Abstract
G-protein gamma subunit 2 (GNG2) plays a vital role in various cellular processes, yet its specific function in colorectal cancer (CRC), particularly in highly invasive cases and brain metastasis, remains unclear. This study identifies GNG2 as a key regulator in metastatic colorectal cancer (mCRC) through bioinformatics analysis and experimental validation. Functional enrichment analyses reveal that GNG2 is related to the PI3K/AKT/mTOR signaling pathway and cell cycle regulation. These findings were further confirmed by in vitro and in vivo experiments. The overexpression of GNG2 significantly induced G0/G1 arrest and further inhibited the PI3K/AKT/mTOR axis in CRC cell lines, including suppressed proliferation, migration, and invasion and metastasis ability. In vivo studies using an orthotopic xenograft model demonstrated that GNG2 overexpression reduced brain metastasis and extended overall survival in mice. Immunohistochemistry and multiplex immunofluorescence confirmed the association between GNG2 overexpression, the PI3K/AKT/mTOR signaling pathway, and G0/G1 arrest. Our study suggests that GNG2 contributes to tumor suppression in CRC, particularly in preventing brain metastasis, and could serve as a promising biomarker and treatment target for mCRC, offering fresh insights into the molecular processes driving cancer progression and metastasis.
Similar content being viewed by others
Introduction
Colorectal cancer has seen a recent rise in incidence, with approximately 3% of patients with advanced disease developing brain metastases (BM) during CRC treatment1,2,3. BM tend to correlate with negative outcomes due to tumor heterogeneity and varying degrees of extracranial involvement, resulting in an average overall survival of merely 5–6 months in CRC patients4,5. The treatment options include local treatments such as surgery, whole brain radiation therapy (WBRT), chemotherapy, and best supportive care.
Despite the poor outcomes for CRC patients with BM, there are still limited research on the pathogenesis and clinical studies. A study investigating the gene expression of 82 CRC patients with BM identified variables such as RAS mutations, the presence of lung metastases, and PD-L1 expression have been linked to BM in CRC patients6. Another study found that in metastatic colorectal cancer (mCRC) patients, a high frequency of RAS mutations (62%) is associated with a poor response to anti-EGFR treatment7. Recently, research on profiles indicating homologous recombination deficiency (HRD) and mismatch repair deficiency (MMRD) has also been conducted in CRC patients with BM. Jing Sun et al. performed whole-exome sequencing (WES) and whole-genome sequencing (WGS) on 19 BM CRC patients, discovering that unstable features, including DDR deficiency (HRD and MMRD), appeared in the CRC initial stages. This discovery indicates that immunotherapy could be a viable option for preventing and treating BM in patients with DDR deficiency8.
Guanine nucleotide-binding protein subunit gamma-2 (GNG2) is a member of the G-protein family, crucial for transducing extracellular signals into intracellular responses9,10. GNG2 contributes to various cellular functions, including proliferation, differentiation, and metastasis11,12,13. For instance, Yuan Feng found that polymorphisms in GNG2 genes are implicated in IgA nephropathy (IgAN) susceptibility in the Chinese Han population14. Siyuan Chen et al. found that interference with GNG2 could partially affect the influence of KCNJ2 in controlling proliferation, migration, and epithelial-to-mesenchymal transition (EMT) process of papillary thyroid carcinoma cells15. However, its specific role and mechanisms in highly invasive colorectal cancers remain largely unknown.
This research thoroughly analyzed the significance of GNG2 in metastatic CRC. Using bioinformatics tools such as TCGA, we evaluated GNG2 expression levels and their prognostic significance. Additionally, we conducted functional assays to explore GNG2’s influence on cell proliferation, migration, invasion and cell cycle regulation within CRC cell lines. Our results indicate that GNG2 may serve as a viable biomarker and treatment target for mCRC, providing new insights into its molecular basis driving tumor progression and metastasis.
Results
High-expression of GNG2 was linked to better prognosis in the metastatic colorectal cancer
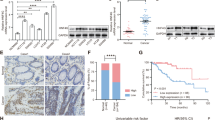
Our study investigated the mRNA expression patterns in normal tissues and metastatic tumors to identify the regulatory molecules of invasive colorectal cancer based on TCGA datasets. Among the 5133 differentially expressed genes (DEGs), 2453 were upregulated, while 2680 were downregulated (Fig. S1). The STRING database was utilized to examine the correlations among DEGs, and Cytoscape was employed to build a wide-ranging co-expression system (Fig. 1A). The degree, representing the quantity of interacting proteins associated with each molecule, was calculated with strongly linked molecules being displayed. As depicted in Fig. 1B, the highest protein among the top five molecules was GNG2 (degree 31), followed by PRKACB (degree 23), CYP3A4 (degree 19), AOX1 (degree 17), and CCL5 (degree 14). These molecules may play pivotal roles in CRC development.
High GNG2 expression is associated with better prognosis in CRC. (A) The Protein–Protein interaction (PPI) network map of differentially expressed genes (DEGs) between metastatic tumor tissue and normal tissue. Node size was proportional to its degree. Red nodes represented upregulated genes, and blue nodes represent downregulated genes. (B) The sequence of the top 30 genes with the highest degrees. (C) GNG2 expression levels were obtained from the TCGA database between metastatic tumor tissue and the normal tissue. (D) The pairing difference analysis of GNG2 expression between metastatic tumor tissue and normal tissue. (E) Kaplan-Meier survival for predicting GNG2 expression related overall survival (p = 0.015) using TCGA database. (F) GNG2 expression profiles in multiple cancers using TCGA database.
GNG2 gene expression patterns in both normal and metastatic CRC samples were explored in our study. A paired difference analysis was conducted to compare GNG2 expression between the two tissue types (Fig. 1C, D). Kaplan-Meier survival analysis demonstrated that elevated GNG2 levels are linked to improved overall survival (OS) in CRC patients (Fig. 1E). ROC curve analysis using TCGA data was also performed (Fig. S2). Additionally, we evaluated the differential expression of GNG2 between cancerous and normal samples in various cancers (Fig. 1F). GNG2 expression was found to be downregulated in multiple cancers, including colon adenocarcinoma (COAD), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), and kidney chromophobe (KICH).
Univariate and multivariate cox regression models indicated that lower GNG2 expression acted as an independent predictor of prognosis in CRC (hazard ratio [HR] = 0.6527, p = 0.04004) (Table 1). The baseline characteristics of the TCGA cohort (n = 88) were presented in Fig. 2A. Pie charts in Fig. 2B, C showed that increased GNG2 level were connected to superior overall survival (OS) in CRC patients.
Establishment of a prognostic nomogram associated with GNG2
A predictive nomogram model built on independent factors was developed using GNG2 expression, age, disease stage, and other clinical characteristics (Fig. 3A). The nomogram’s robustness and precision were assessed using calibration diagrams, demonstrating its effectiveness in estimating 1-, 3-, and 5-year overall survival (Fig. 3B–D). The model’s performance in outcome discrimination was evaluated using the concordance index (C-index) and AUC (Fig. 3E, F). Decision curve analysis (DCA) underscored the substantial clinical benefit in TCGA cohorts (Fig. 3G). These analysis results indicated that GNG2 had high prognostic value.
Construction of a prognostic nomogram. (A) Nomogram of TCGA colorectal carcinoma model between metastatic tissue and adjacent normal tissue. (B–D) Calibration curves of the nomogram model for 1-year, 3-year, and 5-year overall survival. (E, F) Kaplan-Meier survival and ROC curves of the nomogram model using TCGA database. (G) Decision curve analysis (DCA) for the clinical benefits and application of the nomogram.
The overexpression of GNG2 inhibits progression, migration and invasion in CRC
Five CRC cell lines were examined to evaluate their GNG2 protein levels in vitro. Low GNG2 expression was found in HCT116 and SW480 cells, while high GNG2 expression was found in HT29 and RKO cells (Fig. 4A, B). Furthermore, HCT116 and SW480 were identified as the most invasive cell lines among colorectal cancer cell lines16,17. Thus, HCT116 and SW480 cells were selected for the next round of in vitro and in vivo examinations.
GNG2 overexpression suppressed the proliferation and migration of CRC. (A, B) GNG2 protein expression in five CRC cell lines were examined. (C–E) The overexpression efficiency of GNG2 in CRC cells at protein and mRNA levels. (F, G) EdU assays suggest that overexpression of GNG2 suppressed the proliferation of CRC cells (scale bar = 50 μm). (H, I) CCK8 assays suggest that overexpression of GNG2 suppressed the proliferation of CRC cells. (J, K) Colony formation assays suggest that overexpression of GNG2 suppressed the proliferation of CRC cells. (L-N) Transwell assays suggest that overexpression of GNG2 suppressed the migration and invasion of CRC cells (scale bar = 100 μm).
The effectiveness of HCT116 and SW480 cell line overexpression was validated using western blotting and qRT-PCR (Fig. 4C–E). The results of EDU (Fig. 4F, G) assays showed the proliferation capacity was greatly suppressed in GNG2 overexpression groups. The EdU-positive cells were 14.51% in the GNG2 high expression group, compared to 41.17% in the untreated control group in SW480 cells (p < 0.05). Comparable outcomes were also noted in the HCT116 cell line. As shown in Fig. 4H, I, CCK8 assays confirmed a notable decline in CRC cell proliferation capacity after overexpressing GNG2 protein at 48 and 72 h. Colony formation assays (Fig. 4J, K) demonstrated a reduction in the average colony count for SW480 cells from 188 ± 9 in the control to 122 ± 10 in the GNG2 overexpression group (p < 0.05). However, in the HCT116 cell line, the mean number of colonies did not show a statistically significant difference. Transwell assays indicated that overexpression of GNG2 significantly suppressed CRC cell migration and invasion capacities (Fig. 4L, M, N).
GNG2 overexpression induces G0/G1 arrest and inhibits the PI3K/AKT/mTOR signaling pathway in CRC
In order to gain deeper insights into the molecular mechanisms by which GNG2 suppresses carcinogenesis, functional analyses, including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Set Enrichment Analysis (GSEA), were carried out. GO analysis demonstrated notable enrichment in several areas: signal transduction in biological processes, the plasma membrane in cellular components, and protein binding in molecular functions (Fig. S3). According to the analyzation of the KEGG pathway, the DEGs were primarily enriched in cancer tissue between the normal tissues and metastatic tumors, Neuroactive ligand − receptor interaction, Calcium signaling pathway, PI3K/AKT signaling pathway, etc. (Fig. 5A). GSEA analysis further demonstrated DEGs were related to the PI3K/AKT signaling and the cell cycle pathway (Fig. 5B-C).
GNG2 overexpression sensitizes CRC to G0/G1 arrest via PI3K/AKT/mTOR signaling pathway. (A) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis between GNG2 control and OE group in CRC cells. (B, C) GSEA revealed the enrichment of differential expressed genes in the RNA-seq dataset. (D–G) GNG2 overexpression induced G0/G1 arrest of CRC cells. (H) GNG2 overexpression significantly suppressed the PI3K/AKT/mTOR signaling pathway and metastasis ability in CRC cells.
After identifying the crucial role of GNG2 in cancer using bioinformatics methods, we validated our findings with in vitro experiments. As demonstrated in Fig. 5D–E, overexpression of GNG2 induced G0/G1 arrest in CRC cells. The quantitative analysis of the cell cycle was shown in Fig. 5F, G. Similarly, in the subsequent western blotting experiments, GNG2 overexpression was found to suppress key molecules of G0/G1 arrest, CDK4 and CDK6, thereby inhibiting cell entry into the S phase for division. For mechanistic experiments, our results showed that the GNG2 overexpression group suppressed the PI3K, p-AKT, and p-mTOR expression levels in HCT116 and SW480 tumors compared to the control group (Fig. 5H, Fig. S4).
Previous studies have shown that high levels of MMP expression are closely associated with tumor invasion and metastasis18,19. Among the matrix metalloproteinase (MMP) family, MMP2 plays a crucial role in the progression and drug resistance of colorectal cancer20. Additionally, the SMAD4 mutation has gradually become recognized as a marker for an increased risk of distant metastasis in colorectal cancer21. In stages I to IV colorectal cancer, 13–22% of patients with SMAD4 deletion are significantly associated with a higher risk of recurrence and metastasis22,23. Thus, we selected MMP-2 and SMAD4 as metastasis-related molecules for testing. In the GNG2 overexpression group, lower expression of MMP-2 and higher expression of SMAD4 were observed compared to the control group (Figs. 5H and S4).
GNG2 inhibits colorectal cancer brain metastasis in vivo
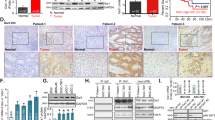
To examine the impact of GNG2 on tumor development, HCT116 cancer cells were intracranially injected into the brains of mice to build the orthotopic xenograft model. HCT116 cells consisted of high-expression and control groups (n = 5). The bioluminescence imaging was recorded and quantitative analysis of tumor bioluminescence imaging is presented in Fig. 6A, B. Body weights were also monitored (Fig. 6C). No notable variations in weight were observed between both sets of mice. As a tumor suppressor gene, GNG2 overexpression in HCT116 tumors resulted in a significantly reduced growth rate compared to the control group. The fluorescence intensity of HCT116 xenografts in the negative control group was 1.5 times higher than that in the GNG2 overexpression group. The overexpression of GNG2 prolonged the overall survival time of the mice.
GNG2 overexpression suppresses CRC progression in vivo. (A) Bioluminescence images of HCT116-luc cells with different GNG2 expression in Balb/c nude mice at different time points. (B, C) body weight and bioluminescence change curves in mice with different GNG2 expression levels. (D) Immunohistochemistry staining for PI3K/AKT/mTOR signaling pathway and G0/G1 arrest in whole-brain tumor tissue sections (scale bar = 100 μm). (E) Multiplex immunofluorescence staining image of PI3K/AKT signaling pathway (PI3K and p-AKT) and G0/G1 arrest marker (CDK4) in the tumor tissue (scale bar = 200 μm).
The results of the histological analyses were consistent with the in vitro findings from the orthotopic xenograft tumors. The representative protein markers of Immunohistochemistry and multiplex immunofluorescence were stained in whole-brain tumor section. Specifically, lower expression levels of PI3K, p-AKT, p-mTOR, CDK4, and CDK6 were noted in the GNG2 overexpression group compared to the negative control group in HCT116 tumors (Figs. 6D–E and S4).
Discussion
This research aimed to specify and present an extensive analysis of GNG2 in brain metastases (BM) from colorectal cancer (CRC), offering valuable insights into this largely unexplored area of research. Our primary objective was to discover a marker for diagnosis and prognosis, and by regulating the expression of this molecule, to set the stage for future targeted treatment strategies for this specific CRC subtype. The mechanism might involve G0/G1 cell cycle arrest via the PI3K/AKT/mTOR signaling pathway, which warrants further exploration and holds promise for clinical application.
Our study initially analyzed normal tissues and metastatic tumors using the TCGA CRC datasets, identifying 5,133 differentially expressed genes (DEGs). Through co-expression analysis and degree values, we discovered that GNG2 had the highest degree. Thus, our hypothesis suggests that GNG2 could be crucial in metastatic CRC. Further analysis revealed that GNG2 is expressed at low-levels in metastatic CRC tissue and may serve as a diagnostic biomarker, as supported by survival curve and nomogram analyses. Recently, the function and regulation of GNG2 have been extensively studied in various malignant cancers. Studies have shown that GNG2 overexpression suppresses the migration and invasion abilities of SK-Mel28 human melanoma cells by suppressing FAK phosphorylation13. What’s more, previous investigations also discovered that GNG2 is crucial in triple-negative breast cancer by being involved in TG2 inhibitor-induced apoptosis, regulating cadherin, Wnt, gastrin, and cholecystokinin receptor (CCKR)-mediated pathways, while also impacting the Warburg effect by lowering glycolytic enzyme levels24. Building on previous research, we conducted a further analysis of GNG2 expression in pan-cancer and found that GNG2 was expressed at low levels in most cancer types. Our findings further demonstrated that GNG2 expression was reduced in the datasets. Then, we verified the bioinformatics results through laboratory and animal-based experiments. Western blot (WB) analysis demonstrated that GNG2 was expressed at low levels in highly invasive CRC cell lines. The evidence suggests that GNG2 functions as a tumor inhibitory factor and is crucial for the prognosis and diagnosis of mCRC.
In recent years, the cell cycle has emerged as a focal point of research in metastatic CRC. Cannabis sativa Linne, a potential G0/G1 arrest inhibitor, has been shown to decrease the phosphorylation of MAPKs, including p38, ERK, and JNK, and to activate apoptosis in CRC with lung metastasis25. Prachi Bajpai et al. also found that G0/G1 arrest was induced by the combination of JAK/HDAC inhibitors and regorafenib. This treatment synergistically reduced PDX tumor growth and inhibited CRC lung metastasis, while increasing the infiltration of CD45 + cells and cytotoxic immune cells, thereby enhancing the antitumor immune response in a syngeneic mouse model26. Moreover, researchers have also found that CDK inhibitors delivered via nanotherapies promote CD8 + T cell infiltration and reduce the production of immunosuppressive cytokines in tumor-associated myeloid cells, thereby sensitizing colon cancer liver metastasis to immune checkpoint blockade (ICB) therapy27. Our studies first showed that high GNG2 expression significantly inhibited tumor proliferation, migration, invasion and induced G0/G1 cell cycle arrest.
In terms of mechanism, KEGG and GSEA analyses suggested that GNG2 appears to participate in the modulation of the PI3K/AKT/mTOR pathway. Moreover, we discovered that overexpression of GNG2 suppressed CRC progression and caused notable G0/G1 phase arrest via the PI3K/AKT/mTOR signaling pathway. The expression of MMP-2 and SMAD4, as metastasis-related molecules, was also assessed. And GNG2 can suppress the metastasis ability of colorectal cancer. These findings align with earlier studies suggesting that the GNG2/PI3K/AKT/mTOR signaling pathway plays a role in high invasiveness and metastatic tumors. Anjiang Zhao et al. have found that GNG2’s interaction with MRAS appears to suppress AKT and ERK phosphorylation, thereby facilitating apoptosis and reducing proliferation in breast cancer cells, particularly in triple-negative breast cancer (TNBC), which is known for its high invasiveness and metastatic potential12. GNG2 is regulated by two lncRNAs through the AMPK, AKT, mTOR, and TGF-β signaling pathways, subsequently modulate autophagy and affect temozolomide resistance, tumor recurrence, and metastasis in glioblastoma28. Recent research has found that the PI3K/AKT pathway is activated by ITIH4, resulting in the inhibition of apoptosis and, consequently, reducing the sensitivity of mCRC to 5-FU29. From bench to bedside, clinical testing is ongoing for PI3K/AKT/mTOR pathway inhibitors. The list of drugs includes inhibitors targeting PI3K, dual PI3K/mTOR, and AKT pathways30. PIK3CA mutations are present in approximately 13% of mCRC cases and are associated with right-sided CRC, older patients, metastatic disease, and poor prognosis31,32. During an initial dose-escalation and extension study (NCT01219699), alpelisib, an oral phosphatidylinositol 3-kinase a (PI3Ka)–selective inhibitor was evaluated in 35 mCRC patients, comprising 26.1% of the study population. Among them, two patients achieved PR. The observed DCR was 34.3%, while the clinical benefit rate (CBR: including complete response, partial response, and stable disease for over 24 weeks) was 8.6%33.
Although we observed that GNG2 overexpression may regulate CRC progression and G0/G1 arrest through the PI3K/AKT/mTOR signaling pathway, the precise mechanisms remain unclear, necessitating further investigation. Our study conducted animal experiments and found that GNG2 overexpression significantly inhibited brain metastasis of CRC cells compared to the control group, with no significant effects on body weight. Further immunohistochemistry (IHC) and multiplex immunofluorescence (mIF) experiments suggested that the PI3K/AKT/mTOR signaling could be the potential mechanisms through which GNG2 exerts its effects. Thus, high-level expression of GNG2 is closely related to better prognosis and could serve as a predictive marker and treatment target in the brain metastases of CRC.
Conclusion
CRC BM with GNG2 low-expression is associated with highly invasiveness. GNG2 may suppress CRC progression via the PI3K/AKT/mTOR axis in the brain metastases of CRC.
Methods
Data collection and processing
Data on RNA sequencing and associated clinical outcomes related to CRC patients were obtained from The Cancer Genome Atlas (TCGA) database(https://gdac.broadinstitute.org/), including normal tissues and tumor tissues. Univariate and multivariate cox regression was performed to identify genes among the differentially expressed genes (DEGs) between normal tissues and metastatic tumor tissue in the TCGA dataset. Using the optimal cutoff value, the patient population was stratified into low-risk and high-risk GNG2 segments. Kaplan-Meier survival curves along with time-dependent receiver operating characteristic (ROC) plots were used to determine the predictive effectiveness of our risk model. The R package “ggplot2” was employed to analyze the GNG2 mRNA expression levels in multiple cancer types.
Functional and pathway enrichment analysis
The R package was utilized to carry out functional and pathway enrichment analysis, investigating the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases34,35,36. Terms with a p-value below 0.05 and a false discovery rate (FDR) less than 0.05 were selected for further functional clustering. Gene set enrichment analysis was performed via the GSEA software.
Protein–protein Interaction (PPI) Network Construction
The STRING10.5 online database (https://string-db.org/) was utilized to analyze all differentially expressed genes (DEGs) and build a protein-protein interaction (PPI) network. Cytoscape software was used for statistical analysis of nodes, edges, and degrees, while R software was utilized to visualize the statistical results.
Nomogram model
Based on the results of multivariate cox regression analysis, a predictive nomogram model was constructed through the ‘rms’ and ‘survival’ R packages. This nomogram projects the overall survival probabilities at 1, 3, and 5 years for CRC patients, and calibration curves were drawn to confirm its precision. In addition, we performed decision curve analysis (DCA) to determine the net clinical advantages provided by the nomogram model.
Cell Culture and stable transfection
HT29, HCT116, Caco2, RKO, and SW480 human CRC cell lines were acquired from Biomedical Sciences Institutes and maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 °C in a humidified 5% CO2 incubator. Then lentivirus overexpressing GNG2 (OE-GNG2 group) or a negative control (NC group) was transfected into HCT116 and SW480 cells using TurboFect (ThermoFisher, Carlsbad, CA). Stably transfected cell clones were selected using 2 µg/mL puromycin (ThermoFisher, Carlsbad, CA) over a period of two weeks.
Real-time quantitative PCR (RT-qPCR)
TRIzol reagent (ET101-01, Transgen biotech, China) was utilized to isolate total RNA. cDNA synthesis was carried out with Thermo Scientific RevertAid RT kits (Thermo Fisher Scientific, Carlsbad, CA) according to the manufacturer’s guidelines. qRT-PCR was conducted with a SYBR Green mix (Thermo Fisher Scientific, Carlsbad, CA) on the ViiA™ 7 RT-PCR system. Real-time PCR was conducted using the following primers: GNG2: forward: CTCCGTGGGCAACTCCTACT; reverse: ACTGGCAGATCTTAGCTCCC. GAPDH: forward: TGTGGGCATCAATGGATTTGG; reverse: ACACCATGATGGGGGGGGGATCAAT. GAPDH was used as the normalization gene. The relative mRNA expressions were quantified through the 2-ΔΔCt method.
Western blotting assay
Total protein was isolated using RIPA buffer (89900, Thermo Fisher Scientific, Carlsbad, CA) containing a protease inhibitor (P1006, Beyotime, Shanghai, China).After separation by 10% SDS-PAGE, proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (88518, Thermo Fisher Scientific, Carlsbad, CA). After a 2-hour incubation in 5% skim milk at 37 °C to block non-specific binding, the membranes were exposed to the target proteins: GNG2 (ab198225, Abcam, UK), PI3K (ab151549, Abcam, UK), AKT/phospho-AKT (ab8805/ab38449, Abcam, UK), CDK4 (ab108357, Abcam, UK), CDK6 (19117-1-AP, Proteintech, China), mTOR/phospho-mTOR (66888-1-Ig/ 67778-1-Ig, Proteintech, China), MMP-2 (ab86607, Abcam, UK) and SMAD4 (17387-1-AP, Proteintech, China) at 4 °C. Following a 2-hour blocking step in 5% skim milk at 37 °C, the membranes were exposed to specific primary antibodies: Goat Anti-Mouse IgG H&L (HRP) (ab6789, Abcam, UK) or Goat Anti-Rabbit IgG H&L (HRP) (ab6721, Abcam, UK) for 30 min at room temperature. The detection was carried out with the ChemiDocXRS + System, and Image Lab software was used to analyze protein expression, normalizing the levels to β-actin (ab210083, Abcam, UK).
Cell proliferation assays
Each well of the 96-well plates was seeded with 3,000 cells with 100 µl of complete medium and cultured during the night. At 0, 24, 48, and 72 h, 10 µl of CCK-8 solution (Biosharp, China) was added to each well, followed by a 2-hour incubation at 37 °C. Absorbance was read at 450 nm using a Microplate Reader. Subsequent absorbance readings were used to generate the cell growth curve.
EdU assays
A total of 3,000 cells in 100 µl of complete medium were plated into each well of the 96-well plates and cells were left to incubate across the night. After 24 h, 20 µM EdU labeling medium was used to grow with cells for 3 h. Following fixation with 4% paraformaldehyde, the cells were incubated with 100 µl of Apollo dye solution (C10310-1, RiboBIO, China) for 30 min. Afterward, the samples were washed with 0.5% Triton X-100 for 10 min. Finally, cells were stained with DAPI, and positive cells were subsequently counted using an N2-DMi8 microscope (Leica, Wetzlar, Germany).
Colony formation assay
For colony formation, 500 cells were plated per well in 6-well plates and cultured for 14 days. After fixation with methanol for 20 min, crystal violet (C0121, Beyotime, Shanghai, China) was applied for 20 min. Colonies were quantified using ImageJ software.
Cell cycle test
In 6-well plates, 100,000 cells were seeded in 2 ml of complete medium and were allowed to incubate for 12 h. Then, The application of DNA staining and permeabilization solution (CCS012, Multi Sciences, China) lasted for 30 min at 37 °C. Flow cytometry was utilized to analyze the results.
Cell migration and invasion assay
After twenty-four hours post-transfection, 4 × 104 HCT116 or SW480 cells were collected and resuspended in 100 µL of serum-free medium. Cells were subsequently transferred to the upper side of a Transwell insert (Millipore). For the invasion assays, Matrigel was used to coat the Transwell chambers, while 800 µL of 10% FBS medium was added to the lower compartment. In contrast, no Matrigel was applied for the migration assays. After a 48-hour cultivation at 37 °C, the cells on the inferior side were then rinsed with phosphate-buffered saline, fixed with 4% paraformaldehyde, and then dyed with basic violet. Photographs were subsequently taken to analyze stained cells using an N2-DMi8 microscope.
In vivo orthotopic xenograft studies
All experimental procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals (NIH). The study is reported in accordance with ARRIVE guidelines (https://arriveguidelines.org). The study was approved by the Experimental Animal Ethics Committee (ID: SJA2023037-2). Balb/c nude mice were administered with 1% pentobarbital sodium for anesthesia and placed on a stereotaxic apparatus for immobilization (RWD Life Science) in the orthotopic implantation tumor model. Then, 4 µL of HCT116-luc cells (5 × 105 ) were inserted into the brain parenchyma (right lateral: 2.0 mm, bregma: 2.0 mm, depth: 3 mm) stereotactically. Intracranial colorectal cancer cells growth was monitored by bioluminescence imaging (AniView100, Biotechnology Co., Ltd, China).
Hematoxylin-eosin (HE) and immunohistochemistry (IHC)
At the end of the study, the animals were euthanized through intraperitoneal injection of an overdose of ketamine (100 mg/kg) and xylazine (10 mg/kg), adjusted according to the animal’s response and condition. And all tumors were collected for further investigation. Then 10% formalin was utilized to fix the whole-brian xenograft tumor tissues, which were then dehydrated, encased in paraffin, and sectioned into 4 μm slices. The sections were dewaxed, hydrated, and underwent antigen retrieval and endogenous peroxidase blocking. The sections were maintained with primary antibodies at 4 °C throughout the night: PI3K (20584-1-AP, Proteintech, China), AKT/phospho-AKT (ab8805/ab38449, Abcam, UK), CDK4 (ab108357, Abcam, UK), CDK6 (19117-1-AP, Proteintech, China) and mTOR/phospho-mTOR (66888-1-Ig/ 67778-1-Ig, Proteintech, China). Subsequently, secondary antibodies were applied to the slides and kept at room temperature for one hour. After DAB and hematoxylin staining, positive cells were counted using a microscope.
Multiplex immunofluorescence (mIF)
FFPE tissue sections were stained using the mIHC Fluorescence kit (RC0086-34RM, RecordBio, Shanghai, China) following the manufacturer’s guidelines. The process involved deparaffinization with xylene, hydration through a series of gradient ethanol solutions, antigen retrieval using sodium citrate buffer (0.01 M, pH 6.0), removal of endogenous peroxidase with H2O2, blocking with 10% goat serum, and staining with specific antibodies followed by TSA-RM. Each staining cycle began with antigen retrieval and concluded with TSA labeling. The antibodies used were: PI3K (20584-1-AP, Proteintech, China) with TYR-620, p-AKT (80455-1-RR, Proteintech, China ) with TYR-690, CDK4 (ab108357, Abcam, UK) with TYR-570. Nuclei were stained with DAPI. All slides were then scanned and analyzed using the Pannoramic MIDI II pathology imaging microscope (3D HISTECH, Hungary).
Statistical analysis
Group differences were assessed using T-tests. The statistical analysis was done using SPSS 17.0 and significance was defined as a p-value below 0.05. Data were presented as average values ± SD.
Data availability
The datasets used and analysed during the current study available from the corresponding author on reasonable request.
References
Mongan, J. P. et al. Brain metastases from colorectal cancer: Risk factors, incidence, and the possible role of chemokines. Clin. Colorectal Cancer 8, 100–105. https://doi.org/10.3816/CCC.2009.n.016 (2009).
Michl, M. et al. Brain metastasis in colorectal cancer patients: Survival and analysis of prognostic factors. Clin. Colorectal Cancer 14, 281–290. https://doi.org/10.1016/j.clcc.2015.05.009 (2015).
Thurmaier, J. et al. Patients with colorectal cancer and brain metastasis: The relevance of extracranial metastatic patterns predicting time intervals to first occurrence of intracranial metastasis and survival. Int. J. Cancer 148, 1919–1927. https://doi.org/10.1002/ijc.33364 (2021).
Cagney, D. N. et al. Incidence and prognosis of patients with brain metastases at diagnosis of systemic malignancy: A population-based study. Neuro-oncology 19, 1511–1521. https://doi.org/10.1093/neuonc/nox077 (2017).
Roussille, P. et al. Prognostic factors of colorectal cancer patients with brain metastases. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 158, 67–73. https://doi.org/10.1016/j.radonc.2021.02.006 (2021).
Roussille, P. et al. Pathological and molecular characteristics of colorectal cancer with brain metastases. Cancers 10. https://doi.org/10.3390/cancers10120504 (2018).
Douillard, J. Y. et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N. Engl. J. Med. 369, 1023–1034. https://doi.org/10.1056/NEJMoa1305275 (2013).
Sun, J. et al. Genomic signatures reveal DNA damage response deficiency in colorectal cancer brain metastases. Nat. Commun. 10, 3190. https://doi.org/10.1038/s41467-019-10987-3 (2019).
Leung, T. et al. Zebrafish G protein gamma2 is required for VEGF signaling during angiogenesis. Blood 108, 160–166. https://doi.org/10.1182/blood-2005-09-3706 (2006).
Lai, W. S. & Ding, Y. L. GNG7 silencing promotes the proliferation and differentiation of placental cytotrophoblasts in preeclampsia rats through activation of the mTOR signaling pathway. Int. J. Mol. Med. 43, 1939–1950. https://doi.org/10.3892/ijmm.2019.4129 (2019).
Yajima, I., Kumasaka, M. Y., Tamura, H., Ohgami, N. & Kato, M. Functional analysis of GNG2 in human malignant melanoma cells. J. Dermatol. Sci. 68, 172–178. https://doi.org/10.1016/j.jdermsci.2012.09.009 (2012).
Zhao, A. et al. GNG2 acts as a tumor suppressor in breast cancer through stimulating MRAS signaling. Cell Death Dis. 13, 260. https://doi.org/10.1038/s41419-022-04690-3 (2022).
Yajima, I. et al. GNG2 inhibits invasion of human malignant melanoma cells with decreased FAK activity. Am. J. Cancer Res. 4, 182–188 (2014).
Feng, Y. et al. 3’UTR variants of TNS3, PHLDB1, NTN4, and GNG2 genes are associated with IgA nephropathy risk in Chinese Han population. Int. Immunopharmacol. 71, 295–300. https://doi.org/10.1016/j.intimp.2019.03.041 (2019).
Chen, S., Huang, M. & Hu, X. Interference with KCNJ2 inhibits proliferation, migration and EMT progression of apillary thyroid carcinoma cells by upregulating GNG2 expression. Mol. Med. Rep. 24 https://doi.org/10.3892/mmr.2021.12261 (2021).
Berg, K. C. G. et al. Multi-omics of 34 colorectal cancer cell lines—A resource for biomedical studies. Mol. Cancer 16 https://doi.org/10.1186/s12943-017-0691-y (2017).
Heck, K. A. et al. Characterisation of colorectal cancer cell lines through proteomic profiling of their extracellular vesicles. Proteomes 11 https://doi.org/10.3390/proteomes11010003 (2023).
Hernandez-Barrantes, S., Bernardo, M., Toth, M. & Fridman, R. Regulation of membrane type-matrix metalloproteinases. Sem. Cancer Biol. 12, 131–138. https://doi.org/10.1006/scbi.2001.0421 (2002).
Castro-Castro, A. et al. Cellular and molecular mechanisms of MT1-MMP-dependent cancer cell invasion. Annu. Rev. Cell Dev. Biol. 32, 555–576. https://doi.org/10.1146/annurev-cellbio-111315-125227 (2016).
Sun, J., Zhang, Z., Chen, J., Xue, M. & Pan, X. ELTD1 promotes invasion and metastasis by activating MMP2 in colorectal cancer. Int. J. Biol. Sci. 17, 3048–3058. https://doi.org/10.7150/ijbs.62293 (2021).
Huang, D. et al. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev. 37, 173–187. https://doi.org/10.1007/s10555-017-9726-5 (2018).
Niu, B. et al. CCL9/CCR1 axis-driven chemotactic nanovesicles for attenuating metastasis of SMAD4-deficient colorectal cancer by trapping TGF-β. Acta Pharm. Sin. B 14, 3711–3729. https://doi.org/10.1016/j.apsb.2024.05.009 (2024).
Wasserman, I. et al. SMAD4 loss in colorectal cancer patients correlates with recurrence, loss of immune infiltrate, and chemoresistance. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 25, 1948–1956. https://doi.org/10.1158/1078-0432.Ccr-18-1726 (2019).
Ancona, P. et al. Transcriptomics studies reveal functions of transglutaminase 2 in breast cancer cells using membrane permeable and impermeable inhibitors. J. Mol. Biol. 436, 168569. https://doi.org/10.1016/j.jmb.2024.168569 (2024).
Mun, J. G. et al. Supercritical extract of cannabis sativa inhibits lung metastasis in colorectal cancer cells by increasing AMPK and MAPKs-mediated apoptosis and cell cycle arrest. Nutrients 14. https://doi.org/10.3390/nu14214548 (2022).
Bajpai, P. et al. Combination of dual JAK/HDAC inhibitor with regorafenib synergistically reduces tumor growth, metastasis, and regorafenib-induced toxicity in colorectal cancer. J. Exp. Clin. Cancer Res. CR 43, 192. https://doi.org/10.1186/s13046-024-03106-8 (2024).
Ding, D. et al. Nanodrug modified with engineered cell membrane targets CDKs to activate aPD-L1 immunotherapy against liver metastasis of immune-desert colon cancer. J. Controll. Release Off. J. Controll. Release Soc. 369, 309–324. https://doi.org/10.1016/j.jconrel.2024.03.052 (2024).
Nayak, R. & Mallick, B. LncRNA-associated competing endogenous RNA network analysis uncovered key lncRNAs involved in temozolomide resistance and tumor recurrence of glioblastoma. J. Mol. Recognit. JMR 36, e3060. https://doi.org/10.1002/jmr.3060 (2023).
Zhao, Y. et al. Discovery, identification and mechanism of chemosensitivity-relate biomarker inter-α-trypsin inhibitor heavy chain 4 in metastatic colorectal cancer. Heliyon 10, e33571. https://doi.org/10.1016/j.heliyon.2024.e33571 (2024).
Martínez-Pérez, J., Torrado, C. & Domínguez-Cejudo, M. A. Valladares-Ayerbes, M. Targeted treatment against cancer stem cells in colorectal cancer. Int. J. Mol. Sci. 25 https://doi.org/10.3390/ijms25116220 (2024).
Sastre, J. et al. Influence of BRAF and PIK3CA mutations on the efficacy of FOLFIRI plus bevacizumab or cetuximab as first-line therapy in patients with RAS wild-type metastatic colorectal carcinoma and < 3 baseline circulating tumour cells: The randomised phase II VISNÚ-2 study. ESMO Open 6, 100062. https://doi.org/10.1016/j.esmoop.2021.100062 (2021).
Wang, Q. et al. PIK3CA mutations confer resistance to first-line chemotherapy in colorectal cancer. Cell Death Dis. 9 https://doi.org/10.1038/s41419-018-0776-6 (2018).
Juric, D. et al. Phosphatidylinositol 3-kinase α-selective inhibition with alpelisib (BYL719) in PIK3CA-altered solid tumors: Results from the first-in-human study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 36, 1291–1299. https://doi.org/10.1200/jco.2017.72.7107 (2018).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–d592. https://doi.org/10.1093/nar/gkac963 (2023).
Acknowledgements
All the authors are thankful to the support from Third Xiangya Hospital, Central South University.
Author information
Authors and Affiliations
Contributions
CH.L. and WL.Y. designed the study, processed bioinformatics data, exhibited experimental validation, and wrote the main manuscript text. ZM.X. analyzed the data and revised the full text. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Luo, C., Xiao, Z. & Yang, W. GNG2 inhibits brain metastases from colorectal cancer via PI3K/AKT/mTOR signaling pathway. Sci Rep 15, 1787 (2025). https://doi.org/10.1038/s41598-025-85592-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-85592-0
Keywords
This article is cited by
-
Rap2B-mediated reprogramming of the PI3K/AKT signaling axis drives resistance to cetuximab-targeted therapy in colorectal carcinoma
Biological Procedures Online (2025)
-
Investigating the effects of miR-526b and miR-655 on doxorubicin sensitivity in breast cancer
Scientific Reports (2025)