confusion matrix comparison
The confusion matrices for both the centralized (CLS) and federated (FLS) models reveal differences in their classification behaviour. The CLS correctly identified 162 true negatives (No-MEL correctly classified), 14 true positives (MEL correctly classified), while committing 10 false positives and 7 false negatives. In contrast, the FLS identified 150 true negatives and 16 true positives, with 22 false positives and only 5 false negatives.
These results indicate that while the CLS reduced the number of false positives, offering slightly better precision, the FLS achieved a higher number of true positives (16 vs. 14), reflecting better sensitivity to actual melanoma cases. This trade-off highlights the FLS’s ability to generalize and prioritize recall, which is particularly valuable in clinical screening where missing melanoma cases can have serious consequences.
Ethical and scalability considerations
While the experimental results demonstrate strong overall performance, several factors may influence the generalization and ethical implications of the proposed system. One important limitation is the potential bias in the ISIC dataset, which, despite being a large and multi-institutional benchmark, contains limited demographic and skin-type diversity25. Most images originate from lighter-skinned populations and specialized dermatology centers in high-income regions26. Although metadata such as age, sex, and anatomical site are available, information on ethnicity or Fitzpatrick skin phototype is not consistently included Footnote 3. This restriction could impact the system’s performance on underrepresented groups and should be addressed in future work by incorporating datasets with broader demographic coverage and diverse skin tones.
From an ethical and scalability perspective, the FL framework provides intrinsic advantages by preserving privacy data remains anonymized and local to each participating institution, and all communications are encrypted during model synchronization. This structure aligns with data protection regulations such as GDPR and HIPAA and supports responsible AI deployment. However, large-scale clinical adoption still faces challenges related to heterogeneous hardware, asynchronous participation, communication overhead, and the need for robust, secure aggregation mechanisms at scale.
Final assessment
The overall findings confirm that FL constitutes a practical and effective framework for medical applications, enabling collaborative model development without compromising patient privacy. Despite being trained on smaller, decentralized datasets, the federated model achieved performance levels comparable to the centralized configuration and even surpassed it in key diagnostic aspects such as recall (0.7619 vs. 0.6667) and ROC AUC (0.9321 vs. 0.9251). These improvements suggest that FL can enhance the detection of melanoma cases in distributed clinical environments, which is especially relevant for early-stage screening where sensitivity is critical.
Beyond numerical performance, the study demonstrates that FL provides a viable balance between diagnostic accuracy and data protection. By maintaining local data ownership and employing encrypted communication, the approach aligns with privacy regulations (e.g., GDPR and HIPAA) while supporting scalable multi-institutional collaboration. This makes FL a promising pathway for the development of trustworthy AI systems in healthcare, capable of leveraging diverse sources of medical data without breaching confidentiality.
Conclusion
This research work presents an AI-based system for melanoma detection, integrating a convolutional neural network, an FL framework, and an intuitive web application for clinical use. The proposed model demonstrated strong performance in distinguishing between melanoma and non-melanoma lesions, with a particular emphasis on recall to support early detection in clinical screenings. Compared to the centralized model, the federated approach increased recall from 0.6667 to 0.7619 (+9.5%) and reduced false negatives from 7 to 5 cases, as reflected in the confusion matrices. This means the FL model correctly identified two additional melanoma cases per 200 screened images, providing greater sensitivity while maintaining balanced performance.
The implementation of FL proved to be a viable strategy for training robust AI models across decentralized datasets, achieving results comparable to centralized learning while preserving patient privacy. Although the federated model slightly reduced specificity (0.872 vs. 0.942) due to an increase in false positives, it achieved a slightly higher ROC AUC of 0.9321 compared to 0.9251 for the centralized model. This improvement indicates that the model preserved and even enhanced its overall discriminative capacity between malignant and non-malignant lesions, despite variations in operating thresholds. The web and API components further enhanced usability and adaptability, providing healthcare professionals with secure access to diagnostic tools.
Future work will focus on expanding the dataset to include more diverse skin tones and demographic groups, and on integrating multimodal data such as dermoscopic and clinical images, patient history, and genetic risk factors to enhance diagnostic precision. In addition, prospective clinical trials will be conducted to validate the system’s real-world performance, usability, and its impact on early melanoma detection and workflow efficiency in dermatology practice. Furthermore will also focus on analyzing possible security vulnerabilities in the FL framework and implementing mechanisms to enhance data protection during model aggregation.
Data availibility
The source code utilized and analyzed in our research is publicly accessible at the DOI 10.5281/zenodo.15805545. The leveraged dataset from ISIC is specifically ISIC Challenge Datasets 2018, which is also available at https://challenge.isic-archive.com/data/#2018. The dataset was acquired as-is, leveraging the existing annotations from the dataset’s metadata. The dataset’s quality was analyzed considering the balance among the MEL and No-MEL classes, and the system was designed with this imbalance in consideration.
References
for Research on Cancer (IARC), I. A. Global Cancer Observatory: Cancer Today, Melanoma of Skin, 2022. https://gco.iarc.fr/today (2022). Accessed: October 2025.
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017).
Haenssle, H. A. et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 29, 1836–1842 (2018).
Oyeniyi, J. & Oluwaseyi, P. Emerging trends in ai-powered medical imaging: enhancing diagnostic accuracy and treatment decisions. International Journal of Enhanced Research In Science Technology & Engineering 13, 81–94 (2024).
Talpur, M. S. H. et al. Illuminating healthcare management: A comprehensive review of iot-enabled chronic disease monitoring. IEEE Access 12, 48189–48209 (2024).
Ghadi, Y. Y. et al. Enhancing patient healthcare with mobile edge computing and 5g: challenges and solutions for secure online health tools. J. Cloud Comput. 13, 93 (2024).
Litjens, G. et al. A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017).
Kaissis, G. A., Makowski, M. R., Rückert, D. & Braren, R. F. Secure, privacy-preserving and federated machine learning in medical imaging. Nat. Mach. Intell. 2, 305–311 (2020).
Flores-Martin, D., Laso, S., Berrocal, J. & Murillo, J. M. Towards digital health: Integrating federated learning and crowdsensing through the contigo app. SoftwareX 28, 101885 (2024).
Sandhu, S. S., Gorji, H. T., Tavakolian, P., Tavakolian, K. & Akhbardeh, A. Medical imaging applications of federated learning. Diagnostics 13, 3140 (2023).
Haggenmüller, S. et al. Federated learning for decentralized artificial intelligence in melanoma diagnostics. JAMA Dermatol. 160, 303–311 (2024).
Xing, H. et al. Achieving flexible fairness metrics in federated medical imaging. Nat. Commun. 16, 3342 (2025).
Haq, I. et al. Lung nodules localization and report analysis from computerized tomography (ct) scan using a novel machine learning approach. Appl. Sci. 12, 12614 (2022).
Saqib, S. M. et al. Cataract and glaucoma detection based on transfer learning using mobilenet. Heliyon 10, 17 (2024).
Asif, R. N. et al. Brain tumor detection empowered with ensemble deep learning approaches from mri scan images. Sci. Rep. 15, 15002 (2025).
Khan, M. A. et al. Automatic melanoma and non-melanoma skin cancer diagnosis using advanced adaptive fine-tuned convolution neural networks. Discov. Oncol. 16, 645 (2025).
ISIC Challenge. Isic challenge datasets (2024). Retrieved July 30, 2024.
Ain, Qu. et al. Privacy-aware collaborative learning for skin cancer prediction. Diagnostics 13, 2264 (2023).
Yaqoob, M. M. et al. Symmetry in privacy-based healthcare: A review of skin cancer detection and classification using federated learning. Symmetry 15, 1369 (2023).
Yaqoob, M. M. et al. Federated machine learning for skin lesion diagnosis: An asynchronous and weighted approach. Diagnostics 13, 1964 (2023).
Rieke, N. et al. The future of digital health with federated learning. npj Digit. Med. 3, 119 (2020).
Tosi, S. Matplotlib for Python developers (Packt Publishing Ltd, 2009).
Laso, S., Herrera, J. L. & Flores-Martin, D. Code: Medical support platform for melanoma analysis and detection based on federated learning, https://doi.org/10.5281/zenodo.15805545 (2025).
Ain, Qu. et al. Privacy-aware collaborative learning for skin cancer prediction. Diagnostics 13, 2264 (2023).
Alipour, N., Burke, T. & Courtney, J. Skin type diversity in skin lesion datasets: a review. Curr. Dermatol. Rep. 13, 198–210 (2024).
Bissoto, A., Fornaciali, M., Valle, E. & Avila, S. (de) constructing bias on skin lesion datasets. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, 0–0 (2019).
Funding
This work was partially funded by the grant DIN2020-011586 funded by MICIU/AEI/10.13039/50100011033 and the “European Union NextGenerationEU/PRTR”, by the Department of Education, Science and Professional Training of the Government of Extremadura (GR24099), and by the European Regional Development Fund. The European Union also funds this under the MSCA project RENOS (contract number 101205037). Views and opinions expressed are however, those of the authors only and do not necessarily reflect those of the European Union or the European Research Executive Agency. Neither the European Union nor the granting authority can be held responsible for them.
Author information
Authors and Affiliations
Contributions
Sergio Laso conceived the framework; Juan Luis Herrera and Daniel Flores-Martin conducted the experiments. Sergio Laso, Juan Luis Herrera, and Daniel Flores-Martin analyzed the results. All authors contributed equally to this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Laso, S., Herrera, J.L. & Flores-Martin, D. Medical support platform for melanoma analysis and detection based on federated learning. Sci Rep 16, 2571 (2026). https://doi.org/10.1038/s41598-025-32453-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-32453-5