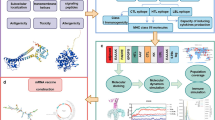
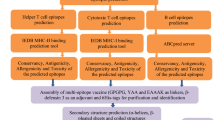
Abstract
Development of a strategy to combat Staphylococcus aureus is a high priority for the World Health Organization. B cell and helper T lymphocyte (HTL) epitopes of leukotoxin ED (LukED) were predicted using computational tools. The predicted epitopes were screened for conservancy, allergenicity, toxicity, autoreactivity, and population coverage. The immunogenic regions of LukED were linked together and to Human β-defensin 3 (hBD3) as adjuvant with appropriate linkers. The predicted 3D structure of the vaccine validated by molecular dynamics (MD) simulations. Subsequently, the 3D structure was docked with the Toll-like receptor (TLR)1/2 to evaluate the binding capacity of the adjuvant. Finally, MD simulation was employed to characterizing the conformational dynamics and stability of this interaction. The predicted epitopes were found to be non-toxic and non-allergenic, with no homology to the human proteome. The vaccine demonstrated a population coverage of 65.15% globally. It was composed of the immunogenic regions of LukED. Molecular docking and MD simulation indicated a stable interaction between hBD3 in the vaccine and TLR1/2 during the simulation period. We have designed vaccine against S. aureus LukED that targets epitope-rich regions, which helps maintain a native-like conformation. This work lays the groundwork for further experimental studies to evaluate the vaccine’s neutralizing effects.
Similar content being viewed by others
Introduction
Diabetic foot ulcers are a common complication in individuals with diabetes, and infection of these wounds can lead to increased morbidity and mortality rates. Studies have consistently shown that Staphylococcus aureus (S. aureus) is the most frequently identified pathogen in diabetic foot infections, with virulence factors such as toxic shock syndrome toxin-1 (TSST-1), leucocidins, and exfoliative toxins playing crucial roles in determining the severity and outcome of wounds1,2,3,4. Antibiotic-resistant S. aureus is a pressing issue that has sparked much debate among the scientific and clinical communities3,5. Despite the initial effectiveness of antibiotics such as penicillin and methicillin against S. aureus, the emergence of penicillin-resistant S. aureus in the 1940s and methicillin-resistant S. aureus (MRSA) strains has since become a growing concern6,7,8. These reports were associated with only hospital infections. While, community-associated MRSA (CA-MRSA), which affects seemingly healthy individuals, has also emerged in the past few decades9,10,11,12. Currently, vancomycin is the preferred antibiotic for the treatment of severe MRSA infections. However, reduced susceptibility to vancomycin has been observed in some S. aureus isolates, referred to as vancomycin intermediate-resistant S. aureus13,14,15.
Efforts are underway to develop novel preventive and therapeutic strategies to combat infections in patients with diabetic foot ulcers. These include biocidal nanomolecules, passive immunotherapy16,17,18, and vaccination19,20. Vaccine strategies that involve whole antigen proteins may lead to low safety or efficacy21,22. This difficulty in vaccine development can be overcome by designing a fusion protein containing short immunogenic peptide fragments called epitopes23. Several studies have employed bioinformatics approaches for vaccine design against S. aureus by targeting essential virulent and antigenic proteins, such as glycosyltransferases, elastin-binding proteins, staphylococcal secretory antigens, cell wall-anchored serine aspartate repeat-containing proteins D and E, alpha-hemolysin, manganese transport protein C, alpha-enolase, TSST-1, and clumping factor surface proteins24,25,26,27,28,29. These antigens were selected on the basis of their function, such as contributing to bacterial adherence to host cells or high conservation among all strains24,25,26,27,28,29.
We have recently conducted a systematic review and meta-analysis that highlights the importance of S. aureus LukED in diabetes foot infections30. LukED is a bicomponent pore-forming toxin consisting of two subunits, LukE and LukD. This toxin is one of the most prevalent virulence factors of S. aureus in diabetic foot infections31. The chemokine receptors CCR5, CXCR1, and CXCR2 are LukED receptor which is express on various immune cells, including neutrophils, T lymphocytes, macrophages, and dendritic cells32,33. Therefore, as a result of killing these cells, LukED disarms both the innate and adaptive immune responses. By developing a vaccine that generates an Ab-mediated immune response against this toxin, the toxin will be neutralized and prevent it from binding to its receptor. Blocking this immune evasion pathway, may reverse the weakened immune status resulting from toxin secretion, and subsequently leading to efficient immune responses and elimination of infection. Hence, the aim of the present study was to design a multi-epitope vaccine against the LukED of S. aureus.
Method
Sequence and structure retrieval
The protein sequences of LukD (O54082) and LukE (O54081) were extracted from UniProt at http://www.uniprot.org. To develop a universal vaccine, conserved epitopes were selected from multiple strains and isolates of S. aureus. To evaluate conservancy, the FASTA formatted amino acid sequences of LukD and LukE from all S. aureus isolates were retrieved from the NCBI protein database at https://www.ncbi.nlm.nih.gov/protein. We then narrowed the list of sequences by clustering those that were 100% identical via the CDhit server (http://weizhong-lab.ucsd.edu/cdhit_suite)34. To determine the conserved regions of the sequences, we aligned them using CLC workbench software. Epitopes located in regions with conservancy greater than 90% were selected for further analyses. The PDB files of the crystal structures of LukD (resolution: 1.7 Å, PDB Id: 4Q7G) and LukE (resolution: 1.9 Å, PDB Id: 7P8S) were retrieved from RCSB at http://www.rcsb.org.
Immunoinformatics analyses
Linear B cell epitope prediction
Iinear B cell epitopes were predicted using sequence- based prediction tools, namely, BepiPred-2.0 (https://services.healthtech.dtu.dk/services/BepiPred-3.0) and bcepred (http://crdd.osdd.net/raghava/bcepred)35. Three dimensional (3D) structure-based linear B cell epitope prediction was performed by submitting the 3D structure of each protein to the ElliPro web tool (http://tools.iedb.org/ellipro). Common linear epitopes that these tools agreed upon were selected as the final B cell epitopes35.
HTL epitope prediction
To predict HTL epitopes, 15-mer peptides that were capable of binding to Human Leukocyte Antigen (HLA) class II molecules were predicted using IEDB (http://tools.iedb.org/mhcii). For this study, reference allele set of HLA-II were used, including 15 HLA-DR alleles (HLA-DRB1*01:01, *03:01, *04:01, *04:05, *07:01, *08:02, *09:01, *11:01, *12:01, *13:02, *15:01, DRB3*01:01, DRB3*02:02, DRB4*01:01, DRB5*01:01), 6 HLA-DQ alleles (DQA1*05:01/DQB1*02:01, DQA1*05:01/DQB1*03:01, DQA1*03:01/DQB1*03:02, DQA1*04:01/DQB1*04:02, DQA1*01:01/DQB1*05:01, DQA1*01:02/DQB1*06:02) and 6 HLA-DP alleles (DPA1*02:01/DPB1*01:01, DPA1*01:03/DPB1*02:01, PA1*01:03/DPB1*04:01, DPA1*03:01/DPB1*04:02, DPA1*02:01/DPB1*05:01, DPA1*02:01/DPB1*14:01. These super type alleles provide maximum population coverage up to 99% of global population36. We selected epitopes with percentile ranks lower than 2% from the results of the IEDB consensus analysis tool37,38. Peptides with lower percentile ranks are defined as higher-affinity epitopes. Furthermore, the selected epitopes with the ability to bind to at least the three most frequent HLA-II alleles were chosen for further analyses.
Allergenicity, antigenicity and toxicity
To determine the allergenicity of the predicted epitopes, two prediction tools, AllerTOP v. 2.0 (https://www.ddg-pharmfac.net/AllerTOP) and AllergenFP 1.0 (https://www.ddg-pharmfac.net/AllergenFP), were utilized39,40. Utilizing predictions from both servers provides a form of cross-validation and strengthens our confidence in the results. To evaluate toxicity, the toxin pred server (https://webs.iiitd.edu.in/raghava/toxinpred/design.php) was used. Non-allegen and non-toxin antigenic epitopes were selected for further steps.
BLAST screening and population coverage
To prevent the risk of autoimmunity, a screening process was carried out on the chosen HTL and B cell epitopes to eliminate peptides that may show similarity with the human proteome. The screening was conducted using the BLASTp algorithm on the NCBI protein‒protein BLAST website (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins). A non-redundant protein sequence database and human (taxid:9606) organism was chosen. Epitopes with six or more sequential amino acid homologs to human proteins were removed from further analysis. We used the IEDB population coverage tool (http://tools.iedb.org/population) for the analysis of the population coverage of the epitopes and their respective HLA-binding alleles41.
Multi-epitope vaccine construction and structural bioinformatics
Multi-epitope vaccine design
As an adjuvant, hBD3 was chosen in our vaccine design42. To assemble the complete multi-epitope vaccine sequence, 17 distinct arrangements of B-cell, HTLepitopes and hBD3were connected by GPGPG and KK, and EAAAK linkers, respectively. Six histidine were added to the C-terminal end to aid protein purification by Ni-NTA chromatography. In all structures, adjuvant and B cell epitopes were located at C- or N-terminal to be more expose for interaction with their receptors.
As this strategy failed to fine a stable 3D structure after MD simulation, to create an effective and near native vaccine construct, two epitope-rich regions of LukE (including 72 amino acid) and LukD (including 169 amino acid), were joined together by KK linkers. To join the adjuvant, a number of flexible and rigid linkers, such as GGGGGGGG, EAAAK, (EAAAK)3, GGGGS and (GGGGS)3, were evaluated to find the best conformation43.
Antigenicity and physicochemical features of vaccine construct
The vaccine construct were analyzed for antigenicity using the VaxiJen v2.0 server (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html)35. VaxiJen considers values greater than the threshold of 0.4 as an antigen when the bacterial dataset is selected.
The ProtParam server (https://web.expasy.org/protparam) was used to assess the physiochemical properties of the vaccine construct44. These properties include the theoretical isoelectric point (PI), molecular weight (MW), stability characteristics, and grand average of hydropathicity (GRAVY). The GRAVY value ranges from around − 2 to + 2, with negative values indicating a more hydrophilic protein and positive values indicating a more hydrophobic protein. The commonly used cut-off value for the instability index is 40.
Tertiary structure prediction
The ColabFold server v1.5.5 was used to generate 3D protein models of vaccine constructs with different epitope and region arrangements45. This server by using Alphafold 2 provides high-quality 3D protein models in two steps: homology search in databases using Many-against-many sequence searching (MMSeqs2) and 3D structure inference using a Python library to communicate with MMSeqs2. The study adopted the standard parameter setup provided by the ColabFold server. Chimera 1.11 software was used to visualize the 3D protein structure models46.
Tertiary structure refinement and validation
Tertiary structures obtained from different arrangements of epitopes, regions, adjuvants and linkers were refined by galaxy refine 2 server (https://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE2)47,48. To identify any errors in the predicted tertiary structures, ProSA-web (https://prosa.services.came.sbg.ac.at/prosa.php) was employed49,50. A more negative Z score suggests a better model. Moreover, native proteins fall within the range of X-ray crystallography (light blue) or NMR spectroscopy (dark blue) with respect to their length. To confirm that the structure of the proteins is energetically favorable, a Ramachandran plot (PROCHECK) was used (https://saves.mbi.ucla.edu)51. In a good model 90% residues in the high-resolution structure would be expected in the favored regions. The tertiary vaccine structure with the best arrangement was subjected to molecular dynamics (MD) simulation to refine and obtain the near-native state. This validated structural model was then used as the basis for molecular docking studies.
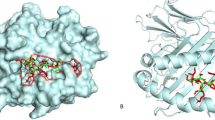
Molecular docking
To analyze the interaction pattern of the adjuvant part of the multi-epitope vaccine with the TLR1/2 heterodimer (PDB code: 2z7x, resolution: 2.1 Å), molecular docking analysis was carried out using HADDOCK server52. To identify the residues involved in the interaction, we selected hBD3 from the vaccine as well as the binding site from the TLR receptor. The ligand binding site of the TLR1/2 heterodimer is formed by specific regions of both TLR1 and TLR2 proteins. For TLR1, the binding site is composed of the amino acid residues from 255 (Tryptophan) to 339 (Valine). For TLR2, the binding site is made up of the amino acid residues from 266 (unspecified) to 355 (Tyrosine)53.
MD simulation
Initially, MD simulation was conducted to refine the vaccine structure to near-native state conformations. This approach demonstrates the ability, to a certain degree, to refine distorted protein conformations. First, a 100 ns MD simulation was performed on the freely designed vaccine and free TLR1/2 heterodimer structure. The final obtained structures were subsequently used for docking via the HADDOCK server (https://wenmr.science.uu.nl)52. The adjuvant residues of the vaccine ranged from 230 to 304 amino acids. the ligand binding groove is formed by specific regions of both TLR1 and TLR2 proteins. For the TLR1 subunit, the binding site is comprised of amino acid residues spanning from 255 to 339 amino acid. In contrast, the binding site for the TLR2 subunit is composed of residues ranging from 266 to 355 amino acid53.
The best structure obtained from docking (complex) was subsequently used for 100 ns MD simulation at 310 K. MD simulation was subsequently performed to understand the interaction stability between the receptor (TLR) and the multi-epitope vaccine construct at the microscopic level using GROMACS 2020 package under the G43A1 force field54. The vaccine structure was solvated in a 15,685 spc216 water model, and 47 Na + and 66 Cl- ions and the TLR1/2 heterodimer were solvated in 43,117 water molecules and 137 Na + and 133 Cl- ions. The complex system was solvated in 55,205 water molecules and 171 Na + and 186 Cl- ions. The minimal distance of 10 Å from any edge of the box along with the periodic boundary conditions was applied in all dimensions.
The particle mesh Ewald (PME) method was used to evaluate the long-range electrostatic interactions. The LINCS algorithm was also used to constrain all hydrogen bonds. The non-bonded cutoff of 10 Å was used as well. To remove steric clashes between the proteins and solvent molecules, energy minimization was performed using the steepest descent algorithm. The systems were equilibrated at the NVT ensemble for 500 ps and at the NPT ensemble for 1000 ps. After equilibration, a 100 ns MD simulation was performed with a time step of 1 fs. Finally, the mm/pbsa binding energy was calculated via the g_mmpbsa script for the binding free energy calculation55,56.
Codon adaptation, in silico cloning and solubility analysis
The Java codon adaptation tool (JCat) (http://www.jcat.de) was utilized to perform codon optimization for expression in a prokaryotic organism and reverse translation of the amino acid sequence57. The Escherichia coli (E. coli) K12 strain was selected to express the multi-epitope construct. Bacterial ribosome binding sites, rho-independent transcription terminators, and cleavage sites of Xhol and Ncol restriction enzymes were excluded to prevent unnecessary errors in protein expression in adopted cells. JCat calculates the codon adaptation index (CAI value) and GC content percentage to ensure high-level protein expression in the adopted cell. Ideally, the CAI should be greater than 0.8 and equal to 1.0, and the percentage of GC content should be ranged between 30% and 70%. The transcriptional and translational efficiencies may be unfavorable if the value is not within this range.
SnapGene software was subsequently used to insert the optimized DNA sequence of the multi-epitope vaccine into the pET-28a (+) vector, the most popular expression plasmid on the market used for recombinant protein production in E. coli. The fragment was inserted into the region between the restriction sites NcoI and XhoI because a previous study has shown that the cloning of proteins via these restriction sites resulted in soluble and functional recombinant proteins58. Initially, restriction sites were not present at the ends of the insert sequence. Therefore, the sequences for the restriction enzymes XhoI and NcoI, were added at the N-terminal and C-terminal ends of the fragment. To determine the solubility of the protein vaccine after overexpression in E. coli, SOLpro at http://scratch.proteomics.ics.uci.edu was utilized59.
Results
Sequence and structure retrieval
The protein sequences of LukD (O54082) and LukE (O54081) were extracted from UniProt to be submited to epitope prediction servers. However, as there are several S. aureus strains and isolates, epitopes located in conserved region of LukE and LukD were selected. To identify the conserved region, from different S. aureus isolates, 11,009 LukE sequences and 12,170 LukD sequences were retrieved using NCBI database. After removing unrelated, partial and ambiguous amino acids-containing sequences, 7113 and 11,227 complete protein sequences were retrieved for the LukE and LukD proteins, respectively. Then, we clustered 100% identical sequences to reduce the number of sequences before alignment. A total of 107 and 144 clusters were generated for the LukE and LukD proteins, respectively. The proteins retrieved from different isolates share levels of similarity and identity after alignment by CLC workbench software (Fig. 1). Epitopes located in conserved regions of proteins among all S. aureus isolates were selected.
Identification of linear B-cell epitopes
The common sequence-based and structure-based B cell epitopes between the bcepred, BepiPred and ElliPro servers were selected from each antigenic protein.
The sequences with homology between the predicted B cell epitopes and the human proteome were excluded to avoid autoimmunity. A total of five B cell epitopes were selected from the antigens, including four epitopes from LukD and one epitope from LukE (Table 1). We then excluded epitopes that were likely allergens or toxins. All epitopes were located in the conserved regions of the target protein.
Identification of HTL epitopes
HTL epitopes identified as strong binder by IEDB consensus method, which was able to bind to at least three frequent HLA-II alleles, were selected. Antigenic epitopes which were non-allergen, non-toxin and located in the conserved regions were selected for further analysis. To avoid autoimmunity, sequences that revealed homology with the human proteome were excluded. Based on the inclusion criteria, selected epitopes must be able to bind to multiple HLA alleles. According to these inclusion and exclusion criteria, LukD did not have a suitable T cell epitope for vaccine construction. (Table 2). The four epitope in LukE with the ability to bind to more than one frequent HLA allele were chosen as HTL epitopes (Fig. 2).
Population coverage of vaccines
The IEDB population coverage tool was used to calculate the population coverage of the predicted HTL epitopes and the related HLA-DR, -DQ and DP supertype alleles. Population coverage is defined as the fraction of individuals in a particular area who can potentially respond to a given epitope. The results revealed that 48.38% in North Africa, 39.21% in West Africa, 39.75% in Central Africa, and 39.16% in East Africa are potentially able to respond to the selected HTL epitopes. In Asia, the epitopes presented 61.49%, 27.2%, 35.39%, and 54.97% coverage in South Asia, Southwest Asia, Southeast Asia, and East Asia, respectively. The epitopes showed 71.36%, 34.27%, 32.12%, and 72.08% population coverage in Europe, Oceania, South America, and North America, respectively. The highest scores were observed for Europe and North America. The average population coverage for the worldwide population was calculated to be 65.15%.
Multi-epitope vaccine sequence construction
To design the multi-epitope vaccine construct, 17 arrangements of adjuvant, HTL, and B cell epitopes were joined using EAAAK linkers60, KK linkers61,62 and GPGPG linkers63, respectively. All the arrangement sequences were submitted to ColabFold v1.5.5 to model the 3D structure. Each structure was refined by the galaxy server. The quality of the refined 3D structure was assessed by applying ProSA and a Ramachandran plot. Although the refined protein model displayed favorable characteristics based on analysis using bioinformatics tools, including being classified as a stable protein by ProtParam, having the lowest energy level, and having the highest percentage (over 90%) of residues located in the favored regions of the Ramachandran plot, the construct surprisingly became linear after the molecular dynamics (MD) simulation was performed.
To solve this problem, we choose epitope-rich regions within the protein, rather than simply linking individual epitopes together to generate a near-native protein vaccine construction. In this regard, the immunogenic regions of LukE and LukD, consisting of 72 amino acids of LukE and 169 amino acid LukD, were joined together by KK linkers (Fig. 2). The KK linker is an appropriate linker for B cell and T cell epitopes because it provides a target sequence for lysosomal proteases and prevents the induction of junctional epitopes61,62.
A number of flexible and rigid linkers, such as GGGGGGGG, EAAAK, (EAAAK)3, GGGGS and (GGGGS)3, were applied to join the adjuvant. ColabFold v1.5.5 was used to model the 3D structure of these sequences. The lowest energy level and highest rama flavor were obtained when a flexible linker (GGGGS)3 was used to join the adjuvant64. Linkers consisting of Gly and Ser residues (“GS” linkers) are the most commonly used flexible linkers43. After MD simulation, the protein attained its three-dimensional structure, suggesting good stability, with favorable thermodynamic properties. The final construct consisted of 311 amino acid residues, five B cell epitopes, four HTL epitopes with the ability to bind to multiple HLA alleles and three HTL epitopes with the ability to bind to one frequent HLA allele. The Ramachandran results for the vaccine model revealed that 82.5, 13.8 and 3.7 residues were found in the favored, allowed and outlier regions, respectively (Fig. 3.a). The result of ProSA revealed a Z score of -5.98, which was within the range of scores commonly reported for native proteins of similar size (Fig. 3.b).
The molecular weight and pI of our multi-epitope vaccine, were calculated to be 34578.76 Da and 9.49, respectively. The protein contained 49 positively charged amino acids (i.e., Arg and Lys) and 31 negatively charged amino acids (i.e., Asp and Glu). The half-life of the protein was estimated to be more than 20 h in yeast (in vivo), 4.4 h in mammalian reticulocytes (in vitro), and more than 10 h in E. coli (in vivo). The aliphatic index was 63.40, indicating that the construct could be stable for a wide range of temperatures. Based on ProtParam tool, proteins with an aliphatic index greater than 40 are generally considered to be thermostable44. The protein was found to be hydrophilic, with a GRAVY of -0.849. Although, the vaccine was classified as an unstable protein based on the instability index, which was assessed to be 40.14, MD simulations recognize it stable. MD simulation provide a more comprehensive and dynamic assessment of protein stability compared to the sequence-based analysis provided by ProtParam, because MD simulations model the actual behavior of the protein in a simulated environment, with regards to the 3D structure, interactions, and conformational changes over time.
The main goal of this part was to obtain a stable protein structure. The exposure of B cell epitopes is also important to induce immune responses. By evaluating final vaccine construct in Ellipro severe, it was demonstrated that four chosen B cell epitopes were still exposed, as shown in Fig. 4. According to the result of VaxiJen, the antigenicity of the whole protein vaccine was 0.910, which was defined as the probable antigen.
Three-dimensional representation of exposed regions in vaccine construct that B cell epitopes were located. The exposed regions are represented by yellow surface, and the bulk of the polyprotein is represented in grey sticks; the specific position of the exposed regions in vaccine construct are as follows: (a) residues 238–263, (b) residues 127–146, (c) residues 156–162, (d) residues 1–32.
Molecular docking
The interaction between TLR1/2 and adjuvant in final vaccine construct was analyzed by molecular docking analysis using HADDOCK server. Among the HADDOCK docking outputs, the best results in terms of the docking interaction pattern between the receptor and ligand were selected for further analysis. The selected docking result had the lowest energy weighted score of -13 ± 9.2 kcal/mol (Table 3).
Molecular dynamics simulation of the immune receptor-vaccine complex
First, the vaccine and the TLR1/2 heterodimer were separately used for 100 ns MD simulation, and the results of the MD simulation are presented in Table 4. The obtained structures from the MD simulation were subsequently used for docking investigations (Table 3).
To evaluate the stability of interaction between the receptor and ligand, the docking output were assessed by MD simulation. The standard deviations of all the parameters in the free and complex forms were trivial, which means that all the systems reached thermal and structural equilibrium during the last 50 ns of the MD simulation (Table 4).
The backbone root mean square deviation (RMSD) was monitored to assess the structural stability of the proteins during the 100 ns MD simulation. The RMSD of free vaccine construct and in complex with TLR are shown in Fig. 5a. The RMSD of free TLR chains and in complex with vaccine are shown in Fig. 5b. The initial fluctuations persist in the vaccine and TLR in two forms (free or complex) for up to 50 ns, but after 50 ns, all the structures become stable. The results shown in Fig. 5; Table 4 indicate that the backbone RMSD of the vaccine and TLR decreases after complex formation because of the generation of hydrophobic and hydrogen bonds between them.
The root mean square fluctuation (RMSF) provides direct insight into structural fluctuations as well as the flexibility of protein residues during the last 50 ns of MD simulation. The RMSF of free vaccine construct and in complex with TLR shows in Fig. 6a. The RMSF of free TLR chains and in complex with vaccine shows in Fig. 6b. After vaccine and TLR binding, the average RMSF of vaccine residues does not change (Table 4), but these values decrease for TLRs, which means that complex formation leads to a decrease in the flexibility of TLRs compared with that of vaccine residues. In addition, the RMSF decrease was greater for TLR2.
The radius of gyration (Rg) of a protein is the distribution of atoms around its center of mass. The protein solvent accessible surface area (SASA) is critical for studying conformational changes within the structure of proteins. This parameter is especially vital in investigating the interaction of proteins with the solvent molecules around them. The Rg and SASA of the vaccine and TLR1 increase slightly after complex formation. However, these parameters decrease for TLR2 after complex formation, which is in agreement with the greater decrease in the RMSF of TLR2 (Table 4). We can conclude that the binding of the vaccine induces structural changes in the TLR receptor that can lead to internal signaling and finally an immunological response.
The average number of hydrogen bonds within the vaccine was almost constant, and the changes were not significant. The average number of hydrogen bonds between TLR2 and the vaccine during the last 50 ns of the MD simulation was 10 ± 2.6, but this parameter for TLR1 was 2.3 ± 1.4. Therefore, the vaccine binds to TLR2 and leads to reduces its SASA, RMSF, and internal hydrogen bonds. Visual inspection of the final structure of the complex (Fig. 6) confirmed this finding.
The average number of hydrogen bonds between protein and solvent in vaccines and TLR1/2 decreased. In other words, the increase in the number of internal hydrogen bonds between the vaccine and TLR1/2 led to a decrease in the number of hydrogen bonds between TLR1/2 and solvent. The percentage of residues that contribute to the secondary structure in the vaccine and TLR1/2 before and after binding was almost constant, and complex formation did not change the secondary structure (Table 1). The hydrogen and hydrophobic interactions of the vaccine and TLR1/2 residues were calculated via the DIMPLOT module of LigPlot software at the final structure of the complex (Fig. 7).
Finally, the MM/PBSA binding of the vaccine during the last 50 ns of the MD simulation was calculated. The average electrostatic interaction plus polar solvation energy was − 8802.8 ± 421 kJ/mol, the average van der waals interaction plus non-polar solvation energy was − 673.6 ± 43.5 kJ/mol, and the average binding free energy during the last 50 ns of the MD simulation was − 9476.44 ± 419 kJ/mol. Thus, the vaccine strongly interacts with TLR1/2, and electrostatic interactions are greater than van der Waals interactions. These results were compatible with the docking results (Table 4).
Codon adaptation and in silico cloning
The multi-epitope vaccine construct was expressed through in silico cloning in the commonly utilized E. coli K12 host. The amino acid sequence was first converted into its corresponding DNA codons via the JCAT server. The GC content and CAI of the optimized DNA sequence were 47.70% and 0.94%, respectively, indicating high potential for expression in the adopted host. Next, the NcoI and XhoI restriction sites were added to the 3’ and 5’ ends of the optimized nucleotide sequence and inserted into the pET28a (+) vector (Fig. 8). The optimized nucleotide sequence contained 933 nucleotides, and the total length of the final cloning vector was 6,170 base pairs. The protein was also predicted to be soluble, with a probability of 0.882897, upon overexpression in E. coli.
Discussion
Staphylococcus aureus is a common pathogen that causes various infections in humans, ranging from mild skin infections to severe life-threatening diseases such as pneumonia, endocarditis, and sepsis22. In particular, S. aureus is associated with diabetic foot infections, which often lead to amputations and contribute significantly to morbidity and mortality in diabetic patients. Traditional treatments for diabetic foot ulcers include surgical debridement, antibiotics, and wound care; however, these treatments are often ineffective, and the prevalence of antibiotic resistance in S. aureus is increasing, which limits treatment options3,4.
Given the growing concern over antibiotic-resistant S. aureus, vaccination has emerged as an effective strategy to prevent S. aureus infection in diabetic patients. However, the major obstacles in developing a durable and effective vaccine are safety issues and a lack of significant protection. For example, the StaphVAX vaccine developed by Nabi against capsular polysaccharide 5/8 antigens was discontinued in phase III clinical trials because there was no significant difference between the vaccine and the placebo65. V710 is another vaccine that was discontinued in phase III clinical trials, because it is associated with a higher mortality rate than the placebo in vaccinated subjects after cardiothoracic surgery66. The SA4Ag vaccine developed by the Pfizer company for patients undergoing elective orthopedic surgery, which is composed of four antigens, has shown no efficacy and has stopped in phase II67. Despite the number of ongoing trials, no successful phase III clinical trials for safe and effective S. aureus vaccines have been completed21.
The aim of our study is designing vaccine that potentially be able to generate antibodies against LukED. Developing a vaccine that generates Ab against LukED could neutralize the toxin by preventing it from binding to its receptors and blocking pore-forming ability in immune cells. This neutralization is critical, as it helps restore the functionality of both innate and adaptive immune responses that the toxin otherwise diminishes.
A recent systematic review and meta-analysis highlighted the importance of the S. aureus LukED in causing diabetic foot infections30. LukED is a bi-component toxin that targets and kills various immune cells, including neutrophils, T cells, macrophages, and dendritic cells, by binding to receptors like CCR5, CXCR1, and CXCR232,33. Studies have shown that LukED targets the cellular receptor CCR5 to eliminate antigen-presenting cells (APC) and S. aureus-specific CCR5+ Th1/Th17 cells, neutrophils, monocytes, and NK cells. CCR5 is the first described cellular receptor that is targeted by LukED to eliminate APCs and S. aureus-specific CCR5+ Th1/Th17 cells33. This allows S. aureus to evade the host innate and adaptive immune responses. Lei Zhang et al., have identified four single-chain fragment variables (scFvs) targeting the LukED produced by S. aureus. These scFvs significantly decreased LukED-induced cell killing by inhibiting the binding of LukED to chemokine receptors (CCR5 and CXCR2) and reduced the death rates of neutrophils and mammary epithelial cells T cells. 68.
The immune response against bacterial toxins typically involves the humoral of the adaptive immune system. This is because Ab can directly neutralize the toxin and prevent its harmful effects. Therefore, in our study we focused on HLA-II epitopes that recognized by HTLs. Providing co-stimulatory signals and cytokines, the activated HTLs help the formation of more efficient humeral responses through efficient proliferation of toxin-specific B cells, generation long-lived memory B cell and plasma cells, Ab class switching and production of higher affinity toxin-specific Abs69.
Although LukE and LukD exhibit little or no sequence diversity among different S. aureus strains, we aligned all the sequences of LukE and LukD registered in NCBI, and only epitopes located in regions with more than 90% conservancy were selected. The IEDB consensus prediction were used to identify HTL epitopes. The specific selection criteria were selecting strong binders that can interact with multiple common HLA-II alleles. BepiPred, Bcepred, and Ellipro were employed for B-cell epitope prediction. This multi-faceted approach aims to enhance the reliability of epitope identification, as consensus among diverse prediction methods can bolster confidence in the results. The rationale is that achieving consensus predictions across diverse methods, each with their own unique training datasets and underlying algorithms, can strengthen the credibility of the identified B-cell epitopes. Finally, four HTL epitope and five linear B cell epitopes of LukE and LukD were selected.Discontinuous epitopes are composed of amino acid residues that are distant in the primary sequence which bring together in the folded structure of the antigen. When using an epitope-reach region instead of whole protein antigen, putting distant amino acid residues together again to form the discontinuous epitope is impossible. Using discontinues epitopes is a suited strategy for vaccine which include whole protein domain or whole protein antigen. To address this issue, we only select linear B cell epitopes. To address this issue, we only select linear B cell epitopes.
To construct highly authentic vaccine, the immunogenic region of LukE and LukD, which contain epitope segments, were linked together via various KK linkers. A lysine linker (-KK-) prevents the formation of junctional epitopes and provides target-specific cleavage in the lysosomal degradation machinery61. Utilizing an immunogenic protein region, rather than simply joining individual epitopes together, can help preserve a more native and stable conformation, and potentially enhance the immunological efficacy. Nimat Ullah and colleagues designed a multi-epitope vaccine by connecting individual B-cell epitope segments from 8 known virulence S. aureus, one of which was LukE. Their results showed that while the vaccine elicited strong IgG Ab titers in immunized mice, the generated antibodies failed to provide protection against infection. The researchers concluded that the computationally-derived, multi-epitope vaccine approach had limitations. They suggest that utilizing an immunogenic protein domain, rather than simply joining individual epitopes together, can help preserve a more native and stable conformation, potentially enhancing the immunological efficacy70. Hence, in the present study, epitope rich region was join together instead of joining individual epitopes by linkers.
The 3D structure which acquired acceptable quality using the Ramachandran plot and ProSA, was selected for MD simulation. MD simulations can help identify potential issues or instabilities in the protein structure that may not be apparent from static structural analyses. It can also help refine the protein structure by allowing it to explore different conformations and reach a more stable, energetically favorable state71.
We used hBD3 as an adjuvant, which is an antimicrobial peptide that inhibits antibiotic-resistant Staphylococcus biofilm formation72, , promotes wound healing73, and is involved in innate immune activation through binding to TLRs (e.g., TLR1 and 2)74. Molecular docking with TLR1/2 revealed potential interactions of adjuvant with TLR2. To assess the structural stability of the vaccine interaction with the TLR1/2 heterodimer, the RMSD and RMSF were monitored. The results of the MD simulations suggested that the vaccine and TLR interaction had acceptable stability and thermodynamic properties. In the final 3D structure, four B cell epitopes were exposed in the vaccine 3D structure. Finally, in silico cloning analysis revealed that the vaccine could be cloned and expressed in E. coli for further in vitro and in vivo experiments.
Recent studies have identified CXCR1 and CXCR2 as LukED targets, which are largely expressed on neutrophils, monocytes, and NK cells30. The region Gln210-Ala219 of LukE, located in loop L3 of the LukE rim domain, has been shown to interact with CXCR1/CXCR2 and be responsible for the lysis of neutrophils and monocytes32. If our multi-epitope vaccine induces an Ab against the B cell epitope KDKKYNK, which is located at residues 65–71 of LukE, this interaction might be blocked by steric hindrance.
The critical dimerization residues involved in the LukED toxin complex formation are Asp286 and Asn289 in the LukE and Arg286 and Asp289 in the LukD33. Inducing blocking antibodies against EVDWQNHTV as a B cell epitope which is located at residues 305–314 and DRKTNHK which is located at residues 180–186 might inhibit dimerization by steric hindrance. Blocking these immune evasion pathways can combat the weakened immune status results from toxin secretion, and an efficient immune response can eliminate the infection.
LukED has also been found to mediate erythrocyte lysis by binding to Duffy antigen receptor for chemokines (DARC) and enabling S. aureus to acquire iron75. Two divergent regions (DRs) of LukE, namely, residues 57–75 (DR1) and residues 182–196 (DR4), have been found to contribute to hemolysis. LukE promotes the recruitment of LukD to erythrocytes by binding DR1 to DARC and could facilitate LukED oligomer formation75. If our multi-epitope vaccine induces an Ab against KDKKYNK as a B cell epitope, which is located at residues 65–71 of LukE, the interaction of LukE with DARC could be blocked.
In conclusion, using epitope-rich region or domain of protein can preserve a more native-like conformation, which could be advantageous for generating efficient and stronger immune response, compared to a simple arrangement of individual epitopes. The results obtained in this study show that the multi-epitope vaccine construct against LukED could be a promising candidate for protection against S. aureus. The immunological, structural, and biochemical features of the vaccine were evaluated. The selected epitopes were arranged with appropriate linkers and adjuvants to increase their immunogenicity and stability. Moreover, the vaccine was found to be able to clone and express in E. coli. However, further in vitro and in vivo experiments are needed to validate the effectiveness of this vaccine candidate and assess its safety and efficacy.
Data availability
The data that support the findings of this study are available on request from the corresponding author.
References
Messad, N. et al. Distribution of edin in Staphylococcus aureus isolated from diabetic foot ulcers. Clin. Microbiol. Infect. 19(9), 875–880 (2013).
Gardner, S. E., Hillis, S. L., Heilmann, K., Segre, J. A. & Grice, E. A. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes 62(3), 923–930 (2013).
Hatipoglu, M. et al. Causative pathogens and antibiotic resistance in diabetic foot infections: a prospective multi-center study. J. Diabetes Complicat. 30(5), 910–916 (2016).
Shettigar, K. & Murali, T. S. Virulence factors and clonal diversity of Staphylococcus aureus in colonization and wound infection with emphasis on diabetic foot infection. Eur. J. Clin. Microbiol. Infect. Dis. 1–12. (2020).
Lompo, P. et al. Antibiotic susceptibility of Staphylococcus aureus and Streptococcus pneumoniae isolates from the nasopharynx of Febrile Children under 5 years in Nanoro, Burkina Faso. Antibiotics 10(4), 444 (2021).
Plough, H. H. Penicillin resistance of Staphylococcus aureus and its clinical implications. Am. J. Clin. Pathol. 15(10), 446–451 (1945).
Rammelkamp, C. H. & Maxon, T. Resistance of Staphylococcus aureus to the action of Penicillin. Proc. Soc. Exp. Biol. Med. 51(3), 386–389 (1942).
Ayliffe, G. The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 24(Supplement_1), S74–S9 (1997).
Klein, E., Smith, D. L. & Laxminarayan, R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg. Infect. Dis. 13(12), 1840 (2007).
Chambers, H. F. & DeLeo, F. R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7(9), 629–641 (2009).
Control CfD Prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997–1999. MMWR Morbidity Mortal. Wkly. Rep. 48(32), 707–710 (1999).
Hussain, F. M., Boyle-Vavra, S. & Daum, R. S. Community-acquired methicillin-resistant Staphylococcus aureus colonization in healthy children attending an outpatient pediatric clinic. Pediatr. Infect. Dis. J. 20(8), 763–767 (2001).
Kaatz, G. W., Seo, S. M., Dorman, N. J. & Lerner, S. A. Emergence of teicoplanin resistance during therapy of Staphylococcus aureus endocarditis. J. Infect. Dis. 162(1), 103–108 (1990).
Chang, S. et al. Infection with Vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348(14), 1342–1347 (2003).
Control & CfD Prevention. Vancomycin-Resistant Staphylococcus aureus—Delaware, 2015. Ann. Emerg. Med. 67(3):386-7. (2016).
Sause, W. E., Buckley, P. T., Strohl, W. R., Lynch, A. S. & Torres, V. J. Antibody-based biologics and their promise to combat Staphylococcus aureus infections. Trends Pharmacol. Sci. 37(3), 231–241 (2016).
Siddiqi, K. S., Husen, A. & Rao, R. A. A review on biosynthesis of silver nanoparticles and their biocidal properties. J. Nanobiotechnol. 16(1), 1–28 (2018).
Oliveira, W. et al. Staphylococcus aureus and Staphylococcus epidermidis infections on implants. J. Hosp. Infect. 98(2), 111–117 (2018).
Levy, J. et al. Safety and immunogenicity of an investigational 4-component Staphylococcus aureus vaccine with or without AS03B adjuvant: results of a randomized phase I trial. Hum. Vaccines Immunotherapeutics 11(3), 620–631 (2015).
Chen, W. H. et al. Safety and immunogenicity of a parenterally administered, structure-based rationally modified recombinant staphylococcal enterotoxin B protein vaccine, STEBVax. Clin. Vaccine Immunol. 23(12), 918–925 (2016).
Amandine, G-B., Gagnaire, J., Pelissier, C., Philippe, B. & Elisabeth, B-N. Vaccines for healthcare associated infections without vaccine prevention to date. Vaccine: X 11, 100168 (2022).
Kzhyshkowska, J., De Berardinis, P. & Bettencourt, P. Research and Development: The Past, Present and Future, Including Novel Therapeutic Strategies. Fighting an Elusive Enemy: Staphylococcus aureus and its Antibiotic Resistance (Immune-Evasion and Toxic Mechanisms, 2022).
Fleri, W. et al. The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front. Immunol. 8, 278 (2017).
ul Qamar, M. T. et al. Designing multi-epitope vaccine against Staphylococcus aureus by employing subtractive proteomics, reverse vaccinology and immuno-informatics approaches. Comput. Biol. Med. 132, 104389 (2021).
Chatterjee, R. et al. Development of a conserved chimeric vaccine for induction of strong immune response against Staphylococcus aureus using immunoinformatics approaches. Vaccines 9(9), 1038 (2021).
Ahmadi, K. et al. Epitope-based immunoinformatics study of a novel Hla-MntC-SACOL0723 fusion protein from Staphylococcus aureus: induction of multi-pattern immune responses. Mol. Immunol. 114, 88–99 (2019).
Dey, J. et al. B and T cell epitope-based peptides predicted from clumping factor protein of Staphylococcus aureus as vaccine targets. Microb. Pathog. 160, 105171 (2021).
Kolla, H. B., Tirumalasetty, C., Sreerama, K. & Ayyagari, V. S. An immunoinformatics approach for the design of a multi-epitope vaccine targeting super antigen TSST-1 of Staphylococcus aureus. J. Genetic Eng. Biotechnol. 19(1), 1–14 (2021).
Shahbazi, M., Haghkhah, M., Rahbar, M. R., Nezafat, N. & Ghasemi, Y. In silico sub-unit hexavalent peptide vaccine against an Staphylococcus aureus biofilm-related infection. Int. J. Pept. Res. Ther. 22(1), 101–117 (2016).
Shahrokh, S., Tabatabaee, A., Yazdi, M. & Siavash, M. Proportion of toxin and non-toxin virulence factors of Staphylococcus aureus isolates from diabetic foot infection: a systematic review and meta-analysis. BMC Microbiol. 24(1), 1 (2024).
Nocadello, S. et al. Crystal structures of the components of the Staphylococcus aureus leukotoxin ED. Acta Crystallogr. Sect. D: Struct. Biology 72(1), 113–120 (2016).
Reyes-Robles, T. et al. Staphylococcus aureus leukotoxin ED targets the chemokine receptors CXCR1 and CXCR2 to kill leukocytes and promote infection. Cell. host Microbe 14(4), 453–459 (2013).
Alonzo, I. I. I. F. et al. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 493(7430), 51–55 (2013).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13), 1658–1659 (2006).
Saha, S. & Raghava, G. P. S. (eds) BcePred: prediction of continuous B-cell epitopes in antigenic sequences using physico-chemical properties. Artificial Immune Systems: Third International Conference, ICARIS 2004, Catania, Sicily, Italy, September 13–16, 2004 Proceedings 3. (Springer, 2004).
Greenbaum, J. et al. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics 63(6), 325–335 (2011).
Wang, P. et al. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinform. 11(1), 1–12 (2010).
Wang, P. et al. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol. 4(4), e1000048 (2008).
Dimitrov, I., Bangov, I., Flower, D. R. & Doytchinova, I. AllerTOP v. 2—a server for in silico prediction of allergens. J. Mol. Model. 20(6), 1–6 (2014).
Dimitrov, I., Naneva, L., Doytchinova, I. & Bangov, I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics 30(6), 846–851 (2014).
Bui, H-H. et al. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinform. 7(1), 1–5 (2006).
Lei, Y. et al. Application of built-in adjuvants for epitope-based vaccines. PeerJ 6, e6185 (2019).
Chen, X., Zaro, J. L. & Shen, W-C. Fusion protein linkers: property, design and functionality. Adv. Drug Deliv. Rev. 65(10), 1357–1369 (2013).
Gasteiger, E. et al. Protein Identification and Analysis Tools on the ExPASy Server. (Springer, 2005).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19(6), 679–682 (2022).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25(13), 1605–1612 (2004).
Lee, G. R., Won, J., Heo, L. & Seok, C. GalaxyRefine2: simultaneous refinement of inaccurate local regions and overall protein structure. Nucleic Acids Res. 47(W1), W451–W5 (2019).
Lee, G. R., Heo, L. & Seok, C. Simultaneous refinement of inaccurate local regions and overall structure in the CASP12 protein model refinement experiment. Proteins Struct. Funct. Bioinform. 86, 168–176 (2018).
Wiederstein, M. & Sippl, M. J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35(suppl_2), W407–W10 (2007).
Sippl, M. J. Recognition of errors in three-dimensional structures of proteins. Proteins Struct. Funct. Bioinform. 17(4), 355–362 (1993).
Laskowski, R., MacArthur, M. & Thornton, J. PROCHECK: validation of protein-structure coordinates. (2006).
Honorato, R. V. et al. The HADDOCK2. 4 web server for integrative modeling of biomolecular complexes. Nat. Protoc. 1–23. (2024).
Jin, M. S. et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130(6), 1071–1082 (2007).
Abraham, M. J. et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25 (2015).
Mahnam, K. & Rajaee, S. M. A theoretical survey to find potential natural compound for inhibition of binding the RBD domain to ACE2 receptor based on plant antivirals. J. Biomol. Struct. Dynamics 41(23), 14540–14565 (2023).
Kumari, R., Kumar, R., Consortium, O. S. D. D. & Lynn, A. g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 54(7), 1951–1962 (2014).
Grote, A. et al. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 33, 526–531 (2005).
Tham, H. Y., Song, A. A., Yusoff, K. & Tan, G. H. Effect of different cloning strategies in pET-28a on solubility and functionality of a staphylococcal phage endolysin. Biotechniques 69(3), 161–170 (2020).
Magnan, C. N., Randall, A. & Baldi, P. SOLpro: accurate sequence-based prediction of protein solubility. Bioinformatics 25(17), 2200–2207 (2009).
Arai, R., Ueda, H., Kitayama, A., Kamiya, N. & Nagamune, T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 14(8), 529–532 (2001).
Sarobe, P. et al. Enhancement of peptide immunogenicity by insertion of a cathepsin B cleavage site between determinants recognized by B and T cells. Res. Immunol. 144(4), 257–262 (1993).
Yano, A. et al. An ingenious design for peptide vaccines. Vaccine 23(17–18), 2322–2326 (2005).
Guan, X., Zou, J., Gu, H. & Yao, Z. Short amyloid-β immunogens with spacer-enhanced immunogenicity without junctional epitopes for Alzheimer’s disease immunotherapy. Neuroreport 23(15), 879–884 (2012).
Bai, Y. & Shen, W-C. Improving the oral efficacy of recombinant granulocyte colony-stimulating factor and transferrin fusion protein by spacer optimization. Pharm. Res. 23, 2116–2121 (2006).
Shinefield, H. et al. Use of a Staphylococcus aureus conjugate vaccine in patients receiving hemodialysis. N. Engl. J. Med. 346(7), 491–496 (2002).
Fowler, V. G. et al. Effect of an investigational vaccine for preventing Staphylococcus aureus infections after cardiothoracic surgery: a randomized trial. Jama 309(13), 1368–1378 (2013).
Gurtman, A. et al. The development of a staphylococcus aureus four antigen vaccine for use prior to elective orthopedic surgery. Hum. Vaccines Immunotherapeutics 15(2), 358–370 (2019).
Zhang, L. et al. Single-chain fragment variables targeting leukocidin ED can alleviate the inflammation of Staphylococcus aureus-Induced mastitis in mice. Int. J. Mol. Sci. 23(1), 334 (2021).
Miller, L. S., Fowler, V. G. Jr, Shukla, S. K., Rose, W. E. & Proctor, R. A. Development of a vaccine against Staphylococcus aureus invasive infections: evidence based on human immunity, genetics and bacterial evasion mechanisms. FEMS Microbiol. Rev. 44(1), 123–153 (2020).
Ullah, N. et al. In silico designed Staphylococcus aureus B-cell multi-epitope vaccine did not elicit antibodies against target antigens suggesting multi-domain approach. J. Immunol. Methods 504, 113264 (2022).
Salo-Ahen, O. M. et al. Molecular dynamics simulations in drug discovery and pharmaceutical development. Processes 9(1), 71 (2020).
Zhu, C. et al. Human β-defensin 3 inhibits antibiotic-resistant Staphylococcus biofilm formation. J. Surg. Res. 183(1), 204–213 (2013).
Takahashi, M. et al. The antimicrobial peptide human β-defensin-3 accelerates wound healing by promoting angiogenesis, cell migration, and proliferation through the FGFR/JAK2/STAT3 signaling pathway. Front. Immunol. 12, 712781 (2021).
Funderburg, N. et al. Human β-defensin-3 activates professional antigen-presenting cells via toll-like receptors 1 and 2. Proc. Natl. Acad. Sci. 104(47), 18631–18635 (2007).
Vasquez, M. T. et al. Identification of a domain critical for Staphylococcus aureus LukED receptor targeting and lysis of erythrocytes. J. Biol. Chem. 295(50), 17241–17250 (2020).
Acknowledgements
We would like to thank Karim Mahnam (Associate Professor of Biophysics, Shahrekord University) for MD simulation and computational support of this research as well as constructive guidance for writing this part. Additionally, we would like to acknowledge Samaneh Shahrokh Esfahani (research assistant at Isfahan Endocrine & Metabolism Research Center) for constructive feedback during the conception of this article.
Author information
Authors and Affiliations
Contributions
PSh, PB and MS: conception and design. PSh and PB: data collection and computational analysis. PSh,and RK: paper writing and preparing figures. PB, PSh and RK: paper review & editing. All authors approved the submitted version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shahraki, P.K., Kiani, R., Siavash, M. et al. Design of a multi-epitope vaccine against Staphylococcus Aureus lukotoxin ED using in silico approaches. Sci Rep 15, 14517 (2025). https://doi.org/10.1038/s41598-025-85147-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-85147-3