Abstract
Genotype-informed anticancer therapies such as BRAF inhibitors can show remarkable clinical efficacy in BRAF-mutant melanoma; however, drug resistance poses a major hurdle to successful cancer treatment. Many resistance events to targeted therapies have been identified, suggesting a complex path to improve therapeutics. Here, we showed the utility of a piggyBac transposon activation mutagenesis screen for the efficient identification of genes that are resistant to BRAF inhibition in melanoma. Although several forward genetic screens performed in the same context have identified a broad range of resistance genes that poorly overlap, an integrative analysis revealed a much smaller functional diversity of resistance mechanisms, including reactivation of the MAPK pathway, PI3K-AKT pathway, and Hippo pathway, suggesting that a relatively small number of therapeutic strategies might overcome resistance manifested by a large gene set. Moreover, we illustrated the pivotal role of the Hippo pathway effector TAZ (encoded by the WWTR1 gene) in mediating BRAF inhibition resistance through transcriptional regulation of receptor tyrosine kinases and through interactions with the E3 ubiquitin ligase NEDD4L.
Similar content being viewed by others
Introduction
A vast array of genetic and nongenetic events, such as point mutation, overexpression, translocation, and alternative splicing, occurring across many genes have been shown to cause resistance to BRAF inhibitors in melanoma and other tumor types1,2,3,4,5,6,7,8,9. Although the analysis of clinical samples provides unique insight into therapeutically relevant resistance tumor heterogeneity, limitations in the availability of clinical samples and functional characterization of the human genome combined with a high number of passenger mutations limit the potential to identify low occurrence resistance events10,11,12,13. To prospectively identify genes that can promote resistance to BRAF and MEK inhibition several landmark studies using genetic screens in the context of BRAFV600E mutant melanoma have been performed. These include gain-of-function (GoF) open reading frame (ORF) and aptamer-mediated dCAS9-activator screens, and loss-of-function (LoF) CRISPR knockout and shRNA screens14,15,16,17. These screens leverage collections of individually made elements, are rather labor-intensive and are associated with relatively high costs. Even the most advanced of these screening approaches have limitations leading to incomplete saturation; for example, an ORF library is typically limited to one isoform per gene, and noncoding GoF events are poorly covered by most screens. Interestingly, these screens and several transposon mutagenesis screens that have been previously performed in the BRAFV600E melanoma cell lines offer insight into the complexity and diversity of the genes that can mediate resistance to a given therapeutic agent in a specific genomic context. Comparison of the results of these screens revealed different gene sets identified even by screens that were functionally analogous (gain- or loss-of-function) and well validated18,19,20. This raises several interesting questions such as whether screens that are genome-wide by design have indeed reached a substantial level of saturation, how many genes in total can mediate resistance and whether they fall into common resistance pathways.
We previously developed an in vitro piggyBac (PB) transposon activation mutagenesis screen with unbiased genome-wide insertions that activated, and in some cases disrupted, endogenous genes and demonstrated its applications in cancer cell tolerance to the cytotoxic reagent paclitaxel and in host cell resistance to Ebola or coronaviruses21,22. Here, we used this approach to further study resistance to BRAF inhibition in BRAFV600E melanoma.
Results
A transposon screen to identify BRAF inhibitor resistance genes
To identify genes conferring resistance to BRAF inhibition in melanoma, ten individual PB mutagenesis libraries were constructed by transfecting independently cultured PLX4720-sensitive BRAFV600E A375 melanoma cells with the PB and transposase (PBase) plasmids (Fig. 1A, B). PLX4720 is an analog of vemurafenib (PLX4032), the first small molecule inhibitor drug clinically approved to treat BRAF-mutant advanced malignant melanoma1. Each library was treated with PLX4720 (Fig. 1C), and insertion sites were identified from each resistant pool by targeted deep sequencing.
PB transposon mutagenesis screen. (A) Scheme of the PB mutagenesis screening process. Parental cancer cells were transfected with transposition (PB) and transposase (PBase) plasmids, passaged with puromycin to generate mutagenized libraries, and treated with PLX4720 to generate resistant colonies or pools. Insert sites were detected using linker-mediated PCR and NGS, with the shown amplicon structure of the transposon (PB), host genomic DNA (gDNA), linker, and PCR primers (arrows). (B) The PB transposition plasmid includes a cassette with an antibiotic selection marker (puroR) for transposon-mutagenized cell maintenance, a CMV promoter (CMV) and a splice donor (SD) for host gene transcription and alternative splicing, and two inverted repeats (IRs) for transposition. (C) Clonogenic assays showing the sensitive parental cells and PLX4720-resistant cells from transposon screen. (D) High confidence hits identified from resistant pools are displayed by genomic location (x-axis: chrs.1–22,x). The inserts for each gene were summarized, read numbers of each gene normalized to the total number of reads were plotted as y-coordinates, and the dot sizes represent the numbers of insertion events. Genes with a predicted activation effect are shown as orange bubbles, and candidates for follow-up validation assays are circled and labeled.
To first gain insight into the breadth of target drug resistance events that can be captured with this approach, the overall distribution of insertions across the genome was analyzed. This analysis revealed that inserts were evenly distributed across the genome without major local preference, highlighting the genome-wide coverage ability of this approach (see also a previous report21). Supporting the mechanistic specificity of the insertion events, a strong enrichment of sequencing reads for a small number of loci was observed. The top 100 inserts represented most (> 99.5%) of the sequencing reads (Fig. S1A) of each pool and corresponded to 902 genomic loci associated with 767 genes (Table S1) based on most proximal genes (see below). We considered the top 132 genes of this aggregated list constituting 95% of sequencing reads, and including all 128 genes with multiple inserts, as high-confidence hits (Fig. 1D). We note however that some pathologically relevant genes (such as PTEN, ranked 134) might have been omitted by this prioritization strategy.
To confirm that the analytical approach identified driver rather than passenger events, 224 colonies, including 23 isolated directly from the primary screening plates and 201 derived by limiting dilution of the multipassaged individual resistant pools, were sequenced (Table S2). On average, 2.2 inserts (ranging from 1 to 12) were found per clone (Fig. S1B), indicating that co-enrichment of passenger inserts in resistant cells was infrequent and thus indicating that candidate resistance-driving genes should be readily identified by considering high read count genes and proximity to redundant insertion events.
To characterize the functional consequence of the insertions, we performed RNA-Seq on 10 clones and plotted the differential gene expression according to the insertion orientation and proximity (Fig. S1C, D, Table S3). This analysis indicated that, as predicted and consistent with our previous report21, expression variations mostly affected the nearest gene (13 of 16 inserts, including LPP, the second nearest gene after an undetectable antisense isoform LPP-AS2, p < 0.05, q < 0.05) and that upstream/sense-strand inserts result in activation of the host gene transcription. We also noted that an insertion as far as 48 kb robustly upregulated the expression of the nearest gene (LPP) (20.8-fold, p = 0). While genome structural and regulatory complexity are likely to impact how far a PB insertion can activate a gene, alternative splicing involving splice donor (SD) and acceptor (SA) site selection is affected by promoter activity and the transcription elongation rate23,24, and several human genes, such as dystrophin (DMD), indeed contain introns well over 100 kb. Thus, the capacity for distal activation demonstrated here is likely due to CMV-initiated strong transcriptional readthrough and the coupling of the ectopic SD to an endogenous SA and may therefore substantially reduce the number of clones needed for genome-wide coverage compared to the proximal effect alone. In addition to upstream sense-strand events, intragenic sense-strand inserts at the 5’ end of the gene or the protein coding sequence, effectively analogous to 5’ alternative splicing isoforms, were also observed to activate transcription in clones containing NEDD4L, BRAF, or MCF2.
Of the 132 high-confidence hits, 52 were associated with multiple incidences of nonidentical sense-strand, upstream or proximal, insertion events and thus most likely resulted in host gene overexpression (Fig. 1D, Table S1). Interestingly, the PB approach might also be capable of identifying resistance through LoF in addition to GOF events. Conceptually, LoF could occur either through insertion in the gene body or through insertion downstream of a gene in the opposite direction to transcription, leading to gene disruption or antisense RNA expression, among other mechanisms. In line with this possibility and with the previous identification of disruption of the NPC1 gene in a similar transposon-mediated screen for viral invasion22, several potentially disruptive insertions were found in genes such as GPC6 and PLS3, and one was discovered in the well-characterized resistance gene PTEN, a lipid phosphatase that acts as a tumor suppressor to counteract the activity of phosphoinositide 3-kinases (PI3K) with PI3K activity promoting proliferation and survival (Table S1-S3). However, robust identification of such LoF driven resistance events is complicated by the broad range of insertion positions and disruption of gene structures that could underlie LoF. In addition, in the context of a diploid genome, LoF might only involve haploinsufficient genes, although these could actually constitute a large fraction of human genes25. Furthermore, cancer cell genomes are frequently not simply diploid due to whole genome duplication or other complex genomic rearrangements. For these different reasons, below, we focus on GoF events.
Confirming the ability of the PB approach to identify functionally relevant and drug target-related resistance events, BRAF (BRAFV600E, the direct target of inhibition, and in which A375 is homozygous for that mutation), was among the top hit genes with 8 insertion events (Fig. 1D, Table S1). Interestingly, although not the most prominent target in the primary screening, the majority (140/201) of subclones isolated by limiting dilution of the pools contained BRAF insertions (Table S2), implying that BRAFV600E overexpression itself promotes a strong proliferative advantage, an observation in line with a previously published study18. Two insertion events were mapped to RAF1 (CRAF), a paralog of BRAF, and a predicted activating insert was also found 2.7 kb upstream of the KRAS gene, the upstream activator of RAF kinases in the mitogen-activated protein kinase (MAPK) signaling cascade (Table S1). Interestingly, among the high-confidence hits, the only other kinase predicted to be upregulated was the SRC family tyrosine kinase gene YES1. SRC kinases, particularly SRC, FYN and YES (encoded by YES1), are indeed positive regulators of RAF kinases and are known to be upstream activators of the MAPK pathway in many contexts, including BRAF inhibitor resistance26. Other candidate resistance genes belong to a broad variety of functional classes. Multiple guanine exchange factors, including three RAP exchange factors (RAPGEF2, RAPGEF4, and RAPGEF6) and two Rho exchange factors (VAV1 and ARHGEF28), were among the high-confidence hits, and another Rho exchange factor (MCF2) ranked 540 from the pools (Table S1), whose RNA expression was also induced 33-fold by an intragenic sense-strand insert in a clone (Fig. S1D, Table S3). Several transcriptional regulators were found among the most redundant hit genes such as EYA1 (11 inserts), POU2F1 (9 inserts), and the zinc finger proteins ZFHX3 and ZFHX4 (7 inserts each). Chromatin binding proteins (ARID1A, SMC2, ACIN1, and PDS5A) and other gene family members, such as sorting nexins (SNX6 and SNX14), semaphorins (SEMA3D and SMA6A) and potassium (Kv) channel interacting proteins (KCNIP1 and KCNIP4), were also identified. Notably, these genes do not colocalize in the host genome, suggesting functionally meaningful enrichment.
Validation of resistance genes
To validate the results of the transposon screen we selected 15 gene candidates based on the predicted activation effect for single-gene ORF-mediated overexpression (circled and labeled in Fig. 1D). The expression of these genes was mediated through lentiviral infection in A375 cells, and cellular proliferation was measured in the presence of the BRAF inhibitor PLX4720, the pan-RAF inhibitor AZ628, and the MEK inhibitor AZD6244 (MEK is immediately downstream of RAF in the MAPK pathway) (Fig. S2). Seven candidates, NEDD4L, WWTR1 (encoding TAZ, a Hippo pathway component), ESRRG, RAPGEF6, ARHGEF28, YES1, and POU2F1 were further confirmed to enhance viability in the presence of inhibitors using dose titration assays (Fig. 2A), proliferation rate assays (Fig. 2B) and long-term clonogenic assays (Fig. 2C). As a frame of reference, the expression of these genes conferred a level of resistance to RAF and MEK inhibition comparable to that of the previously reported and well-validated resistance gene COT27. Supporting the specificity of the findings for BRAF inhibition, none of these genes induced resistance to the microtubule-disrupting agent paclitaxel while expression of the drug exporter ABCB1 gene did, as expected (Fig. S2B)21.
Effect of candidate resistance genes on cell proliferation evaluated by cDNA overexpression. (A) Half-maximal inhibition concentration (IC50) values of cells expressing the vector controls, candidate resistance genes, and a previously known resistance gene, COT. Experiments were performed on two sets of lentiviral vectors (left: blasticidin; right: puromycin). Values are means ± 95% confidence intervals (CIs). (B) Proliferation rate of cells transfected with candidate genes, either untreated (ND) or treated with 0.5 µM PLX4720. CellTiter-Glo measurements 4 days after drug treatment were normalized to the Day-0 values. Data are presented as mean ± standard error of the mean (SEM), n = 3. (C) Clonogenic assay with cells treated with the corresponding drugs at twofold serial dilutions.
We note that among the genes that were not validated, some were likely due to the limitations of the ORF clones. For example, AKAP13 was only modestly overexpressed (1.5- to 2.2-fold greater than basal expression, Fig. S2C) but did induce partial resistance to PLX4720; BRAF wild-type cDNA also did not induce resistance, consistent with previously reported ORF screening results that did not identify wild-type BRAF as a resistance gene15, likely because only the V600 mutation allele overexpression can drive resistance3, which is what occurs upon transposon insertion in A375 cells. Furthermore, the available NEDD4L ORF construct used in the ORF screen and initially tested in our validation studies corresponded to a WW2 domain deletion isoform that failed to induce resistance (named herein NEDD4L-dWW2 to avoid confusion and further described below)28, highlighting the possibility that specific ORF inactivity or particular isoforms can contribute to false negatives in ORF based genetic studies. Overall, the results show a high rate of validation for the top transposon screen hits.
Concordance of multiple genetic screens
The exact model of melanoma used here (the A375 cell line) was previously used to identify resistance genes via multiple screening modalities14,15,16,17. Surprisingly, the overlap between the lists of genes identified previously and the top 132 genes we identified was limited to 6 genes in the case of a previously reported ORF screen (ESRRG, FOXP2, RAF1, RAPGEF4, VAV1, and WWTR1) and 6 different genes in the case of a dCAS9-activator screen (ATP10A, BCAS3, GLIS3, MECOM, PCDH7, and ZFHX4). Although this could be explained at least in part by incomplete genome coverage of each library with some events functionally tested in only one of the screens, this very limited overlap across likely true positive hits (based on validation rates in the different studies) was still somewhat unexpected. For example, the genome-wide ORF screen should, in theory, capture the majority of predicted activated genes mapped to coding sequences in our screen. Interestingly, among the seven genes that we confirmed to induce resistance by overexpression experiments, WWTR1 and ESRRG were successfully identified in the ORF resistance screen (z score ≥ the threshold level of 2.5 in that report); RAPGEF6 narrowly missed the hit status (z score = 2.43), but YES1 (z score = − 0.13) was not a hit. Finally, three (NEDD4L, POU2F1, and ARHGEF28) were among the genes that did not pass quality-control filters prior to screening. This illustrates the challenge of large genetic screens both in terms of functional coverage due to genomic bias or technical factors as well as inherently somewhat arbitrary choices of thresholds for hit calling.
To better understand whether the lack of overlap points to a higher-than-expected rate of false positive discovery or to low functional saturation and thus corresponding more to lack of screen sensitivity, we analyzed the functional relationship between the hit gene lists from the various GoF and LoF screens performed to uncover resistance events to BRAF inhibition in A375 cells. Genes identified by GoF ORF15 and dCAS9 activator14 screens and LoF CRISPR knockout16 and shRNA knockdown17 screens were mapped to a global functional cellular network using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (Fig. 3A, Table S4)29. Between the transposon and ORF sets, we found 139 STRING connections, which was significantly greater than random (p < 10–6) and more significant than other gene set pairs tested (Fig. S3A). The overall network constituted by genes from both screens was highly interconnected, and some of the genes identified in one screen were connected only to genes from the other screen but not to genes found within their own screen (circled in red in Fig. 3B). Similarly, 66 STRING database gene–gene connections were found between the transposon and dCAS9 activator hit gene lists (p = 0.046). The STRING connections between the LoF CRISPR and shRNA screens were also significant (p < 10–5), but neither matched the GoF screens except for the CRISPR-to-ORF pair, albeit presenting no actual gene overlap. We further sought to leverage the results of all the screens together to identify core programs contributing to resistance. We identified a core set of 133 genes by globally searching for all RefSeq genes with at least 10 connections to each of the screened gene sets (excluding the small gene set of the shRNA screen) (Table S5) with the top functional clusters of MAPK and PI3K pathways, RAS-related genes, and receptor tyrosine kinases (RTKs) (Fig. 3C). We further specifically analyzed the STRING connections between the screened gene sets and major MAPK pathway components and found significant functional connections from the GoF gene sets (p-values ranging from 0.01 to 10–5) (Fig. S3B).
Interconnections between different screens. (A) Summary of STRING connections between different genetic screens. Each node represents a screen, with the gene set size indicated within parentheses. Edges represent STRING correlations with arrows pointing from the query gene set to the reference gene set. Numbers separated by commas denote numbers of overlapping genes, total STRING connections, and p values. (B) STRING network connections between the transposon and ORF gene sets. Nodes are color-coded according to their gene list of origin and edges. Nodes with red circles (interlinked hits) are those genes from a given list that only have connections to the other list. Edges are color-coded according to whether the connection is between genes from the same list or not. Edges connecting genes found in both screens to other genes (inter-and intra-list connections incident to solid pink nodes) are distinguished from those connecting nonoverlapping genes to the other list (only inter-list connections). (C) Top functional groups identified by DAVID Gene Functional Classification analysis of the core gene set highly connected to all screens. Enrichment scores are indicated within parentheses. (D) MAPK and PI3K signaling. Cells were treated with the indicated doses of PLX4720 for 24 h, and the levels of the indicated phosphorylated (pGENE) and total (tGENE) proteins were measured by western blotting.
Consistent with our STRING network analysis results and with other studies indicating that reactivation of MAPK activity is a major route for resistance in BRAF-driven models2,27,30,31,32,33, among the 7 genes validated through ORF overexpression, 5 (NEDD4L, WWTR1, RAPGEF6, ARHGEF28 and YES1) led to maintenance of the MAPK signaling pathway upon BRAF inhibition (Fig. 3D). Overall, these results show that there is much better concordance at the functional network level across genes discovered by different screens than suggested by the lack of actual overlap of the gene lists. Moreover, many of the genes found to induce resistance appear to coalesce on a relatively narrow set of mechanisms, suggesting that a few core programs of resistance are repeatedly reached through different paths and could potentially be targeted to counter resistance.
The Hippo pathway modulator TAZ as a global effector
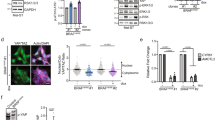
The Hippo pathway effector TAZ as a transcriptional activator was by far the most prevalent hit for resistance gain among all genes tested in long-term drug treatment assays (Fig. 2C). Activation of YAP, the paralog of TAZ, has previously been shown to promote resistance to BRAF inhibition34,35,36. Hippo pathway inhibition (inhibition of MST1/2 kinases corresponding to Hippo in drosophila), leads to YAP/TAZ transcriptional program activation by nuclear translocation. Hippo pathway inactivation was previously shown to effectively resensitize cells to RTK inhibition37. Although the baseline level of TAZ protein did not predict the response to BRAF inhibitors in a panel of BRAFV600E melanoma cell lines, ectopic overexpression increased resistance in several BRAFV600E melanoma cell lines (Fig. S4A, B). Similarly, knocking down WWTR1 induced further sensitization of A375 cells and antagonized their resistance to PLX4720 (Fig. 4A, B). This is also reminiscent of the sensitization of BRAF and other MAPK pathway activation tumor models to MAPK pathway inhibitors via the suppression of YAP134,35,38. WWTR1 was identified as a candidate resistance gene in the previous ORF screen along with YAP115, and interestingly, as stated above, the SRC family tyrosine kinase YES1 validated in our screen was previously shown to regulate YAP1, which indeed was named after its association with YES1 (Yes Associated Protein 1)39. Consistent with these findings, we observed that the SRC family kinase inhibitor dasatinib strongly sensitized WWTR1-overexpressing cells to PLX4720 (Fig. 4C). We also observed that YAP1 knockdown increased PLX4720 sensitivity in A375 parental cells but did not affect WWTR1 overexpressing cells (Fig. 4D, E), consistent with the redundancy between the paralogs YAP1 and TAZ. Furthermore, knockdown of TEADs (TEAD1-4), the transcriptional partners that mediate the DNA binding of YAP1 and TAZ40,41, sensitized both parental and WWTR1 overexpressing A375 cells to PLX4720 (Fig. 4F, G).
TAZ regulates gene expression. (A) PLX4720 IC50 values of control (bsd-VEC) or WWTR1-overexpressing cells following siRNA mediated knockdown of NEDD4L or WWTR1. SCR, scramble. Values are means ± 95% CIs, n = 3. (B) NEDD4L and TAZ protein expression in cells treated with siRNA for 48 h and then treated with 0.5 µM PLX4720 for 24 h. (C) Cell viability of the indicated cells treated with 300 nM PLX4720 (PLX) and 12.5 nM dasatinib (Das). Error bars denote the SEM, n = 3. (D) Effect of YAP1 knockdown on protein expression and phosphorylation status in NEDD4L and WWTR1 overexpressing cells. (E) PLX4720 IC50 values of control (bsd-VEC), WWTR1-, or NEDD4L-overexpressing cells with YAP1 knockdown (siYAP) or scramble (SCR). Values are means ± 95% CIs, n = 5. (F) TEAD1 protein expression in TEAD1 knockdown in control (VEC), WWTR-, and NEDD4L-overexpressing cells. (G) PLX4720 IC50 values of control (bsd-VEC), WWTR1-, or NEDD4L-overexpressing cells with TEAD1 knockdown. Values are means ± 95% CIs, n = 3. (H) Intersect of TAZ-regulated targets and DEGs from PLX4720-sensitive and PLX4720-resistant melanoma cell lines. (I) Expression heatmap of the 50 overlapping genes. In the first column, the regulatory effects of TAZ are denoted either as repression (blue) or activation (red). The second column represents differential expression in two groups of melanoma cell lines, with blue indicating high expression in the sensitive group and red indicating high expression in the resistant group. (J) Unsupervised clustering of the expression values of 18 overlapping genes that most varied between the PLX4720-sensitive and PLX4720-resistant melanoma cell lines. Bars on the bottom panel indicate PLX4720 IC50 values.
To elucidate the transcriptional changes induced by TAZ and their relevance across melanoma models, we divided 22 BRAFV600 mutant melanoma cell lines in our collection into two groups based on their sensitivity to PLX4720 and analyzed the differences in global gene expression between the two groups and identified 508 differentially expressed genes (DEGs) (Table S6). We also performed RNA-Seq on parental and WWTR1 ORF-expressing A375 cells and found 315 DEGs with fourfold up- or downregulation (Table S7). Comparison of these two DEG lists revealed a significant overlap of 50 genes out of 269 TAZ target DEGs with measurements available in the cell line panel (chi-square = 284, p < 0.0001, Fig. 4H). This subset included several genes previously implicated in melanoma or tumor malignancy more broadly, such as SOX10, whose downregulation leads to elevated EGFR and PDGF receptor signals and increased resistance to BRAF inhibition9; MIA, which is involved in melanocyte lineage and melanoma development42,43; and ERBB3, which can reactivate MAPK upon MEK inhibition through a feedback mechanism44,45. Importantly, for almost all (46/50) genes, the DEG effect of the WWTR1 ORF was predicted by the cell line PLX4720 sensitivity—39 genes downregulated by TAZ were highly expressed in the sensitive cell line group, while 7 upregulated genes were expressed in the resistant group (Fig. 4I). Furthermore, unsupervised clustering of the 18 most diversely expressed genes divided the melanoma lines according to their response to PLX4720 (Fig. 4J).
TAZ and NEDD4L interconnect
The E3 ubiquitin-protein ligase NEDD4L was a prominent hit in our study with 32 nonidentical insertion events across 20 sites and 12.5% of all sequencing reads in the pools (Fig. 1D, Table S1). Interestingly, NEDD4L was also a hit in a previous BRAF inhibitor resistance study20. It was also implicated in EGFR-mTOR signaling in lung adenocarcinoma46,47 although in a somewhat functionally opposite way to our expectation. At least 8 NEDD4L protein isoforms have been reported (https://www.uniprot.org/uniprotkb/Q96PU5)48,49 and we noticed that two more isoforms at 230 kDa and 180 kDa might be present in A375 (Fig. S4A). To understand whether resistance to BRAF inhibition was limited to a specific isoform of NEDD4L, we initially investigated a prominent A375 cell isoform (PB clone B181 in Fig. S1D, S5A). However, cDNA-mediated (ORF) expression of a canonical NEDD4L isoform (110 kDa) was readily able to induce resistance in A375 and in other cell lines (Fig. S4B). Moreover, PLX4720 sensitivity across cell lines was not correlated with the expression of any specific isoform (Fig. S4A).
The NEDD4L protein contains an N-terminal C2 domain, four WW domains, and a HECT domain with E3 ubiquitin ligase catalytic activity (Fig. 5A)28,50. Previous studies have indicated that NEDD4L mediates the degradation of AMOT proteins, which are known regulators of the Hippo pathway51,52,53. According to these studies, AMOT proteins repress TAZ and YAP1 activity at least in part via cytoplasm retention of TAZ and YAP. We therefore studied the mechanistic underlying of NEDD4L mediated resistance by expressing a catalytically inactive C962A mutant of the HECT domain (DD) or a WW2-domain deletion mutant (dWW2), which was previously shown to be critical for the recognition of a number of NEDD4L substrates. In both cases, we found that resistance was indeed impaired compared to that obtained with wild-type NEDD4L (Fig. 5B, S5A-C).
Functional connection between NEDD4L and TAZ. (A) Diagram of NEDD4L domains and mutations. (B) Clonogenic assay with cells expressing vector (VEC) or NEDD4L variants treated with PLX4720 at a twofold serial dilution. (C) Endogenous and exogenous NEDD4L and TAZ protein expression in control (bsd-VEC), NEDD4L-, and WWTR1-overexpressing cells after PLX4720 treatment for 24 h. (D) Correlation between WWTR1 and NEDD4L mRNA expression across a panel of cancer cell lines of diverse origins. Each dot denotes a cell line. Red lines indicate mean ± SEM, n = 737. (E) WWTR1 and NEDD4L mRNA expression in pre-treatment, on-treatment, and progression patient samples. Values represent means ± SEMs, and p-values were calculated using two-tailed paired t-test. (F) AKT phosphorylation in NEDD4L-overexpressing A375 cells treated with 0.5 µM PLX4720 and the indicated concentrations of MK2206 for 24 h. (G) Viabilities of the indicated cells treated with 300 nM PLX4720 and 1 µM MK2206 for 5 days. Values represent means ± SEMs, n = 10. Cellular activities were measured using CellTiter-Glo assays. (H) Kinase signals in vector control (bsd-VEC), NEDD4L-, or WWTR1-overexpressing cells. Cells were treated for 24 h and phosphorylated (pGENE) or total (tGENE) protein levels were measured by western blotting. (I) Quantification of phosphorylated EGFR (pEGFR) and total EGFR (tEGFR) protein expression from immunoblots of vector control (VEC), NEDD4L- (NEDD4L), and WWTR1 (TAZ)-overexpressing A375 cells either untreated (ND) or treated with PLX4720 (PLX). Parenthesized numbers indicate biological replicates. All values were normalized to the corresponding untreated vector control sample (VEC_ND) within each experiment. (J) Kinase signals in scramble control, NEDD4L, and WWTR1 knockdown cells treated with PLX4720. Cells were transfected with siRNA for 48 h, followed by 0.5 µM PLX4720 treatment for 24 h. (K) Correlation between WWTR1 and EGFR mRNA expression across a panel of cancer cell lines of diverse origins. Each dot denotes a cell line. Red lines indicate mean ± SEM, n = 737. (L) Hypothetical mechanism of TAZ and NEDD4L in mediating resistance to BRAF inhibition. Solid lines indicate known effects while the dashed line represents an indirect effect with a question mark denoting an unidentified component.
NEDD4L has also been reported to regulate SMAD2/3, which are downstream effectors of the TGFβ pathway50. However, the projected effects of such possible role in BRAF-mutant melanoma are contradictory: NEDD4L-mediated SMAD2/3 degradation would lead to the termination of TGFβ signaling rather than its activation28, while previous reports have shown that TGFβ signaling upregulates the EGFR pathway to confer resistance to BRAF inhibition in melanoma9. Nonetheless, we tested this potential involvement by expressing missense mutations (S382A, S468A, or both (SASA)) targeting phosphorylation sites for SGK1, which is critical for SMAD2/3 regulation, and found that while protein expression of the S468A mutant failed despite decent mRNA expression (Fig. S5A), both S382A and SASA were comparable to those of the wild type in inducing resistance (Fig. 5B, S5A–C). Furthermore, knockdown of SMAD2/3 modestly reduced rather than increasing the resistance mediated by both parental A375 and NEDD4L cells (Fig. S5D, E), and the SMAD2/3 protein level did not increase upon NEDD4L knockdown (Fig. S5F), suggesting the independent regulation of sensitivity to BRAF inhibition by NEDD4L and by TGFβ.
WWTR1 expression led to moderate NEDD4L upregulation at the protein (Fig. 5C) and mRNA levels (1.5-fold, p = 3.19E−06, FDR = 3.03E−05; Table S7); conversely, WWTR1 knockdown led to reduced NEDD4L protein expression (Fig. 4B). NEDD4L expression was also positively correlated with WWTR1 expression in a panel of cancer cell lines (Fig. 5D, Table S8), and both were correlated with melanoma treatment outcome (Fig. 5E, with individual patient samples shown in Fig. S6). These findings suggested that NEDD4L might be involved in TAZ-mediated resistance.
Consistent with a role of NEDD4L in the regulation of other signaling pathways, NEDD4L overexpression led to a strong increase in AKT phosphorylation upon BRAF inhibition (Fig. 3D), and correspondingly, the AKT inhibitor MK2206 that strongly reduced AKT phosphorylation (Fig. 5F) effectively resensitized NEDD4L-overexpressing A375 cells to PLX4720 (Fig. 5G). NEDD4L also led to the phosphorylation of EGFR (at residue Y1068) and another RTK, IGF1R, while TAZ upregulated EGFR protein expression (Fig. 5H, I). Supporting these observations, knockdown of NEDD4L reduced EGFR and IGF1R phosphorylation and knockdown of WWTR1 reduced EGFR expression (Fig. 5J). The regulation of EGFR expression by TAZ is also in line with the positive correlation between EGFR and WWTR1 expression in a cell line panel (Fig. 5K, Table S8) and the elevated EGFR mRNA expression in WWTR1 ORF-overexpressing cells (2.0-fold, p = 3.31E−12, FDR = 8.08E−11; Table S7). Therefore, TAZ plays an important role in the maintenance of MAPK pathway activation both through transcriptional regulation of some pathway component genes and likely through other signaling mechanisms (Fig. 5L). In line with this, both the EGFR inhibitor pelitinib (EKB569) and the IGF1R inhibitor BMS754807 countered resistance induced by NEDD4L, WWTR1, and all other resistance genes validated in this study (Fig. S7), suggesting that RTK inhibitors might resensitize cells to MAPK pathway inhibition more broadly than previously described2,6,54.
Discussion
As an alternative to genetic screening platforms consisting of complex mutagen libraries (such as ORFs), transposon mutagenesis has the potential to tackle some of the complexity of host genome harboring both known protein coding genes and unidentified components, non-coding elements, or alternative isoforms. Moreover, activation of endogenous elements might provide more context specific results than ectopically expressed transcription activators. While transposon mutagenesis has been widely used in vivo19,55, its application as an in vitro cell line screening platform results in expandable off-the-shelf libraries that are ideal for applications involving complex treatment conditions, such as comparing resistance to multiple drugs. The low cost and simplicity of this approach, requiring only transient transfection of two plasmids, enables straightforward high-throughput platform deployment20. While not utilized in this study, additional features, such as tagging transposition plasmids with degenerate nucleotide barcodes, should further improve platform scalability. Such barcode diversity can then be sequenced to reveal nonidentical insertions even at the same genomic location, essentially eliminating the reliance on library replicates for identification of high confidence hits.
Our study provided further support for therapies targeting the Hippo pathway37 or protein ubiquitination56,57, and we also demonstrated that many different genes with a broad range of functions implicated in resistance to target inhibition therapies appear to coalesce to the pathway of primary target inhibition (here the MAPK pathway) and a restricted number of additional pathways, suggesting that the staggering complexity of the resistance landscape may not need to be addressed by an equivalently complex array of therapeutic strategies.
Methods
Cell culture reagents
All cell lines were obtained from the collection of the Genomics of Drug Sensitivity in Cancer (GDSC) project13, cultured in either DMEM/F12 or RPMI supplemented with 5% FBS and 1% penicillin/streptavidin, and maintained in a 37 °C/5% CO2 cell culture incubator. Therapeutic compounds were purchased from Selleckchem and ChemieTek and dissolved in dimethyl sulfoxide.
Transposon mutagenesis library construction and screening
The piggyBac activation transposition plasmid pPB-SB-CMV-puro-SD was described previously21. A derivative plasmid containing the degenerate nucleotide barcodes mentioned in the Discussion section was verified by sequencing and the plasmid map and complete sequence are provided in the supplementary information (Fig. S8, Supplementary Methods and Text). Cell line libraries were constructed by transfecting A375 cells with the transposition plasmid and the transposase plasmid pCMV-hyPBase58. After puromycin selection, cells were treated with PLX4720, and surviving cells were pooled or isolated. Insertion site sequencing libraries were prepared using ligation-mediated PCR and identified with next-generation sequencing (NGS) as described previously21.
Gene annotation for insert sites
Illumina sequencing data in FASTQ files were demultiplexed and trimmed to retain only the genomic DNA sequences. Reads of 7 bp or longer were aligned to the human reference genome (hg19) using Bowtie and unique reads were recorded. For the resistant pools, the top sequences by read count were reported based on our estimate of the survivor colony numbers from each library, with the top 100 inserts representing more than 99.5% of the total sequencing reads. For each insert, the three most proximal genes were annotated.
Melanoma patient cohort
The patient cohort with metastatic melanoma containing BRAFV600E mutation enrolled on clinical trials for treatment with a BRAF inhibitor vemurafenib or combined BRAF + MEK inhibitor (dabrafenib + trametinib or LGX818 + MEK162) was reported previously59. Patients provided written informed consent for the collection of tissue and blood samples for research and genomic profiling, as approved by the Dana-Farber/Harvard Cancer Center Institutional Review Board (DF/HCC Protocol 11–181).
Lentiviral cDNA plasmid construction and cell assays
ORF entry clones were subcloned and inserted into two sets of lentiviral expression vectors (blasticidin and puromycin) for cell-based assays. Cells viability, apoptosis, and proliferation were assayed. All assays were performed at least three times.
Immunoblotting
Immunoblotting was described in the Supplementary Methods. Signals were detected using SuperSignal West Femto substrate (Thermo Fisher) and imaged with a G-box imager (Syngene). Original image files were provided in the Supplementary Information. To measure relative intensities, immunoblot images were quantified using ImageJ (https://imagej.net/ij/) and signals were normalized to control gene bands from the same gel.
RNA-Seq
Total RNA was prepared using RNeasy Mini Kit (Qiagen) and processed using a TruSeq RNA Sample Preparation Kit (Illumina) to generate libraries of sequencing molecules. For samples derived from transposon-inserted clones, equal amounts of 12 indexed subsamples were pooled and sequenced using an Illumina sequencer instrument. STAR aligner60 was used to map sequencing reads to transcripts. Read counts for individual transcripts were produced with HTSeq-count61, followed by the estimation of expression values as RPKM (reads per kilobase per million) and the detection of differentially expressed transcripts using EdgeR62. For WWTR1-overexpressing cDNA clones, samples were prepared from four biological replicates of viral infections, indexed, pooled, and sequenced. The Benjamini–Hochberg false discovery rate (FDR) was used to estimate the statistical significance of differences in gene expression.
STRING analysis
Known and predicted protein–protein associations were tested using STRING analysis (http://string-db.org, Homo sapiens: 9606.protein.links.detailed.v9.1.txt.gz). STRING analyses were first performed between pairs of screened gene sets (Table S4). To identify a core set of genes responsible for resistance, all RefSeq genes were queried for their connections to components of every screened gene set (Table S5). The list of genes with at least 10 connections to each of four screens (excluding the small shRNA screen gene set) was used as input for the Database for Annotation, Visualization and Integrated Discovery (DAVID) analyses63.
Clustering of gene expression values
The genes most differentially expressed between BRAF inhibitor-sensitive and -resistant cell lines were selected for unsupervised clustering using Pearson’s correlation across genes and cell lines. Clustering and heatmap generation were performed using Gene-E (http://www.broadinstitute.org/cancer/software/GENE-E/index.html).
Additional protocol details can be found in the Supplementary Methods and Text.
Data availability
Melanoma patient RNA-Seq data were deposited in the NCBI GEO under accession number GSE73470. This RNA-seq dataset can also be accessed from European Genome-phenome Archive (EGA Study ID EGAS00001000992). Cell line data are available through the Genomics of Drug Sensitivity in Cancer (https://www.cancerrxgene.org/) or DepMap (https://depmap.org/portal/) portals.
References
Bollag, G. et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467(7315), 596–599 (2010).
Corcoran, R. B. et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2(3), 227–235 (2012).
Corcoran, R. B., et al. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci. Signal, 2010. 3(149): p. ra84.
Flaherty, K. T. et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 363(9), 809–819 (2010).
Montagut, C. et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res 68(12), 4853–4861 (2008).
Prahallad, A. et al. Unresponsiveness of colon cancer to BRAF (V600E) inhibition through feedback activation of EGFR. Nature 483(7387), 100–103 (2012).
Sanchez-Laorden, B., et al. BRAF inhibitors induce metastasis in RAS mutant or inhibitor-resistant melanoma cells by reactivating MEK and ERK signaling. Sci. Signal 2014. 7(318): ra30.
Shi, H. et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 4(1), 80–93 (2014).
Sun, C. et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 508(7494), 118–122 (2014).
Barretina, J. et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483(7391), 603–607 (2012).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499(7457), 214–218 (2013).
Solit, D. B. et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature 439(7074), 358–362 (2006).
Garnett, M. J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483(7391), 570–575 (2012).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517(7536), 583–588 (2015).
Johannessen, C. M. et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature 504(7478), 138–142 (2013).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343(6166), 84–87 (2014).
Whittaker, S. R. et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov 3(3), 350–362 (2013).
Choi, J. et al. Identification of PLX4032-resistance mechanisms and implications for novel RAF inhibitors. Pigment Cell Melanoma Res 27(2), 253–262 (2014).
Mann, M. B. et al. Transposon mutagenesis identifies genetic drivers of Braf(V600E) melanoma. Nat Genet 47(5), 486–495 (2015).
Zhu, E. Y. et al. Understanding cancer drug resistance with Sleeping Beauty functional genomic screens: Application to MAPK inhibition in cutaneous melanoma. iScience 26(10), 107805 (2023).
Chen, L. et al. Transposon activation mutagenesis as a screening tool for identifying resistance to cancer therapeutics. BMC Cancer 13(1), 93 (2013).
Bruchez, A., et al. MHC class II transactivator CIITA induces cell resistance to Ebola virus and SARS-like coronaviruses. Science, 2020.
Anvar, S. Y. et al. Full-length mRNA sequencing uncovers a widespread coupling between transcription initiation and mRNA processing. Genome Biol 19(1), 46 (2018).
Kornblihtt, A. R. et al. Multiple links between transcription and splicing. RNA 10(10), 1489–1498 (2004).
Davoli, T. et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155(4), 948–962 (2013).
Ruiz-Saenz, A. et al. A reversible SRC-relayed COX2 inflammatory program drives resistance to BRAF and EGFR inhibition in BRAF(V600E) colorectal tumors. Nat Cancer 4(2), 240–256 (2023).
Johannessen, C. M. et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468(7326), 968–972 (2010).
Gao, S. et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol Cell 36(3), 457–468 (2009).
Jensen, L. J., et al. STRING 8: A global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res., 2009. 37(Database issue): D412–6.
Goetz, E. M. et al. ERK mutations confer resistance to mitogen-activated protein kinase pathway inhibitors. Cancer Res 74(23), 7079–7089 (2014).
Long, G. V. et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat Commun 5, 5694 (2014).
Van Allen, E. M. et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov 4(1), 94–109 (2014).
Yadav, V. et al. Reactivation of mitogen-activated protein kinase (MAPK) pathway by FGF receptor 3 (FGFR3)/Ras mediates resistance to vemurafenib in human B-RAF V600E mutant melanoma. J Biol Chem 287(33), 28087–28098 (2012).
Lin, L. et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet 47(3), 250–256 (2015).
Kapoor, A. et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 158(1), 185–197 (2014).
Shao, D. D. et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158(1), 171–184 (2014).
Chapeau, E. A., et al. Direct and selective pharmacological disruption of the YAP-TEAD interface by IAG933 inhibits Hippo-dependent and RAS-MAPK-altered cancers. Nat. Cancer, 2024.
Flaherty, K. T., Wargo, J. A. & Bivona, T. G. YAP in MAPK pathway targeted therapy resistance. Cell Cycle 14(12), 1765–1766 (2015).
Sudol, M. et al. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J Biol Chem 270(24), 14733–14741 (1995).
Zanconato, F., et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. (2015).
Zhao, B. et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22(14), 1962–1971 (2008).
Blesch, A. et al. Cloning of a novel malignant melanoma-derived growth-regulatory protein, MIA. Cancer Res 54(21), 5695–5701 (1994).
Poser, I. et al. Functional role of MIA in melanocytes and early development of melanoma. Oncogene 23(36), 6115–6124 (2004).
Carraway, K. L., 3rd and Cantley, L. C. A neu acquaintance for erbB3 and erbB4: A role for receptor heterodimerization in growth signaling. Cell. 78(1): p. 5–8 (1994).
Turke, A. B. et al. MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer Res 72(13), 3228–3237 (2012).
Ding, K., et al. JAC4 inhibits EGFR-driven lung adenocarcinoma growth and metastasis through CTBP1-mediated JWA/AMPK/NEDD4L/EGFR axis. Int. J. Mol. Sci. 24(10) (2023).
Li, G. et al. Downregulation of NEDD4L by EGFR signaling promotes the development of lung adenocarcinoma. J Transl Med 20(1), 47 (2022).
Dunn, D. M. et al. Common variant of human NEDD4L activates a cryptic splice site to form a frameshifted transcript. J Hum Genet 47(12), 665–676 (2002).
Chen, H. et al. NEDD4L on human chromosome 18q21 has multiple forms of transcripts and is a homologue of the mouse Nedd4-2 gene. Eur J Hum Genet 9(12), 922–930 (2001).
Debonneville, C. et al. Phosphorylation of Nedd4-2 by Sgk1 regulates epithelial Na(+) channel cell surface expression. EMBO J 20(24), 7052–7059 (2001).
Rheinemann, L. et al. Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release. J Biol Chem 297(2), 100975 (2021).
Wang, C. et al. The Nedd4-like ubiquitin E3 ligases target angiomotin/p130 to ubiquitin-dependent degradation. Biochem J 444(2), 279–289 (2012).
Zhao, B. et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev 25(1), 51–63 (2011).
Villanueva, J. et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell 18(6), 683–695 (2010).
Ni, T. K. et al. Low-copy piggyBac transposon mutagenesis in mice identifies genes driving melanoma. Proc Natl Acad Sci USA 110(38), E3640–E3649 (2013).
Alrosan, A. Z. et al. Potential roles of NEDD4 and NEDD4L and their utility as therapeutic targets in high-incidence adult male cancers (Review). Mol Clin Oncol 19(3), 68 (2023).
Li, B., Adam Eichhorn, P. J. & Chng, W. J. Targeting the ubiquitin pathway in lymphoid malignancies. Cancer Lett. 594, 216978 (2024).
Yusa, K. et al. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci USA 108(4), 1531–1536 (2011).
Kwong, L. N. et al. Co-clinical assessment identifies patterns of BRAF inhibitor resistance in melanoma. J Clin Invest 125(4), 1459–1470 (2015).
Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29(1), 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics 31(2), 166–169 (2015).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40(10), 4288–4297 (2012).
Huang da, W., Sherman, B. T., and Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4(1): 44–57 (2009).
Acknowledgements
We thank Pentao Liu for providing the piggyBac transposase plasmid pCMV-hyPBase, Joan Massague for the NEDD4L cDNA plasmids, and Bristol-Myers Squibb for the U219 expression data for the cancer cell lines. We also thank Wilhelm Haas, Keith T. Flaherty, and Adam Lacy-Hulbert for useful discussions.
Author information
Authors and Affiliations
Contributions
L.C.: project coordination, experiments, data analysis, and manuscript preparation. C.H.B.: project design and supervision, data analysis, and manuscript preparation. I.P-M.: STRING and clustering analyses. A.D.: contribution to cell line assays. X.Y.: contribution to cell culture and immunoblotting. D.F.: provision of patient RNA-Seq and treatment data. R.I.S.: sequencing data processing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, L., Pruteanu-Malinici, I., Dastur, A. et al. Transposon mediated functional genomic screening for BRAF inhibitor resistance reveals convergent Hippo and MAPK pathway activation events. Sci Rep 15, 3048 (2025). https://doi.org/10.1038/s41598-025-86694-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-86694-5