Abstract
Advanced additive manufacturing capabilities have enabled a transformational ability to create sophisticated cellular structures using diverse materials. By altering the topology of the unit cell, the mechanical behavior, such as the stress-strain response during compression, can be modulated. Nevertheless, identifying a printable topology within an enormous design space that would precisely deliver the targeted nonlinear material response is challenging. We propose a data-driven generative framework based on a conditional variational autoencoder (cVAE) architecture that can inverse design the cellular structure based on the intended nonlinear stress-strain response. Trained on a dataset of structure-property pairs, the cVAE learns a compact and expressive latent space that enables efficient mapping from targets to feasible geometries. Two inference modes are explored: (1) decoder-only generation, which enables the exploration of diverse designs conditioned solely on the desired mechanical response, and (2) encoder-decoder generation, which further allows for the incorporation of desired topologies, ensuring the generated structure conforms to both mechanical properties and to desired-topology constraints. The results demonstrate that the model can generate structurally plausible and mechanically accurate designs, with the predicted stress-strain curves closely matching the targets. Even under joint conditioning, the model effectively balances geometric fidelity and functional performance.
Similar content being viewed by others
Introduction
The emergence of advanced additive manufacturing technologies has enabled the 3D printing of complex cellular structures using a wide range of materials, including polymers1, metals2, and ceramics3. Cellular structures (or materials) are often used for light-weighing and impact attenuation in the automotive, defense, and aerospace industries4,5,6. These cellular structures typically consist of a periodic arrangement of millimeter-scale building block called a cell, which has substructures that locally experience complex deformation mechanisms such as bending, buckling, and shearing during global compression. With an ability to tailor the topology, these structures can potentially exhibit a combination of energy absorption capability and stiffness-to-weight ratio unrealizable by the bulk material counterpart7,8,9.
The mechanical behavior (e.g., the stress-strain response) of the cellular structures can be effectively predicted through numerical simulation at the mesoscale, i.e. resolving the individual strut and the pore spaces, using non-linear finite element (FE) analysis10,11,12,13. However, addressing the inverse problem, i.e., identifying the topology for a desired mechanical stress-strain response, poses a significant computational challenge. Traditional approaches to inverse design, such as topology optimization14,15, parametric sweeps16, and heuristic-based algorithms17, have been studied extensively; however, they predominantly focus on the linear properties such as Young’s modulus and Poisson’s ratio. The adoption of these methods to nonlinear stress-strain response is either analytically and computationally challenging because these methods suffer from high computational costs and limited scalability.
In recent years, data-driven models and deep generative frameworks have emerged as a powerful alternative, offering the ability to learn complex structure-property relationships, enabling rapid realization of the candidate designs18,19. Generative AI models have shown immense potential in solving inverse design problems across a variety of domains, including image synthesis20,21, material science22,23, drug discovery24, and structural designs25. Among these, variational autoencoders (VAEs)26, Generative Adversarial Networks (GANs)27, and Denoising Diffusion Probabilistic Models (DDPMs)28 have been widely used. These models learn to map complex, high-dimensional design spaces to lower-dimensional latent representations, from which new, plausible data samples–such as mesostructural topologies–can be generated. When extended to conditional variants29,30,31, these models allow for the targeted generation of outputs based on specified conditions such as mechanical properties or topology, making them highly relevant for performance-driven materials design. In the context of cellular or architected materials, several recent works have demonstrated the use of generative models for inverse design of metamaterials. Comprehensive reviews by Lee et al.32 and Zheng et al.33 provide an overview of this growing field. Some notable contributions include VAE34,35 and GAN36,37,38, DDPM-based models39,40 for inverse design of architected materials. These studies demonstrate the growing maturity of generative AI-based inverse design models, especially for 2D or 3D structures.
In this study, we implemented a cVAE model as the generative tool to inverse design the cellular 2D mesostructures from nonlinear stress-strain curves. While we acknowledge the effectiveness of ML models in previous studies, our approach offers several complementary and novel features that distinguish it from models in the existing literature. First, our training dataset comprises over several thousand diverse cellular structures generated using a cellular automata–based process, which produces a wider variety and more intricate set of topologies than conventional strut- or lattice-based approaches18,19,35,38,41. These structures exhibit complex pore arrangements, curved walls, and variable connectivity, thereby enriching the design space and enabling the model to learn a more expressive mapping between geometry and mechanical behavior. Second, unlike most existing studies that condition generative models on a limited set of scalar properties such as Young’s modulus, Poisson’s ratio, or stiffness tensors34,37,42, our framework performs inverse design based on the entire nonlinear stress–strain curve. This functional conditioning enables the generation of structures that not only satisfy elastic performance but also capture complex deformation characteristics such as yielding, buckling and densification commonly encountered in real-world applications. Third, we explored two inference capabilities: generating structures based solely on the stress–strain curves, and generating mesostructures based on both a desired shape and a target stress–strain curve. In the first mode, the model samples mesostructures conditioned solely on the mechanical response, allowing for rapid generation of multiple topologically diverse mesostructures that exhibit similar nonlinear behavior. In the second mode, the model incorporated both the target mechanical response and a reference mesostructure. This enabled morphologically biased inverse design, wherein the generated mesostructures not only matched the desired stress–strain behavior but also retained key geometric features of the reference mesostructure, which was often overlooked in the prior studies.
(a) Topologies of the unit cell structures generated using cellular automata. A total of 60,000 mesostructures were generated. (b) Finite element (FE) simulation of the generated cellular mesostructures under uniaxial compression. (c) Nominal stress-strain plots extracted from the FE simulation. (d) Principal component analysis (PCA) was applied to the stress-strain data, enabling reduction of dimensionality to 20 principal components.
Methods
To train the AI model, we first generated numerous unit-cell topologies using a method based on cellular automata. This method uses a random seed as input and generates structurally viable unit cells while adhering to any other user-provided constraints, such as material symmetry. A detailed explanation of the methodology to create unit cells of cellular structure can be found in our previous paper43. Here, we summarize the workflow for generating a database of mesostructures and the corresponding stress-strain for training the AI model (Fig. 1).
Material design
A total of 60,000 mesostructures were generated, each with a unit cell size of 5 mm \(\times\) 5 mm (50 \(\times\) 50 pixel grid), using Grasshopper (a plugin for Rhinoceros 7)44. Each pixel within the grid has a value of 1 or 0 corresponding to the presence or absence of the material. Grasshopper was used to 43 convert this binary, pixelated information representing a mesostructure’s unit cell to arrive at a mesh, consisting of nodes and elements for the FE simulation. The mesh utilized an element size of 0.1 mm. This mesh size was chosen based on the mesh convergence study detailed in Supplementary S1.
FE simulation
These generated structures were subjected to uniaxial compression simulations to acquire the stress-strain response. The simulations were performed using Abaqus/Explicit (version 2020)45, solving the elastodynamic governing differential equation. A previously calibrated hyperelastic and viscoelastic material properties for Elastic 50A from Formlabs12,43 was used. For hyperelasticity, we chose Mooney-Rivlin model46,47 for which the strain energy potential (U) is given by:
where \(C_{10}\), \(C_{01}\) and \(D_1\) are material constants, \({\bar{I}}_1\) and \({\bar{I}}_2\) are the first and the second invariants of the deviatoric component of the left Cauchy-Green deformation tensor, and J is the Jacobian of the deformation gradient tensor (second order). The values of material constants obtained from the calibration process were \(C_{10}=139019.19\) Pa and \(C_{01}=345441.15\) Pa at room temperature12. We assigned a Poisson’s ratio of 0.48, and consequently \(D_1 = 8.37\times 10^{-8}\) 1/Pa 12.
For viscoelasticity, we chose a two-term Prony series12,45. The time-dependent normalized shear modulus in the form of Prony series (\(g_R\)) is:
Here, \(g_i\) and \(\tau _i\) are normalized shear modulus and relaxation time for \(i^{th}\) term in the series. The calibrated parameters are \(g_1=3.19\times 10^{-2}\), \(\tau _1=20.4\) s, \(g_2=3.8\times 10^{-2}\), and \(\tau _2=3879.2\) s12.
The unit cell was placed between two rigid platens, as shown in Fig. 1(b). Plane strain conditions were assumed along the out-of-plane axis, with periodic boundary conditions (PBC) applied to the lateral walls to constrain the in-plane motion43. Displacements of the bottom platen were fixed in all degrees of freedom while a constant velocity of \(v_2 = -0.02\) m/s (strain rate = 4 s\(^{-1}\)) was applied to the top platen, which was allowed to displace to a final nominal strain of 50%. Frictional sliding was applied to all the nodes in contact with the top and the bottom platens. We used the ‘general contact’ algorithm45 that handles the self-contacts between walls of the cellular material and the contacts of the cellular material with the platens. The default ‘hard’ contact with the pressure overclosure relationship was used to define the normal behavior, and ‘penalty’ friction formulation with a friction coefficient of 0.7512 was used to define the tangential contact behavior between two surfaces. We restricted our simulation to the quasi-static strain rate regime wherein the inertial effect can be ignored. At the selected strain rate, the kinetic energy was less than \(1\%\) of the internal energy43, confirming the validity of the quasi-static approximation. The workflow, including mesostructure geometry creation, mesh generation, and the definition of constitutive material models and boundary conditions, was scripted in Grasshopper44, enabling high-throughput simulation of all 60,000 mesostructures.
The nominal stress was calculated from the total reaction force on the top platen at individual time points. Stresses at every 0.5% strain increment were extracted from the simulation, resulting in a 1D array of stress values with 100 data points. The nominal stress-strain data from the finite element simulations for all the mesostructures were reduced in dimensionality using the principal component analysis (PCA). PCA is a multivariate statistical method that identifies and extracts essential features for a lower dimensional representation. PCA attempts to retain the largest variation in the original data and is expressed in terms of uncorrelated variables called principal components48,49. In this study, only the first 20 principal components were chosen, so the dimensionality was reduced from 100 to 20 principal components to give a compact representation of the data while minimizing the variance loss. Consequently, the generative AI architecture was aimed at generating the topology of the cellular structure from the principal components. The following subsection provides a detailed explanation of the generative AI architecture, including its training process and application in inverse designing structures from the principal components of the stress-strain data.
Schematic of the conditional variational autoencoder (cVAE) architecture used in this work. The binary representation of the topologies (X) and the PCAs (Y) of the cellular structures serve as inputs to the encoder, which learns a latent distribution z. During training, the decoder combines the latent variables with the conditional inputs (Y) to reconstruct the unit cell topologies.
Conditional variational autoencoder (cVAE)
We implemented a conditional variational autoencoder (cVAE)29, which extends the standard VAE framework by incorporating conditional information to guide the encoder to obtain the latent space representation and the decoder to generate the reconstructed image from the latent space. The schematic of the cVAE architecture used for inverse-design is shown in Fig. 2. The cVAE contains the probabilistic encoder and the probabilistic decoder. The inputs to the encoder are the 50\(\times\)50 pixel images of the structures (X) and the corresponding 20\(\times\)1 array representing the PCA obtained from the stress-strain data (referred to as the condition, Y). The image input (X) passes through a series of convolution blocks consisting of convolution and pooling layers that extract features from the image while reducing the spatial information. For each convolution block, the sequence of operations was Conv2D-BN-Dropout-Conv2D-BN-Dropout, wherein the Conv2D is a 2D convolution layer with (3,3) kernel, ReLU activation, and same padding, BN is the batch normalization layer, and Dropout is the dropout layer with rate 0.1. The BN and Dropout layers were added to reduce overfitting50,51. A total of four hierarchical stages of convolution blocks were implemented with increasing filter size (32,64,96,128), each followed by an average pooling layer with a pool size of (2,2) to reduce the spatial resolution. A middle block with 256 filters was then applied to the final pooled feature maps, using a combination of 3\(\times\)3 and 1\(\times\)1 convolutions. Instead of flattening the spatial output directly, a global average pooling layer was used to reduce the spatial dimensions, resulting in a compact feature vector that summarizes the spatial information.
To integrate the conditional information (Y), the compact feature vector was batch-normalized and concatenated with the conditional information. This combined representation was passed through a dense layer to learn an intermediate feature embedding. Finally, two separate dense layers were used to produce the parameters of the latent Gaussian distribution: the mean (\(\mu\)) and the log of variance (\(\sigma ^2\)), each having 20 dimensions. A stochastic latent vector, z, was sampled from this distribution using the reparametrization technique26 to ensure end-to-end differentiability. The final encoder model outputs the mean, log-variance, and sampled latent vector, encapsulating both image content and condition in the latent representation.
The decoder in cVAE aims to reconstruct the input image from the latent representation, while conditioning the generation process with the condition Y. The decoder takes two inputs: the latent vector z and the condition Y. These two inputs were concatenated to form a unified feature vector, which was then passed through a dense layer with 1152 units and ReLU activation. This dense output was reshaped into a spatial tensor of the shape (3,3,128), serving as the initial feature map for the upsampling pathway. The upsampling process was implemented using a sequence of transposed convolution layers to gradually reconstruct the spatial dimensions of the image. At each upsampling stage, the number of filters was progressively decreased (128,96,64,32) in reverse order of the encoder. For each resolution level, two convolution layers with ReLU activation and same padding were used to enhance representational capacity. The final layer with shape (50,50,32) is passed through as Conv2D layer with a single filter and sigmoid activation function, producing the single 50\(\times\)50 pixel reconstructed image (X’).The sigmoid activation ensures that the output values are constrained to the range [0, 1], appropriate for grayscale image reconstruction.
After training, the encoder in cVAE learned to map both the input data X and the conditional information Y to a structured latent space by approximating the posterior distribution \({q_\phi }(z|X,Y)\), where z represents the latent variables and \(\phi\) represents neural network weight for the encoder. The decoder learned to reconstruct the samples based on the learned latent representation and the provided conditions Y by modeling the conditional likelihood \({p_\theta }(X|z,Y)\), where \(\theta\) represents neural network weight for the decoder. The training objective can be formulated as the minimization of the negative conditional evidence lower bound:
The first term represents the negative log likelihood between the reconstructed image and the input image; thus, minimizing the negative likelihood ensures that the generated samples closely align with the input data X. The second term is the Kullback-Liebler (KL) divergence52 which imposes a penalty for deviations of the learned latent distribution from the prior p(z). In our cVAE model, we assume the standard Gaussian distribution N(0, 1) as a prior i.e \(p(z) \sim N(0,1)\) (Equation 4), and use binary cross-entropy loss (\(L_r\), Eq. 5) as the reconstruction loss.
In the implementation, the KL term was averaged over the latent dimension. For the reconstruction loss in Equation 5, \(x_{mn} \in \{0,1\}\) represents the pixel value in the original configuration of the (X)\(_i\) cellular structure, \(p_{mn}\) represents pixel values in the reconstructed cellular structure (X’)\(_i\), and \(t=50\) is the number of pixels in the horizontal and vertical dimensions.
Inference phase of the cVAE wherein only the trained decoder is used for inference. The inputs to the decoder are the latent variable z randomly sampled from a standard Gaussian distribution N(0,1) and the PCA of the target stress-strain curves that serve as the condition. The trained decoder can generate diverse cellular topologies based on the sampled latent variable.
To allow flexible weighting between the reconstruction and regularization terms, we used a scalar coefficient \(\beta\), following the formulation of the \(\beta\)-VAE framework53. This provides control over the trade-off between reconstruction fidelity and latent space disentanglement, and prevents posterior collapse, a condition where the KL divergence terms vanish (\(D_{KL} \approx 0\)). In this work, we refrain from using the KL annealing technique54 and instead implemented a constant \(\beta\) of 0.1 for which we obtained acceptable reconstruction performance. The total loss is defined as the combination of the reconstruction loss and the weighted KL divergence. This combined objective is minimized using the Adam optimizer. The model is trained for a total of 200 epochs with variable batch sizes, namely 100, 20 and 80 epochs with 32, 64 and 128 batch sizes, respectively55. The evolution of the total loss and the reconstruction loss for the training and validation data are plotted in Fig. 3. The validation losses closely track the training losses, suggesting good generalization and the absence of significant overfitting.
Results and discussions
In this section, we evaluated the performance of trained cVAE framework in solving the inverse design problem for architected cellular mesostructures. The central objective was to determine whether the model can reliably generate structurally plausible topologies that correspond to a target stress-strain response. By leveraging a learned mapping between mechanical behavior and topology, the cVAE serves as a generative tool capable of synthesizing mesostructures without the need for computationally expensive iterative optimization. We present two inference pathways for the cVAE, corresponding to different use cases of the trained model during the generation and analysis of the cellular topologies.
Decoder-only inference
In the first case (refer to Fig. 4), only the decoder of the trained cVAE is used for inference. Two inputs are fed to the decoder: (1) a latent vector z, randomly sampled from a multivariate standard Gaussian distribution N(0, 1), and (2) a condition vector, which is the PCA representation of the desired stress-strain curve. The inset scatter plot demonstrating the distribution of the first two latent variables \(z_1\) and \(z_2\) obtained for the training datasets are shown in the figure along with four randomly sampled z values. By varying the sampled z, the decoder can generate a diverse set of plausible cellular topologies that have the stress-strain response similar to the target condition. This approach demonstrates the model’s ability to explore the design space and produce multiple feasible solutions for a single performance specification.
Generated cellular structures for varying values of \(z_1\) and \(z_2\), with all other latent variables set to zero. The corresponding stress-strain data of the generated structures are shown, as well. Within the stress-strain plot, the horizontal axis represents the nominal strain, and the vertical axis represents normalized stress.
Latent space traversal for a target stress-strain curve
We tested the trained ML in its ability to generate cellular structures by feeding the decoder with the PCA of the target stress-strain curve and the latent variable z. For representation, we systematically chose \(z_1\) and \(z_2\) from {-2,-1,0,1,2} while fixing the remaining z to zero. The generated cellular structures are shown in Fig. 5 along with the PCA and stress-strain data. For each combination of \(z_1\) and \(z_2\), the decoder generates the cellular structure, conditioned with the same PCA of the target stress-strain data. We observed a smooth and continuous morphological transition across the grid (\(z_1\), \(z_2\)). Variation along the \(z_1\) axis (left to right) tends to modify the shape of the structure, resulting in relatively more overall transformation at the unit-cell level affecting the distribution of material. As \(z_1\) increases, the connectivity of the internal elements evolves noticeably, with new voids appearing, and the overall structure becomes increasingly intricate. In contrast, variation along \(z_2\) (top to bottom) seems to preserve the overall shape of the structure, while modulating relatively more localized geometrical attributes.
For all the AI-generated structures, the stress-strain responses were computed using the previously developed ML predictor43, which mapped the mesostructure geometry to the stress-strain response. These predicted responses were compared with the target stress-strain curve that was used as conditional input during generation. Across the generated samples, the resulting stress-strain curves exhibited good agreement with the target curves, indicating that the model effectively learned to produce multiple alternative structures that satisfied the desired mechanical response.
Conditional generation of mesostructures from functional targets
We further tested the trained decoder with widely varying targets (stress-strain curves). We used decoder-only inference (refer to Section 3.1) wherein the inputs to the decoder were the PCA of the target stress-strain curves and the latent variables sampled randomly from the standard Gaussian distribution. The generated structures are shown in Fig. 6 along with the PCA and stress-strain curves. These structures exhibit distinct morphological features, ranging from sparse and flexible to dense and stiffer configurations. This morphological diversity aligns with the differing target stress-strain data, indicating that the AI model effectively learns the relationship between the structural form and target mechanical function. For example, structures with greater compliant stress-strain response appear to have lower relative density and thinner interconnected struts, while those matching stiffer targets show bulkier geometries with higher material reinforcement.
While the majority of generated structures’ stress-strain response closely matched their target values, some cases exhibited deviations, particularly in the high strain regime (for example, second row, third column of Fig. 6). These discrepancies may stem from the model’s finite capacity or from regions of the design space that are underrepresented in the training data. They indeed point to the opportunities for improvements in the AI model.
In the majority of the cases, the model generated satisfactory geometries. These structures exhibit well-defined connectivity and consistent feature resolution, making them viable candidates for practical realization. However, some exceptions were observed (for example, third row, fourth column) wherein the generated structures were poorly connected and had structural discontinuities. Such a scenario might require further exploration of the latent space to navigate toward generating more feasible and robust designs. Alternatively, one may impose additional criterion to exclude such structures.
Despite some challenges, the cVAE demonstrated strong performance in generating mesostructures that are both structurally meaningful and mechanically consistent with a wide range of target stress-strain responses. The model effectively captured the complex relationship between geometry and mechanical behavior, enabling inverse design across a diverse design space. Even in cases where some deviations were observed, the generated structures retained the overall characteristics of the targets, underscoring the robustness and generalization capacity of the learned latent representation.
Generated cellular structures for different target stress-strain response. The corresponding principal component analysis (PCA) representations and stress-strain data of the generated structures are compared with the target values. In stress-strain data, the horizontal axis represents the nominal strain, and the vertical axis represents normalized stress.
We also generated the structures beyond the training data domain by providing input stress–strain curves that fall outside the range of the stress-strain curves in the training data (refer to Fig. 7). The generated structures have volume fractions significantly different from those present in the training set (training data: 0.31–0.65, generated cases: 0.25 and 0.75). The model maintains reliable performance across unseen property and structure spaces, underscoring its effectiveness for inverse design beyond the training distribution.
Cellular Structure generation beyond the training data. The model generates cellular structures (left) from target stress–strain curves (right) that lie outside the range of the training dataset. The generated structures exhibit volume fractions (VF) of 0.75 (top) and 0.25 (bottom), which are notably higher and lower, respectively, than the training data range (0.31–0.65).
Large-scale inference and performance evaluation
Figure 8 illustrates representative examples and statistical evaluation of decoder-generated structures from a pool of 3000 samples of stress-strain data. These stress-strain data were generated from 3000 cellular structures from the test data that were not used for training the AI model. The top, middle, and bottom rows show structures corresponding to the lowest, average, and highest root mean squared error (RMSE) values, respectively, based on the deviation between predicted and target stress–strain curves. The lowest-RMSE examples exhibit excellent agreement with the target, indicating high-fidelity reconstruction. In contrast, the highest-RMSE cases reveal significant deviation, highlighting limitations in decoder generalization for certain regions of the latent space. The rightmost panel presents the probability distribution of RMSE across all samples, revealing a positively skewed trend where the majority of predictions yield low error, while a smaller subset contributes to the long tail of higher RMSE values. This analysis underscores the decoder’s general capability to reproduce mechanical responses with reasonable accuracy, while also quantifying prediction uncertainty across a large sample space.
When evaluating the mechanical behavior of cellular structures, stress–strain data are frequently utilized to derive quantitative measures that assess the mesostructures’ ability to absorb energy and endure high stress conditions. In the context of energy-absorbing applications, the mesoscale design of a unit cell is often engineered to maximize energy dissipation while minimizing transmitted peak stress, thereby protecting internal components from damage. To assess the generation capabilities of the AI model, Fig. 9 presents scatter plots comparing the Target vs AI generated structures values of two key mechanical metrics: energy density (ED) and peak stress (PS), each evaluated at a nominal strain of 0.5. These values are computed by analyzing the stress–strain response–specifically, ED is determined as the area under the curve up to 0.5 nominal strain, while PS corresponds to the maximum stress reached at the same strain level. The results demonstrate reasonable agreement between AI generated designs and target values, with coefficient of determination (R\(^2\)) scores of 0.83 for ED and 0.73 for PS.
Representative AI-generated structures selected from a set of 3,000 samples are shown. The first, second, and third rows correspond to the structures with the lowest, average, and highest root mean squared error (RMSE), respectively. The rightmost plot displays the probability distribution of RMSE values across the entire sample set.
Encoder and decoder for inference
In the second case, both the encoder and decoder were used for inference (refer to Fig. 10. This mode is particularly useful for inverse design or reconstruction tasks, where the goal is to generate a cellular structure that closely matches a specific target topology. The encoder processes the input image alongside the associated condition (the PCA of a target stress-strain curve) and encodes this information into a latent representation z. This latent vector captures the complex spatial and conditional relationships necessary to reconstruct the structure. The decoder then takes this latent representation and the same condition vector as input to regenerate the corresponding cellular topology. This dual-module inference process ensures that the reconstructed output is both closer to the input target shape and consistent with the desired mechanical behavior, offering a powerful approach for data-driven structure-property mapping and interpretation.
Performance Evaluation
The test data with 3000 structure and property (i.e. the stress-strain curves) pairs were also used to evaluate the performance of the cVAE model in the dual inference mode. Figure 11 shows the representative samples with the lowest, the average and the highest RMSE determined from the stress-strain curves. The generated samples are compared with the corresponding test data to assess the model’s reconstruction capability. Given the structure-property inputs, the model is able to effectively reconstruct the original topologies. However, certain high-level geometric features are partially lost during reconstruction as these reconstructions are created from the reduced latent space43,56. The averaged structural similarity index (SSIM)57, DICE similarity coefficient (DSC)13,58, and normalized cross-correlation (NCC)59 determined based on 3000 data are 0.70, 0.88, and 0.83, respectively. SSIM measures the perceived structural resemblance between the generated and target images by comparing luminance, contrast, and texture. DICE quantifies the spatial overlap between two structures, indicating how well the generated topology matches the target one. NCC evaluates the linear correlation of intensity patterns between the generated and target images, reflecting overall similarity in spatial distribution. The relatively high DSC and NCC values confirm strong spatial and statistical agreement between the generated and target structures, whereas the moderately lower SSIM arises primarily from the inherent smoothness of cVAE-based reconstructions, which tend to blur fine edges and local textures.
Evaluation of cVAE using test data. The 3000 structure-property pairs in the test data were fed to the trained cVAE to evaluate the reconstruction ability and the performance of the model. The first, second and third rows correspond to the structures with lowest, average and the highest root mean squared error (RMSE), respectively. The rightmost plot displays the probability distribution of RMSE values accross the entire sample set.
Mesostructure realization with desired topology and stress-strain response
Some applications of architected cellular materials— such as mechanical metamaterials, soft robotics, and biomedical implants— require mesostructures that not only exhibit specific mechanical response but also conform to pre-defined topological features. Achieving both structural fidelity and functional performance simultaneously is a non-trivial inverse design challenge. It turns out that the trained cVAE in encoder-decoder inference mode can generate mesostructures with not only the target property but also with the desired topology.
We tested the trained cVAE model by feeding the four desired topologies, each with four target stress-strain curves. The AI-generated structures resulting from the joint shape and property conditioning are presented in Fig. 12. The leftmost column presents some widely popular topologies, namely, square-shaped, X-shaped, honeycomb, and circular void lattice. These structures serve as a reference topology for the generated designs and may represent functional motifs (e.g., symmetry, connectivity, patterns, and voids) necessary for manufacturability, aesthetic integration, or domain-specific requirements. Across the generated samples, the outputs preserve key visual and structural features of the desired topologies while introducing subtle geometric modifications necessary to align with the target stress-strain curves. These modifications include adjustments in the strut thickness and connectivity.
This approach demonstrates a critical advancement in inverse design: the ability to synthesize mesoarchitectures that satisfy both geometric and mechanical specifications. By leveraging the flexibility of deep generative models, the proposed framework enables multi-constraint design automation, significantly broadening the practical utility of data-driven materials design in complex engineering domains.
Generated cellular structures for different target stress-strain response and target shape. The corresponding stress-strain data of the generated structures are compared with the target values. The encoder is used to predict the latent variable for the target shape, which is then fed to the decoder along with the condition to generate the cellular structures.
Printability of AI-generated designs
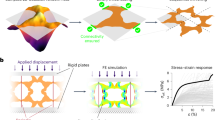
Figure 13 shows the physical realization and mechanical validation of a mesostructure generated using cVAE framework. The left images show the AI-generated topology, designed to match the user-specified nonlinear stress-strain response. The out-of-plane dimension was set to 15 mm. This structure was subsequently printed using Stereolithography (SLA), a vat photopolymerization method12, using a Formlabs Form3+ (Formlabs, Somerville, MA, USA) printer. Elastic 50A, a soft and highly deformable material, was used as the printing resin. The Layer thickness was set to 0.1 mm, and the samples were printed without any support structures. The center photographs show the snapshot of the printed sample. This sample was subjected to uniaxial compression in Instron 3343 (Instron Bluehil, Norwood, MA, USA) with a 1 kN load cell, under a strain rate of 0.2 mm/s. The tests were performed up to a maximum stress of 0.5 MPa. The nominal stress was computed from the load cell force divided by the initial macroscopic cross-sectional area of \(3\times 3\times 3\) specimen: \(\sigma _{exp}=F_{exp}/A_0\) with \(A_0\approx\) 15 mm \(\times\) 15 mm. The comparison between the stress-strain responses of the printed sample and the target stress-strain data is shown in the right plot. Uniaxial compression tests (n = 3) were conducted to assess the reproducibility of the mechanical response. Figure 13 shows the mean stress–strain curve (black line) with gray shaded regions indicating the standard deviation (SD) at each strain value across the samples. The range of SD corresponds to ±0.05 for Fig. 13a and ±0.025 for Fig. 13b, illustrating the experimental variability in the measured responses.
The printed structure satisfactorily exhibited a comparable mechanical response to the target stress-strain response, notwithstanding some deviations particularly in the mid (sample-a) and high-strain regimes (sample-b). We encountered a post-processing issue associated with printing, specifically washing the resin off the printed specimen, the resulting outcome of this challenge is particularly noticeable by comparing the differences in printed geometry with STL sample on the left for Fig. 13-b. These discrepancies may be due to limitations in the resolution of the printer and error of the ML model. Despite these differences, the generated design produced a physically viable structure that successfully exhibited nonlinear behavior quantitatively comparable to the target. This result underscores the immense potential of the cVAE framework for producing manufacturable designs, while also highlighting the need for further refinement in bridging the gap between inverse designed STL and the printed specimen.
Printability of AI-generated design. (Left) AI-generated mesostructures. (Center) 3D-Printing mesostructures. (Right) The stress-strain responses of the printed samples in comparison with the target stress-strain data. For each case, three samples were printed and tested (n=3). The black-dashed curves represented the mean and the dark shaded region represents the range of the standard deviation (SD).
Limitations and future work
In this study, 60,000 finite element (FE) simulations were performed using a \(1\times 1\) unit cell to ensure high-throughput simulations. With access to greater computational resources, this analysis could be extended to larger domains involving multiple unit cells. For instance, training an ML surrogate on a \(3\times 3\) cellular structure may capture a broader range of mechanical instabilities arising from the interactions between adjacent deforming cells–precluded in single-unit-cell simulations. The current FE simulations employed periodic boundary conditions for the \(1\times 1\) structure, chosen to approximate a uniaxial strain response. However, such a configuration may not fully represent the global buckling modes typically observed in larger sizes (e.g., \(12\times 12\times 12\) simple cubic cellular structure), demonstrated in our previous work12.
While the cVAE was effective for rapid inverse design and exploration of the design space, it also inherits several well-known limitations of variational autoencoders. VAEs tend to produce slightly blurred or over-smoothed geometries and can occasionally generate topologies with broken connectivity or narrow struts that are not physically realizable. A few generated samples in this work exhibited such discontinuities, particularly in under-represented regions of the training space. Future work will address these issues by introducing topology-aware loss functions, structural validity filters, and manufacturability constraints that penalize disconnected or impractical features. Recent advances in generative modeling have shown that denoising diffusion models can outperform VAEs in terms of sample quality and physical realism. Consequently, denoising diffusion models can serve as a replacement for, or a refinement stage to, VAEs–albeit at the expense of larger training datasets and increased inference time.
The framework for generating unit cells with cellular automata can be extended into three dimensions. Additional rules can be introduced to enforce geometric constraints that ensure printability–an important consideration for a 3D inverse design framework.
Conclusions
We implemented a generative AI model applied for inverse designing 2D cellular mesostructures for a target nonlinear stress-strain curve. We applied a conditional variational autoencoder (cVAE) for the realization of multiple cellular structure designs given the desired mechanical response. By learning the complex, nonlinear inverse mapping between stress-strain data and the desired topology, the cVAE model enabled efficient and flexible expression of mesostructures that satisfactorily conform to the intended mechanical response for the dataset considered. Our approach contributes towards addressing a key challenge in architected material design: the inverse problem of generating printable topologies that not only achieves the targeted mechanical properties but also retains the desired topology when constraints are imposed.
Through both decoder-only and full encoder-decoder inference modes, we demonstrated the potential to generate diverse, physically realizable topologies. For the simulated 2D structures considered, the decoder-only approach enabled exploration of the design space through conditional sampling of latent variables informed by mechanical performance, while the encoder–decoder configuration allowed prediction of structures consistent with both targeted mechanical responses and imposed topological constraints. The fidelity of the generated structures to the target stress–strain responses was validated across a wide range through direct comparison of the predicted and reference stress–strain data, both in the original and principal component representations.
Moreover, the framework was able to handle dual conditioning scenarios in our examples, allowing for simultaneous satisfaction of geometric and mechanical criteria–a key requirement for many real-world applications in metamaterials, additive manufacturing, and bio-inspired design. While some deviations and impractical topologies were observed in isolated cases, these highlight future avenues for further refinement, using methods such as integrating physics-informed priors, attention layers in the model architecture, higher-resolution data, or post-generation optimization.
Overall, this study used the cVAE as a tool for inverse material design of printable specimens, demonstrating the feasibility of automated, data-driven generation of printable mesostructures tailored to functional and topology requirements. Future work may extend this framework to 3D structures, multi-objective design (e.g., thermal, acoustic properties), and implementing generative feedback from printability studies.
Data availability
Data generated in this manuscript will be made available conforming to the policies of the Los Alamos National Laboratory and upon reasonable request to point of contact Sushan Nakarmi for accessing the data from the study.
References
Bezek, L. B. et al. Effect of part size, displacement rate, and aging on compressive properties of elastomeric parts of different unit cell topologies formed by vat photopolymerization additive manufacturing. Polymers 16, 3166 (2024).
Yang, L. et al. Additive manufacturing of metal cellular structures: design and fabrication. Jom 67, 608–615 (2015).
Lin, H. et al. 3d printing of porous ceramics for enhanced thermal insulation properties. Adv. Sci. 12, 2412554 (2025).
Schaedler, T. A. et al. Designing metallic microlattices for energy absorber applications. Adv. Eng. Mater. 16, 276–283 (2014).
Schaedler, T. A. & Carter, W. B. Architected cellular materials. Annual Rev. Mater. Res. 46, 187–210 (2016).
Boursier Niutta, C., Ciardiello, R. & Tridello, A. Experimental and numerical investigation of a lattice structure for energy absorption: application to the design of an automotive crash absorber. Polymers 14, 1116 (2022).
Mohsenizadeh, M., Gasbarri, F., Munther, M., Beheshti, A. & Davami, K. Additively-manufactured lightweight metamaterials for energy absorption. Mater. Des. 139, 521–530 (2018).
Uribe-Lam, E., Treviño-Quintanilla, C. D., Cuan-Urquizo, E. & Olvera-Silva, O. Use of additive manufacturing for the fabrication of cellular and lattice materials: a review. Mater. Manuf. Process. 36, 257–280 (2021).
Mueller, J., Raney, J. R., Shea, K. & Lewis, J. A. Architected lattices with high stiffness and toughness via multicore-shell 3d printing. Adv.Mater. 30, 1705001 (2018).
Lei, H. et al. Evaluation of compressive properties of slm-fabricated multi-layer lattice structures by experimental test and \(\mu\)-ct-based finite element analysis. Materi. Des. 169, 107685 (2019).
Kumar, A., Collini, L., Daurel, A. & Jeng, J.-Y. Design and additive manufacturing of closed cells from supportless lattice structure. Additive Manuf. 33, 101168 (2020).
Nakarmi, S. et al. The role of unit cell topology in modulating the compaction response of additively manufactured cellular materials using simulations and validation experiments. Model. Simul. Mater. Sci. Eng. 32, 055029 (2024).
Nakarmi, S. et al. Mesoscale simulations and validation experiments of polymer foam compaction-volume density effects. Mater. Lett. 382, 137864 (2025).
Xia, L. & Breitkopf, P. Design of materials using topology optimization and energy-based homogenization approach in matlab. Struct. Multidisciplinary Optim. 52, 1229–1241 (2015).
Radman, A., Huang, X. & Xie, Y. Topology optimization of functionally graded cellular materials. J. Mater. Sci. 48, 1503–1510 (2013).
Bauer, J., Hengsbach, S., Tesari, I., Schwaiger, R. & Kraft, O. High-strength cellular ceramic composites with 3d microarchitecture. Procd. National Acad. Sci. 111, 2453–2458 (2014).
Nguyen, J., Park, S.-I. & Rosen, D. Heuristic optimization method for cellular structure design of light weight components. Int. J. Precision Eng. Manuf. 14, 1071–1078 (2013).
Meier, T. et al. Obtaining auxetic and isotropic metamaterials in counterintuitive design spaces: an automated optimization approach and experimental characterization. npj Comput. Mater. 10, 3 (2024).
Vangelatos, Z. et al. Strength through defects: A novel bayesian approach for the optimization of architected materials. Sci. Adv. 7, eabk2218 (2021).
Ramesh, A. et al. Zero-shot text-to-image generation. In International conference on machine learning, 8821–8831 (Pmlr, 2021).
Ramesh, A., Dhariwal, P., Nichol, A., Chu, C. & Chen, M. Hierarchical text-conditional image generation with clip latents. arXiv preprint arXiv:2204.061251, 3 (2022).
Yao, Z. et al. Inverse design of nanoporous crystalline reticular materials with deep generative models. Nat. Mach. Intell. 3, 76–86 (2021).
Sanchez-Lengeling, B. & Aspuru-Guzik, A. Inverse molecular design using machine learning: Generative models for matter engineering. Science 361, 360–365 (2018).
Zhavoronkov, A. et al. Deep learning enables rapid identification of potent ddr1 kinase inhibitors. Nat. Biotechnol. 37, 1038–1040 (2019).
Liao, W., Lu, X., Fei, Y., Gu, Y. & Huang, Y. Generative ai design for building structures. Autom. Construct. 157, 105187 (2024).
Kingma, D. P., Welling, M. et al. Auto-encoding variational bayes (2013).
Goodfellow, I. J. et al. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 27 (2014).
Ho, J., Jain, A. & Abbeel, P. Denoising diffusion probabilistic models. Adv. Neural Inf. Process. Syst. 33, 6840–6851 (2020).
Sohn, K., Lee, H. & Yan, X. Learning structured output representation using deep conditional generative models. Adv. Neural Inf. Process. Syst. 28 (2015).
Mirza, M. & Osindero, S. Conditional generative adversarial nets. arXiv preprint arXiv:1411.1784 (2014).
Dhariwal, P. & Nichol, A. Diffusion models beat gans on image synthesis. Adv. Neural Inf. Process. Syst. 34, 8780–8794 (2021).
Lee, D., Chen, W., Wang, L., Chan, Y.-C. & Chen, W. Data-driven design for metamaterials and multiscale systems: a review. Adv. Mater. 36, 2305254 (2024).
Zheng, X., Zhang, X., Chen, T.-T. & Watanabe, I. Deep learning in mechanical metamaterials: from prediction and generation to inverse design. Adv. Mater. 35, 2302530 (2023).
Wang, L. et al. Deep generative modeling for mechanistic-based learning and design of metamaterial systems. Comput. Methods App. Mech. Eng. 372, 113377 (2020).
Zheng, L., Karapiperis, K., Kumar, S. & Kochmann, D. M. Unifying the design space and optimizing linear and nonlinear truss metamaterials by generative modeling. Nat. Commun. 14, 7563 (2023).
Tian, J., Tang, K., Chen, X. & Wang, X. Machine learning-based prediction and inverse design of 2d metamaterial structures with tunable deformation-dependent poisson’s ratio. Nanoscale 14, 12677–12691 (2022).
Zheng, X., Chen, T.-T., Guo, X., Samitsu, S. & Watanabe, I. Controllable inverse design of auxetic metamaterials using deep learning. Mater. Des. 211, 110178 (2021).
Challapalli, A., Patel, D. & Li, G. Inverse machine learning framework for optimizing lightweight metamaterials. Materi. Des. 208, 109937 (2021).
Vlassis, N. N. & Sun, W. Denoising diffusion algorithm for inverse design of microstructures with fine-tuned nonlinear material properties. Comput. Methods Appl. Mech. Eng. 413, 116126 (2023).
Bastek, J.-H. & Kochmann, D. M. Inverse design of nonlinear mechanical metamaterials via video denoising diffusion models. Nat. Mach. Intell. 5, 1466–1475 (2023).
Meier, T. et al. Scalable phononic metamaterials: Tunable bandgap design and multi-scale experimental validation. Mater. Des. 252, 113778 (2025).
Kumar, S., Tan, S., Zheng, L. & Kochmann, D. M. Inverse-designed spinodoid metamaterials. npj Comput. Mater. 6, 73 (2020).
Nakarmi, S., Leiding, J. A., Lee, K.-S. & Daphalapurkar, N. P. Predicting non-linear stress-strain response of mesostructured cellular materials using supervised autoencoder. Comput. Methods Appl. Mech. Eng. 432, 117372 (2024).
McNeel, R. et al. Grasshopper-algorithmic modeling for rhino. http://www.grasshopper3d.com (2013).
Dassault Systèmes. Abaqus Analysis User’s Manual, Version 2020 (2020).
Mooney, M. A theory of large elastic deformation. J. Appl. Phys. 11, 582–592 (1940).
Rivlin, R. Large elastic deformations of isotropic materials. i. fundamental concepts. Philosophical Trans. Royal Soc. London. Series A, Math. Phys. Sci. 240, 459–490 (1948).
Abdi, H. & Williams, L. J. Principal component analysis. Wiley Interdisciplinary Rev. Comput. Statist. 2, 433–459 (2010).
Yang, C., Kim, Y., Ryu, S. & Gu, G. X. Prediction of composite microstructure stress-strain curves using convolutional neural networks. Mater. Des. 189, 108509 (2020).
Ioffe, S. & Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Int. Conference Mach. Learn., 448–456 (pmlr, 2015).
Li, X., Chen, S., Hu, X. & Yang, J. Understanding the disharmony between dropout and batch normalization by variance shift. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2682–2690 (2019).
Kullback, S. & Leibler, R. A. On information and sufficiency. Annals Math. Statist. 22, 79–86 (1951).
Higgins, I. et al. Early visual concept learning with unsupervised deep learning. arXiv preprint arXiv:1606.05579 (2016).
Fu, H. et al. Cyclical annealing schedule: A simple approach to mitigating kl vanishing. arXiv preprint arXiv:1903.10145 (2019).
Smith, S. L., Kindermans, P.-J., Ying, C. & Le, Q. V. Don’t decay the learning rate, increase the batch size. arXiv preprint arXiv:1711.00489 (2017).
Liu, Y., Neophytou, A., Sengupta, S. & Sommerlade, E. Relighting images in the wild with a self-supervised siamese auto-encoder. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, 32–40 (2021).
Wang, Z., Bovik, A. C., Sheikh, H. R. & Simoncelli, E. P. Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 13, 600–612 (2004).
Dice, L. R. Measures of the amount of ecologic association between species. Ecology 26, 297–302 (1945).
Zhao, F., Huang, Q. & Gao, W. Image matching by normalized cross-correlation. In 2006 IEEE International Conference on Acoustics Speech and Signal Processing Proceedings, vol. 2, II–II (IEEE, 2006).
Funding
This work received support from the Dynamic Materials Properties (C2) Science Campaign and Advanced Scientific Computing–Physics of Engineered Materials programs. This work was supported by the U.S. Department of Energy through the Los Alamos National Laboratory. Los Alamos National Laboratory is operated by Triad National Security, LLC, for the National Nuclear Security Administration of the U.S. Department of Energy (Contract No. 89233218CNA000001).
Author information
Authors and Affiliations
Contributions
Sushan Nakarmi: Conceptualization; Software; Methodology; Formal analysis; Validation; Visualization; Writing—original draft; Writing—reviewing and editing; Data curation. Nitin Daphalapurkar: Conceptualization; Supervision; Methodology; Validation; Writing—original draft; Writing—reviewing and editing; Funding acquisition. Jeff Leiding: Funding acquisition; Project administration; Writing—reviewing and editing. Jihyeon Kim: Validation; Writing—reviewing and editing. Kwan-Soo Lee: Funding acquisition; Supervision; Writing—reviewing and editing. Dana Dattelbaum: Funding acquisition; Writing—reviewing and editing. Darby Luscher: Funding acquisition; Writing—reviewing and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nakarmi, S., Daphalapurkar, N.P., Lee, KS. et al. Inverse design of cellular structures with the targeted nonlinear mechanical response. Sci Rep 16, 3185 (2026). https://doi.org/10.1038/s41598-025-33184-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-33184-3