Abstract
Cholangiocarcinoma (CCA) is an aggressive cancer originating from bile duct epithelial cells, with a high rate of recurrence following surgical resection. Recurrence is categorized as early linked to aggressive tumor biology than late recurrence. This study aimed to identify novel peptide mass fingerprints (PMFs) and potential biomarker panels in the serum of CCA patients with early and late recurrence using mass spectrometry. Serum samples of 81 CCA patients were analyzed using MALDI-TOF MS and LC–MS/MS, with statistical analysis correlating peptide profiles with clinical outcomes like disease-free survival (DFS) and overall survival (OS). A 365-day DFS cut-off effectively distinguished early from late recurrence, with early recurrence linked to poorer survival outcomes. The PMFs from MALDI-TOF MS differentiated recurrence types based on specific mass signatures. LC–MS/MS analysis identified 95 peptides associated with cancer progression in early recurrence and 60 in late recurrence. Distinct protein associations were found: ATR, POLA1, BLM, SP100, and PPP1R15A for early recurrence, and SERPINA1, TGFB2, SERPING1, and CAD for late recurrence, with strong interactions with chemotherapeutic drugs. This study successfully demonstrated the use of PMFs for rapid discrimination between early and late recurrence in CCA and identified potential serum peptide biomarkers to improve accuracy in recurrence classification.
Similar content being viewed by others
Introduction
Cholangiocarcinoma (CCA) is an aggressive malignancy arising in the biliary tract, often diagnosed at advanced stages due to its asymptomatic early phases. Surgical resection followed by adjuvant chemotherapy is the primary curative treatment; however, outcomes of poor survival remain1. A significant challenge in managing CCA is disease recurrence after initial treatment, categorized as early or late based on the onset post-surgery. Early recurrence occurs within one year and affects 20 to 65% of patients. Early recurrence is often linked to aggressive tumor biology, poor differentiation, and lymphovascular invasion. Late recurrence, is associated with slow-growing tumor cells or the patient’s immune response2,3,4. Therefore, accurately predicting early recurrence for each regimen in individual patients may guide the selection or modification of adjuvant treatment plans. Although carcinoembryonic antigen (CEA) and cancer antigen 19-9 (CA 19-9) have been utilized in screening, diagnosis, treatment monitoring, recurrence detection, and disease progression for CCA, they also have several limitations, including specificity to cancer types, overlap with benign conditions, limited diagnostic values, inconsistent levels of biomarkers, lack of established cut-off values and limited role in early detection5,6. Thus, multiple biomarkers or biomarker panels are discussed as potential tools for improving the diagnosis and management of CCA. These approaches aim to enhance specificity and sensitivity beyond what individual biomarkers like CEA and CA 19-9 can provide7,8.
Peptide biomarkers, small proteins or peptides detectable in biological samples, play a pivotal role in diagnostics by providing insights into physiological and pathological conditions. They are crucial in diagnostics, as they can indicate the presence or progression of diseases, monitor therapeutic responses, or predict disease outcomes. For example, prostate-specific antigen (PSA) is widely used in the early detection and monitoring of prostate cancer9, while amyloid-beta peptides are employed as biomarkers in the diagnosis of Alzheimer’s disease10. In addition, carcinoembryonic antigen (CEA) and cancer antigen 19-9 (CA 19-9), although not highly specific to cancer types and with limitations such as overlap with benign conditions, remain valuable tools for monitoring treatment responses and detecting recurrence in cancers11. In medical practice, peptide biomarkers are increasingly used to enhance early detection, improve diagnostic accuracy, and support personalized treatment approaches. Their specificity and capacity to reflect molecular-level changes in biological processes underpin their utility. Prior research has highlighted the critical role of peptide biomarkers in cancer studies12,13. Numerous studies have identified differentially expressed peptides across various cancers, contributing to the development of diagnostic tools and therapeutic strategies. For example, peptide biomarkers have significantly improved early detection, staging, and the monitoring of treatment responses and recurrence in cancers such as prostate, breast, and ovarian14,15,16. These findings demonstrate the potential of peptidome approaches to drive advancements in personalized medicine and cancer management, with peptide biomarkers offering substantial promise for enhancing detection, diagnosis, and disease monitoring.
Based on information above, the serum peptidome in CCA patients with early and late recurrence has yet to be fully explored. This study aims to identify novel peptide mass fingerprints (PMFs), peptide clusters, and potential biomarkers in the serum of CCA patients with early and late recurrence. We investigated disease-specific peptide profiles by matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry (MALDI-TOF MS) combined with liquid chromatography-tandem mass spectrometry (LC–MS/MS). Additionally, we examined the associations between these peptides and chemotherapy drugs. We anticipated that serum peptide biomarkers could potentially aid in prognosis and inform treatment strategies for CCA.
Results
Clinical characteristics and survival analysis of cholangiocarcinoma (CCA) patients
The clinical characteristics in CCA patients were shown in Table 1. We performed a cut-off value for categorizing CCA patients with recurrent status (early and late recurrence) using 365 days according to previous publications2,4,3. By CCA patients had DFS < 365 days that were categorized early recurrence (42%), while CCA patients had DFS ≥ 365 days that were categorized late recurrence (58%).
Survival analysis using the Log rank test and multivariate analysis through Cox regression revealed that factors such as positive surgical margin, moderately and poorly differentiated histology, late staging, and early recurrence were significantly associated with shorted survival rates. Specifically, multivariate Cox regression analysis indicated that surgical margins, histological differentiation, cancer staging, and recurrent status were significant predictors of survival compared to their referent categories. Notably, early recurrence displayed a markedly high hazard ratio as 6.36 folds, p < 0.001 when compared with late recurrence, underscoring its critical impact on patient prognosis (Table 1). Consequently, this study prioritized recurrence status to enable further peptidome analysis using mass spectrometry.
Serum peptide barcode of CCA patients with early and late recurrence
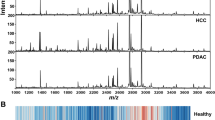
The criteria for categorizing patients with recurrent CCA were based on a time frame of 365 days post-surgery. Specifically, patients who experienced recurrence before 365 days were classified into the early recurrence group, comprising 34 individuals. In contrast, those with recurrence occurring at 365 days or later were assigned to the late recurrence group of 47 individuals. Following this categorization, serum samples from CCA patients underwent peptide profiling using MALDI-TOF MS. The results revealed that the peptide patterns in the serum of early recurrence patients differed significantly, notably, five prominent peptides observed at m/z 2496.908598, 2697.984489, 3034.15095, 3710.089335, and 4288.621730 when compared to those from late recurrence patients (Fig. 1A). Subsequently, the peptide profiles from two groups of patients were subjected to statistical analysis using Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA). OPLS-DA generated a clear separation between early and late recurrent patients indicated that potential peptide biomarkers associated with the differences in patient conditions. (Fig. 1B). The OPLS-DA model demonstrated moderate goodness of fit (R2Y = 0.584) and predictive ability (Q2Y = 0.436) (Fig. 1C). To validate the model, a permutation test with 1000 iterations was performed. The permutation test showed R2Y of 0.947 (p < 0.001) and Q2 of 0.663 (p < 0.001) As shown in Fig. 1D, the results confirmed the validity of the model, with a p-value less than 0.05, indicating that the model’s predictive ability is not due to overfitting.
Peptide mass fingerprint of serum peptides using MALI-TOF MS analysis from CCA patients. (A) The average spectra of serum samples from early recurrent CCA (upper panel) and late redcurrant CCA (lower panel) in the range of 1000–6,000 m/z. The dashed lines highlight the positions of peptide m/z values that demonstrate distinct differences. (B) Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA). (C) Cross-validation of the OPLS-DA model assessed classification performance using accuracy, R2Y (goodness of fit), and Q2Y (predictive ability). (E) A permutation test with 1,000 permutations was performed on the OPLS-DA model, yielding an empirical p < 0.001.
Upon obtaining PMFs of serum peptides to distinguish between early and late recurrence in CCA patients, MALDI-TOF MS provides rapid screening for stratification of recurrent status. Additionally, to improve the power and accuracy of detection between early and late recurrence in CCA patients, we investigated peptide-base biomarkers through LC–MS/MS analysis. We aimed to identify potential peptide biomarkers for integration with PMFs from MALDI-TOF MS, to improve the accuracy and precision in diagnosing recurrence in CCA patients.
Identification of differentially expressed peptides in plasma of CCA patients with early and late recurrence
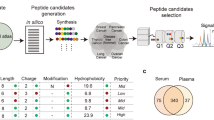
To minimize genetic variation and identify representative peptide biomarkers for early and late recurrence, we pooled the samples from each group in equal protein amounts and performed peptidome analysis using LC–MS/MS17. Pooled serum peptides were analyzed using LC–MS/MS, identified 5,798 proteins. A Venn diagram illustrated the overlap and differences between early and late recurrence groups, with 1,747 shared peptides and 2,327 and 1,724 unique peptides exclusive to early and late recurrence, respectively. A Principal Component Analysis (PCA) revealed distinct patterns, with Component 1 effectively separating the two groups. Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) further confirmed clear separation between the groups, identifying key discriminating peptides. VIP score analysis revealed 1,025 significant peptides, with the top 15 listed. Volcano plot analysis, using a p < 0.05 and fold change > 2 threshold (Fig. 2), identified 155 peptides including 95 upregulated in early recurrence and 60 upregulated in late recurrence, listed in supplementary Tables 1 and 2.
The data analysis and candidate peptide pre-filtration. (A) Venn diagram created using the program Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny), illustrating the total count of proteins that exhibited differential expression of each group. (B) Principal Component Analysis (PCA), the scores plot of PCA discriminate two groups. (C) Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) showing clear separation of two groups. (D) The Variable Importance in Projection (VIP) showed top 15 peptides including KCNS3; Potassium voltage-gated channel subfamily S member 3, OMP; Olfactory marker protein, PRR36; Proline-rich protein 36, FBXO33; F-box only protein 33, DIAPH1; Protein diaphanous homolog 1, DIRAS1; GTP-binding protein Di-Ras1, RASL11A; Ras-like protein family member 11 A, HERC6; Probable E3 ubiquitin-protein ligase HERC6, CCDC70; Coiled-coil domain-containing protein 70, MCTP1; Multiple C2 and transmembrane domain-containing protein 1, THBS2; Thrombospondin-2, FAF2; FAS-associated factor 2, DPT; Dermatopontin, SCNN1G; Amiloride-sensitive sodium channel subunit gamma, KRT1; Keratin. (E) The Volcano plot analysis was performed to filter significant peptides that met the established criteria.
Network analysis of serum peptides in CCA patients with early and late recurrence
A total of 155 peptides were filtered through a stringent screening process, with 95 peptides identified in early recurrence and 60 peptides in late recurrence. Subsequently, candidate peptides from both groups were analyzed for protein-chemical interactions using the STITCH database. Common chemotherapeutic drugs, including Gemcitabine, Cisplatin, Capecitabine, Oxaliplatin, and 5-Fluorouracil (5-Fu) widely used in the treatment of CCA, were incorporated into the interaction list to predict associations and computational interactions. This analysis aimed to present a comprehensive network of interactions between the candidate peptides and these chemotherapeutic drugs (Table 2).
In early recurrent patients, the network analysis performed using STRING revealed intricate interactions between proteins and chemotherapy drugs. The results showed the peptide interaction network of SP100 (Nuclear autoantigen Sp-100), ATR (Serine/threonine-protein kinase ATR), POLA1 (DNA polymerase alpha catalytic subunit) and PPP1R15A (Protein phosphatase 1 regulatory subunit 15 A) showed a strong relationship with the chemotherapy drug, Cisplatin, while we also found less of BLM (RecQ-like DNA helicase BLM) with Cisplatin. In addition, ATR, BLM and CEP164 (Centrosomal protein of 164 kD) also with their predicted functional partner CHEK1 (Checkpoint kinase 1) which had strong associations with several chemotherapeutic drugs such as Gemcitabine, Cisplatin and 5-Fu. In additional to peptide-chemotrophic drugs interactions, we also found peptide-peptide interaction which related with signaling pathways to promote cancer progression, such as cell proliferation, angiogenesis, tumor microenvironment and metastasis. Main nodes including SP100, BLM and ATR proteins have been reported to play a crucial role in numerous pathways in cancer progression in several publications18,19,20,21,22. In this study, SP100 showed a strong association with SUMO1 (Small Ubiquitin-Like Modifier 1), which is predicted to be a functional partner. SUMO1 is an upstream regulator of several proteins involved in signaling pathways identified in this analysis including MAP3K1 (Mitogen-activated protein kinase kinase kinase 1), ZFHX3 (Zinc finger homeobox protein 3)/EHBP1 (EH domain-binding protein 1-like protein 1) signaling, HNRNPH3 (Heterogeneous nuclear ribonucleoprotein H3; hnRNP H3), BLM and UBTF (Nucleolar transcription factor 1)/NCL (Nucleolin) signaling. For BLM exhibited strong interactions with ATR/CEP164/CEP70 (Centrosomal protein of 70 kD) which play roles in cell cycle checkpoint, DNA damage response and DNA repair, thereby protecting against cell death and sustaining cancer cell survival. In addition, BLM also interacted with SETX (Probable helicase senataxin)/ predicted POLR2A (DNA-directed RNA polymerase II subunit RPB1) interaction /INTS5 (Integrator complex subunit 5) as well as SUPT5H (Transcription elongation factor SPT5) which plays roles in RNA processing, transcription regulation, and DNA damage response (Fig. 3). Additionally, non-interaction nodes were identified that have oncogenic roles in cancer progression and recurrence. A total of 95 peptides were reported to have oncogenic functions, as listed in supplementary Table 1.
Protein–chemical interaction using the STRITH software in early recurrence. In the network, proteins were represented as nodes, and the thickness of the connecting lines indicated the degree of association between the proteins or chemicals. Gray line was peptide-peptide interaction, green line was peptide-drug interaction, red line was drug-drug interaction. Circle represented network interaction of peptides that were found in early recurrence.
In late recurrent patients, we found one direct peptide-chemotherapeutic drug and one indirect peptide-chemotherapeutic drug. SERPINA1 (Alpha-1-antitrypsin) had strong association with Cisplatin, while CAD (Carbamoyl phosphate synthetase, aspartate transcarbamylase, and dihydroorotase) showed indirect relationship with Cisplatin, 5-Fu and Capecitabine through strong interaction with predicted partner DPYD (Dihydropyrimidine Dehydrogenase) and medium interaction predicted partner TYMS (Thymidylate Synthase). It had that strong association with Gemcitabine, Cisplatin, Capecitabine, and 5-Fluorouracil (5-Fu). In addition, we also found strong peptide-peptide interaction including SERPINA1, SERPING1 Plasma protease C1 inhibitor and TGFB2 (Transforming Growth Factor Beta 2) which play roles in tumor growth and metastasis. Moreover, we found that CAD was a central node or hub of strong interaction with SLC23A3 (Solute carrier family 23 member 3) which has an essential role in the transport of certain molecules across cell membranes. Predicted partners, including DPYSL3 and 4 (Dipeptidyl Peptidase-Like 3 and 4), DPYS (Dipeptidyl Peptidase I), DPYD (Dihydropyrimidine Dehydrogenase), CRMP1 (Collapsin Response Mediator Protein 1), CPS1 (Carbamoyl-Phosphate Synthetase 1), DHODH (Dihydroorotate Dehydrogenase) and TYMS have functions in cellular metabolism. In addition, we also found a strong relationship of CXXC1 (Receptor-transporting protein 5) and SETD1B (Histone-lysine N-methyltransferase) (Fig. 4). Our result also showed non-interaction nodes that have been reported in several publications in cancer progression as shown in a supplementary Table 2.
Protein–chemical interaction using the STRITH software in late recurrence. In the network, proteins were represented as nodes, and the thickness of the connecting lines indicated the degree of association between the proteins or chemicals. A peptide that was centrally located and interacts with multiple other peptides was referred to as a ‘hub’ peptide. Gray line was peptide-peptide interaction, green line was peptide-drug interaction, red line was drug-drug interaction. Circle represented network interaction of peptides that were found in late recurrence.
Discussion
Our results were based on cut-off values using median of DFS or about 365 days after surgical treatment. This consistent use of a 365-day threshold underscores its potential as a standard marker for assessing recurrence risk. Survival analysis and Cox regression further illustrate that early recurrence was associated with significantly shorter survival compared to late recurrence. In addition, we identified that early recurrence was an independent factor contributing to poor survival outcomes. This cut-off value aligned with findings from several previous studies. They established that this time frame is crucial for understanding recurrence patterns across various cancer types, including bile duct2 pancreatic4 and colorectal cancers3. The consistency of these findings across multiple studies highlights the importance of early detection and intervention strategies for patients at risk of recurrence. Implementing routine surveillance protocols that focus on this critical time frame could lead to improved patient management and outcomes. The use of CEA and CA 19-9 as biomarkers for recurrence is currently being debated due to several issues, especially their limited specificity5,6. Thus, biomarker panels or multiple biomarkers are essential to improve accuracy and specificity predicting these outcomes, providing a valuable tool for identifying patients at higher risk for early recurrence7,8.
Generally, cancer recurrence is considered a result of the cancer progression. Typically, this progression involves the production of peptides and proteins that promote cancer development. These molecules are secreted into the bloodstream to drive tumor growth, immune evasion, metastasis, and intercellular communication23. Many secreted peptides and proteins serve as biomarkers, indicating the presence or progression of cancer, making them valuable for peptide- and protein-based biomarker detection24. In practically, MALDI-TOF MS has been reported as a useful tool for diagnosis and prognosis in several abnormalities, as the peptide signature in serum or PMFs showed specific patterns for several disease, especially cancers16,25,26,27. To explore peptide patterns of recurrent cancer, our study provided novel evidence from serum peptidome to categorize early and late recurrent status using PMFs through MALDI-TOF MS. Our finding showed that the peptide patterns of early recurrence were markedly different from those patterns in late recurrence. Moreover, we also identified peptide signatures that were markedly dominant in early recurrence at m/z values of 2496.9, 2697.9, 3034.1, 3710.1, and 4288.6. These results showed that PMFs MALDI-TOF MS could be useful to discriminate between early and late recurrence. Our study was consistent with previous reports on bile duct cancer, also known as CCA. Our study has revealed that PMFs via MALDI-TOF MS in the serum of 92 bile duct cancer patients at University College Hospital, UK, compared with healthy volunteers, had distinct differences in the peptide profiles of bile duct cancer patients. Analysis of peptide positions on the combined spectrum distinguished eight peptides with statistically significant differences in peak area under the curve, specifically at m/z values of 887.2, 1263.7, 1350.8, 2082.1, 2210.3, 2554.5, 2903.3, and 5805.025. Additionally, in 2023, a study on PMFs in patients with cervical cancer at various stages found clear differences in peptide profiles among the groups, which included healthy volunteers, precancerous lesions, and cervical cancer stages I, II, and III16.
Based on previous reports, PMFs via MALDI-TOF MS could not only differentiate between healthy individuals and cancer patients but also stratify the aggressiveness of the disease by stage. This provides strong evidence supporting our findings in using PMFs to distinguish between early and late recurrent CCA patients. Therefore, due to its high efficiency and sensitivity in detecting PMFs, MALDI-TOF MS could be a primary choice for rapid screening of disease abnormalities, especially recurrent status of cancer. For clinical advantage, rapid diagnosis enables prompt treatment and ensures that improve the treatment plans for patients. However, in addition to the speed of diagnosis, the accuracy of the diagnosis is crucial factor to consider in clinical application. Although MALDI-TOF MS has been effective in distinguishing between early and late recurrence in CCA patients, the single laser MALDI approach yields low ion counts, resulting in the loss of low-abundance or difficult-to-ionize molecules28. The low resolution and associated mass accuracy of linear TOF analyzers may limit the discovery of peptide biomarkers. Furthermore, MALDI-TOF MS does not support de novo peptide sequencing due to its limited resolution and inability to perform peptide fragmentation, hindering the identification of specific peptide peaks. Consequently, the mass spectra linked to the stratification of recurrent status peptides are challenging to identify with MALDI-TOF MS. Therefore, LC–MS/MS was employed to identify and quantify potential peptide biomarkers to enhance the accuracy and precision in diagnosing recurrence in CCA patients.
MALDI-TOF MS and LC–MS/MS have different ionization and detection capabilities, which can result in varying mass detections for the same sample which has been demonstrated by Everley et al.29. MALDI-TOF MS often produces singly charged ions, which may lead to lower ionization efficiency. In contrast, LC–MS/MS typically generates multiply charged ions, allowing for the detection of a broader range of molecules with varying ionization efficiencies, thus providing higher sensitivity and resolution. In this study, the peptides identified by LC–MS/MS were compared with five prominent peptides analyzed by MALDI-TOF MS (at m/z 2496.9, 2697.9, 3034.1, 3710.1, and 4288.6), observed in the serum of early recurrence patients compared to late recurrence patients. The LC–MS/MS analysis identified m/z values of 2496.9, 2697.9, and 3034.1 as predominant in the early recurrence group, consistent with MALDI-TOF MS findings. These values correspond to DIHAAAKEIAEVNEINLEKVWD from Kinetochore-associated protein 1 (KNTC1) (UniProt ID: P50748), VTIIRSGVKPRKAVRILLNKKTAH from Serine/threonine-protein kinase DCLK1 (DCLK1) (UniProt ID: O15075), and LFIVIVPQKLLEFRYFILPYVIYR from Dol-P-Glc: Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase (ALG10) (UniProt ID: Q5I7T1), respectively. Among the three identified m/z values—2496.9 (KNTC1), 2697.9 (DCLK1), and 3034.1 (ALG10)—KNTC1 and DCLK1 were predominant in the early recurrence group. Both KNTC1 and DCLK1 have been reported to play oncogenic roles in promoting cancer progression. KNTC1 enhances proliferation and migration while suppressing apoptosis in gastric cancer (GC) cells via the PI3K/Akt/mTOR pathway and has been identified as a potential biomarker for gastric cancer30. DCLK1, a serine/threonine kinase, regulates cancer cell stemness, epithelial-mesenchymal transition (EMT), and drug resistance in high-grade ovarian cancer31. Although ALG10 has not been reported to promote cancer progression, it may serve as a potential peptide biomarker for distinguishing early and late recurrences in MALDI-TOF MS detection. Unfortunately, these peptides were not detected at significant levels in the LC–MS/MS analysis.
Therefore, to improve the classification of early and late recurrence in CCA patients, LC–MS/MS was performed to identify peptide biomarkers. The pooled samples from each group were subjected to LC–MS/MS analysis to minimize genetic variability and identify peptide biomarkers. These biomarkers, in conjunction with PMFs using MALDI-TOF MS, were used to enhance the accuracy of recurrence classification.
In early recurrence, the identified candidate peptide-based biomarkers in both groups were separately analyzed for peptide-chemotherapeutic drug interaction network using the STITCH database. In early recurrence, we found that 16 peptides had peptide-peptide interactions, while 5 peptides had both peptide-peptide and peptide-drug interactions. This study proposed, ATR, POLA1, BLM, SP100 and PPP1R15A (GADD34) as major candidate peptides that had a significant impact in the interaction network, while the remaining peptides also served as co-biomarkers for this condition (Fig. 3 and supplementary Table 1). This study identified a set of proteins involved in DNA stress response, DNA repair, and the maintenance of genomic instability, which are key features of cancer progression and chemoresistance.
ATR (Ataxia Telangiectasia and Rad3-related protein) is protein kinase family, phosphoinositide 3-kinase–related kinase (PIKKs), a key protein involved in the cellular response to DNA damage. It plays a critical role in the DNA damage response (DDR) by detecting DNA replication stress and activating repair pathways32. ATR also has non-canonical roles in cancer migration and invasion33,34. In addition, a high level of ATR is associated with poor survival in glioblastoma patients. Furthermore, ATR has been shown to play a role in cancer progression through in vitro and in vivo models, promoting cell proliferation, migration, and invasion—abilities that contribute to an increased risk of cancer recurrence34. In 2024, Buchynska et al. demonstrated that elevated ATR expression, along with its interaction and functional partner, checkpoint kinase 1 (CHEK1), is associated with the recurrence of endometrial carcinomas20. ATR also facilitates the function of kinesin-like protein (KIFC1) through phosphorylation, which contributes to recurrence in breast and colorectal cancers21. Consistent with these findings, our study highlights the predominant role of ATR in the early recurrence of CCA patients.
Inhibition of ATR has been reported as a target cancer treatment32,34,35. ATR is also reported co-interaction with POLA1 in DNA replication, the cell cycle and involving DNA repair for maintaining genome stability36. A previous study has indicated a potential role for CEP164 in ATM/ATR-mediated DNA damage response (DDR) and UV-induced nucleotide excision repair pathways37. ATR function in regulated DNA damage response and repair processes has been reported to be mediated through UBTF, a multifunctional architectural protein. UBTF is a multifunctional architectural protein containing multiple HMG boxes or the nucleolar proteins. It has been found that the Pol I transcription factor UBTF plays a dual role in regulating both Pol I and Pol II-mediated transcription. UBTF has also been reported to participate in DNA damage and repair processes, acting through mediators of the ATR/ATM-regulated DNA damage response38. Additionally, UBTF is involved in the cellular response to growth factor stimulation and can regulate cancer progression through the MAPK/ERK signaling pathway39. In addition, UBTF also has been reported to interact with NCL to be nucleolar proteins not only in genotoxic stress sensing but wound healing also40.
BLM is a DNA helicase essential for maintaining genomic stability. In cancer, various mutations can disrupt BLM function, leading to genomic instability and promoting cancer progression. Under these conditions, BLM aids cancer cell survival by supporting mechanisms that adapt to the genomic stress41. In addition, BLM has been reported to correlate with malignant progression and recurrence in pancreatic adenocarcinoma, with high expression levels of BLM associated with shorter disease-free survival (DFS), as shown by the GEPIA database19. In addition, Du et al. demonstrated that high levels of BLM were associated with recurrence in CCA patients, as demonstrated by the GEPIA database. They also confirmed the roles of BLM in contributing to CCA progression through the enhanced proliferation and migration abilities of CCA cell lines. In addition, BLM has been reported as downstream of ATR signaling via phosphorylation at Thr99 and Thr122 42. BLM and ATR work together in cancer progression by regulating human exonuclease V (EXO5) to restart stalled DNA replication forks after stress during cancer treatment. ATR-mediated replication stress responses and BLM helicase partners via EXO5 in tumors correlated with higher mutation rates and poor patient survival, highlighting its oncogenic potential22. Cohen S et al. have revealed that BLM and SETX are recruited to transcription-coupled DNA double-strand breaks (DSBs) during DSB repair43. SETX is involved in various processes related to genome integrity, transcription, RNA metabolism, and DNA damage repair and inflammation in cancer progression44.
SP100 is a nuclear autoantigen that plays a role in several cellular processes. Several studies have reported that SP100 has tumor suppressive functions, and some studies have suggested dual-functions in cancer suppression and cancer progression depended on cancer types. In pancreatic adenocarcinoma, SP100 has been reported to be associated with poorer survival and adverse clinical features45. A study on glioma demonstrated that elevated expression of the Speckled Protein (SP) family, including SP100 and SP140, were associated with poorer prognosis, including high recurrence rate and shorter survival, in glioma patients. Furthermore, the SP family contributes to glioma progression through the TRIM22/PI3K/AKT signaling pathway18. The functions of SP100 in CCA progression is not well understood. Our results for the first time found that SP100 was overexpressed in early recurrent CCA patients.
PPP1R15A (GADD34) is a regulatory subunit of protein phosphatase 1. For HCC, PPP1R15A could promote immunosuppressive in generating tumor micro environment and progression as well as high expression of PPP1R15A associated with poor clinical outcomes in HCC46.
In early recurrent patients, a total of 5 of 16 peptides, including, ATR, BLM, POLA1, SP100 and PPP1R15A, were directly associated with chemotherapeutic drugs for CCA treatment, Cisplatin, while CEP167 showed indirect interactions with three chemotherapeutic drugs, namely, Gemcitabine, Cisplatin and 5-Fu as demonstrated in protein-chemical interaction network (Fig. 3).
In late recurrence, we found that there were 4 peptides and 2 peptides having peptide-peptide and-drug interaction, respectively. Our results showed that SERPINA1, TGFB2, SERPING1 and CAD were major candidate peptides that had a significant impact in the interaction network, while the remaining peptides also served as co-biomarkers for this condition (Fig. 4 and supplementary Table 2).
SERPINA1, also known as alpha-1 antitrypsin (AAT), is a member of the serpin (serine protease inhibitor) superfamily. Its primary function is to inhibit serine proteases, particularly neutrophil elastase, which plays a significant role in inflammatory processes. Ma et al. reported the and role of SERPINA1 in colorectal cancer. They demonstrated that SERPINA1 is associated with clinical risk factors for recurrence, including tumor stage and lymph node metastasis. Additionally, they investigate the role of SERPINA1 in cancer progression by mediating STAT3 signaling in both in vitro and in vivo models of colorectal cancer47. An elevated levels of SERPINA1 have been reported in CCA and are associated with poorer survival outcomes and more advanced tumor staging and shorter disease-free survival (DFS). Furthermore, high SERPINA1 expression is linked to enriched pathways related to the complement system and extracellular matrix interactions, highlighting its potential role in the tumor microenvironment and cancer progression and recurrence48. While, SERPING1 or PAI-1 is a member of the serpin (serine protease inhibitor) superfamily like SERPINA1. It encodes a highly glycosylated plasma protein that plays a critical role in regulating the complement cascade and the immune response. Synthesized in the liver, SERPING1 is important for managing several physiological processes, including complement activation, blood coagulation, fibrinolysis, and the generation of kinins. Diseases linked to SERPING1 include hereditary angioedema and partial deficiency of complement components49. High level of SERPING1 is reported that associated shorter RFS and OS in gastric cancer50. In STICTH database, SERPINA1 has a strong association with TGFB2 and SERPING1. TGFB2 is a member of the transforming growth factor-beta (TGF-β) which plays roles in immunosuppressive tumor microenvironment and promote epithelial-to-mesenchymal transition (EMT), contributing to cancer cell migration and invasion in gastric cancer51. Moreover, high level of TGFB2 has been reported to associate with shorter DFS and OS in triple-negative breast cancer52. The interaction of serpin superfamily (SERPINA1 (alpha-1 antitrypsin) and SERPING1 (PAI-1)) and TGFB family have been reported that serpin superfamily is downstream of TGFB153,54. However, there are no studies relating to SERPINA1, TGFB2 and SERPING.
CAD (Carbamoyl-phosphate synthetase 2, Aspartate transcarbamoylase, and Dihydroorotase) is a multifunctional enzyme critical for the first three rate-limiting steps of pyrimidine nucleotide synthesis, essential for nucleic acids, cell membranes, and active intermediates55. Dysregulation or mutations in CAD are associated with cancer, neurological disorders, and metabolic diseases. It supports growth-related processes by producing precursors like UDP, vital for glycosylation, protein modifications, and synthesis of polysaccharides and phospholipids. CAD is regulated by post-translational phosphorylation by MAPK/cAMP-Dependent PKA/PKC and PI3K-AKT-mTORC1-S6K1 Pathways56. CAD is enriched in a set of cancer types with poor clinical outcomes. Ridder et al. reported that elevated CAD levels are associated with shorter recurrence-free survival and overall survival in liver cancer patients57. Additionally, Wang et al. revealed that analyses of The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets demonstrated a similar association between high CAD levels and reduced recurrence-free survival and overall survival in lung cancer58. These findings support our study’s observation that CAD was dominantly expressed in late recurrence in CCA patients.
In addition, late recurrent patients, a total of 2 of 4 peptides, including, SERPINA1 was directly associated with chemotherapeutic drugs for CCA treatment, Cisplatin, while CAD was showed as a hub of interactions to all chemotherapeutic drugs through DHODH, DPYD and TYMS (Fig. 4). This interaction should be validated in further investigations.
Based on the findings above, we identified strong interactions and significant associations between the selected peptides and recurrence, as predicted through the STITCH database and supported by published evidence. By using PMFs generated by MALDI-TOF MS and peptide-based biomarkers identified via LC–MS/MS from CCA patients with recurrence, these results demonstrate the potential to develop robust diagnostic panels that can precisely classify disease severity. These insights lay the foundation for future research focused on personalized treatment strategies based on recurrence timing, with rapid screening methods like PMFs and peptide biomarker panels playing a key role in guiding therapeutic decisions and improving patient outcomes (Fig. 5).
In conclusion, peptide mass fingerprinting (PMFs) using MALDI-TOF MS and peptide biomarker identification via LC–MS/MS enable precise differentiation between early and late recurrence, enhancing diagnostic accuracy. This study demonstrates the utility of PMFs and peptide biomarkers to identify high-risk recurrence of CCA patients, thereby improving treatment strategies.
Materials and methods
Ethics approval and consent to participate
This study was conducted based on the principles of Good Clinical Practice, the Declaration of Helsinki, and national laws and regulations about clinical studies. In addition, informed consent was obtained from all patients. All processes of this study were accepted and approved by the Khon Kaen University Ethics Committee for Human Research under the reference number HE661318.
Population and sample group
This research was a single-center study conducted as a retrospective-prospective analytical observational study involving a sample of 81 patients diagnosed with cholangiocarcinoma. Data were collected retrospectively from the medical records at Srinagarind Hospital, Faculty of Medicine, Khon Kaen University, including clinical data of patients from January 1, 2017, to December 31, 2021. Serum samples were obtained from the biobank at the Cholangiocarcinoma Research Institute, Khon Kaen University.
Patients with CCA were categorized into early and late recurrence groups based on a disease-free survival (DFS) cut-off of 365 days. By this definition, CCA patients with DFS < 365 days were categorized as having early recurrence, while CCA patients with DFS ≥ 365 days were categorized as having late recurrence. This cut-off value was selected in accordance with previous studies2,4,3, which demonstrated its relevance in recurrence classification.
Prognostic factors were collected using a retrospective data collection form from the patient medical records, utilizing the ISAN Cohort database from the Cholangiocarcinoma Research Institute, Faculty of Medicine, Khon Kaen University. The data collected included age at diagnosis, gender, histological confirmation, tumor size, cancer grade, cancer staging, surgical margin, lymph node metastasis, lympho-vascular invasion, histological grade, and chemotherapy received.
Clinical outcome follow-up
The follow-up period for patients with cholangiocarcinoma extended from the date of surgery for at least 5 years, starting from January 1, 2017. All causes of death were monitored through a life status verification from the database of the Ministry of Interior, and additional data were collected from medical records documented by physicians. Typically, after treatment, patients were scheduled for follow-up at Srinagarind Hospital, Faculty of Medicine, Khon Kaen University, every 6 months for at least 5 years. The variables studied included recurrence, 5-year survival, overall survival (OS), and disease-free survival (DFS).
Sample collection and serum preservation
For serum sample collection from patients with cholangiocarcinoma prior to surgical treatment, blood was drawn from a vein (venipuncture) with a volume of 5 milliliters into a clot blood tube. It was ensured that clot formation was complete before centrifugation. Serum was then separated from red blood cells using a centrifuge at 3,000–3,500 RPM at 4 °C for 10 min. The serum was aspirated and aliquoted into 1 µl portions in Eppendorf tubes to avoid repeated thawing of samples, and then stored at -80 °C in the biobank of the Cholangiocarcinoma Research Institute, Khon Kaen University, until further analysis. protein quantification was performed using the Lowry assay.
Peptide barcode analysis using MALDI-TOF MS
Serum samples were mixed with a matrix solution (MALDI solution: α-cyano-4-hydroxycinnamic acid (CHCA) in 50% acetonitrile (ACN) with 0.1% trifluoroacetic acid) at a sample-to-matrix ratio of 1:5. The resulting mixture was applied onto a MALDI target plate (MTP 384 ground steel, JEOL, Japan), with each sample spotted 30 times on the plate. The plate was allowed to dry at ambient temperature before analysis using the JMS-S3000 SpiralTOF (JEOL, Japan) in linear positive mode, focusing on the detection of peptide barcodes with molecular weights between 1,000 and 10,000 Da. Each sample was subjected to 1,500 laser shots. The MALDI-TOF data was acquired using JEOL msTornado Control version 1.16 (JEOL, Japan) and processed with JEOL msTornado Analysis version 1.15 (JEOL, Japan). The spectra were processed using default parameters for smoothing, variance stabilization, baseline correction, and peak detection, and then exported in CSV format for further data preparation. Mass binning at 1.0 Da was performed with a mass range set from 1,000 Da to 10,000 Da. Prior to initial experiments, an independent external mass calibration for positive-ion mode was established. External calibration was conducted using peptides with known mass-to-charge ratios (m/z), including Angiotensin II (m/z = 1,046), P14R (m/z = 1,533), human ACTH fragment 18–39 (m/z = 2,465), bovine insulin oxidized B chain (m/z = 3,465), and bovine insulin (m/z = 5,731). Mass calibration of the instrument was performed by manual peak assignment to the reference list by JEOL msTornado Control version 1.16 with mass accuracy ± 100 ppm.
Peptidome analysis in serum using LC–MS/MS
To reduce genetic variation and identify peptide biomarkers representative of early and late recurrence, we analyzed pooled samples from each group using LC–MS/MS, ensuring equal protein amounts in each pool17. Serum samples were purified using C18 ZipTip and analyzed for peptide content via LC–MS/MS on the Q-TOF Impact II™ system. Specifically, one microliter of peptide digests was enriched on a µ-Precolumn (300 μm i.d. × 5 mm, C18 Pepmap 100, 5 μm, 100 Å; Thermo Scientific, UK) and subsequently separated on a 75 μm i.d. × 15 cm column packed with Acclaim PepMap RSLC C18, 2 μm, 100 Å (nanoViper, Thermo Scientific, UK). The C18 column was maintained at 60 °C in a thermostatted oven. Solvents A and B, containing 0.1% formic acid in water and 0.1% formic acid in 80% acetonitrile, respectively, were used to elute peptides with a gradient of 5–55% solvent B over 30 min at a constant flow rate of 0.30 µL/min. Electrospray ionization was performed at 1.6 kV using the CaptiveSpray, with nitrogen as the drying gas at a flow rate of approximately 50 L/h. Collision-induced dissociation (CID) was conducted with nitrogen as the collision gas, and both MS and MS/MS spectra were acquired in positive-ion mode at a frequency of 2 Hz over an m/z range of 150–2200, with collision energy adjusted to 10 eV according to m/z. Each sample was analyzed in triplicate, and MaxQuant version 2.5.0.0 (Tyanova et al., 2016) was used for peptide quantification and sequencing, employing the Andromeda search engine to match MS/MS spectra against the Uniprot Homo sapiens database. The enzyme specificity was set at unspecific digestion. Variable modifications were set to oxidation of methionine residues and N-terminal protein acetylation. In the settings of advanced identifications, matching between runs was enabled (with standard settings). All other settings were kept as standard. The protein false discovery rate (FDR) was controlled at 1%, estimated using reverse-sequence searches, with a maximum of five modifications allowed per peptide for data analysis.
Bioinformatics analysis of peptidomics data
Visualization and statistical analysis of the LC–MS data, including Principal Component Analysis (PCA), differential analysis (one-way ANOVA, volcano plots, boxplots, and heatmaps), were performed using MetaboAnalyst version 6.0, applying a significance threshold of p < 0.05 (Pang et al., 2022). Functional analysis of proteins containing identified peptides was conducted with the Panther database (Mi et al., 2019) and ShinyGO (Ge et al., 2020). Additionally, associations between peptides, their source proteins, and related proteins or chemicals were investigated using the STITCH database (Szklarczyk et al., 2016).
The overall survival (OS) was calculated using the Kaplan-Meier method, where disease-free survival was defined as the time from surgery to recurrence, and overall survival was defined as the time from surgery to death. Patients who survived beyond the study period had their median DFS, median survival time, and survival rates calculated with a 95% confidence interval. Comparisons between groups were analyzed using the Log-rank test, while univariate and multivariate analyses to identify prognostic factors were performed using Cox regression models. A p-value of < 0.05 was considered statistically significant. All analyses were conducted using IBM SPSS Statistics version 26.
Data availability
The raw MS/MS spectra data are available in ProteomeXchange: JPST003447 and PXD057396 (preview URL for Reviewers: https://repository.jpostdb.org/preview/1049237987672425a2e1cca, Access key: 6186).
References
Banales, J. M. et al. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 17, 557–588. https://doi.org/10.1038/s41575-020-0310-z (2020).
Nassar, A. et al. Factors of early recurrence after resection for Intrahepatic Cholangiocarcinoma. World J. Surg. 46, 2459–2467. https://doi.org/10.1007/s00268-022-06655-1 (2022).
Yamauchi, N. et al. Clinical significance of early recurrence after curative resection of Colorectal Cancer. Anticancer Res. 42, 5553–5559. https://doi.org/10.21873/anticanres.16061 (2022).
Groot, V. P. et al. Defining and Predicting Early recurrence in 957 patients with resected pancreatic ductal adenocarcinoma. Ann. Surg. 269, 1154–1162. https://doi.org/10.1097/sla.0000000000002734 (2019).
Kim, H. S. et al. Serum carcinoembryonic antigen and carbohydrate antigen 19-9 as preoperative diagnostic biomarkers of extrahepatic bile duct cancer. BJS Open. 5 https://doi.org/10.1093/bjsopen/zrab127 (2021).
Kang, J. S. et al. Limits of serum carcinoembryonic antigen and carbohydrate antigen 19-9 as the diagnosis of gallbladder cancer. Ann. Surg. Treat. Res. 101, 266–273. https://doi.org/10.4174/astr.2021.101.5.266 (2021).
Tanase, C., Albulescu, R. & Neagu, M. Proteomic approaches for Biomarker panels in Cancer. J. Immunoass. Immunochem. 37, 1–15. https://doi.org/10.1080/15321819.2015.1116009 (2016).
Liu, J. et al. Proteomics-derived Biomarker Panel facilitates distinguishing primary lung adenocarcinomas with intestinal or mucinous differentiation from lung metastatic colorectal Cancer. Mol. Cell. Proteom. 23 https://doi.org/10.1016/j.mcpro.2024.100766 (2024).
Antonarakis, E. S. et al. The natural history of metastatic progression in men with prostate-specific antigen recurrence after radical prostatectomy: Long-term follow-up. 109, 32–39 https://doi.org/10.1111/j.1464-410X.2011.10422.x (2012).
Ostrowitzki, S. et al. A phase III randomized trial of gantenerumab in prodromal Alzheimer’s disease. Alzheimers Res. Ther. 9, 95. https://doi.org/10.1186/s13195-017-0318-y (2017).
Kanda, M. et al. The combination of the serum carbohydrate antigen 19-9 and carcinoembryonic antigen is a simple and accurate predictor of mortality in pancreatic cancer patients. Surg. Today. 44, 1692–1701. https://doi.org/10.1007/s00595-013-0752-9 (2014).
Petricoin, E. F., Belluco, C., Araujo, R. P. & Liotta, L. A. The blood peptidome: A higher dimension of information content for cancer biomarker discovery. Nat. Rev. Cancer. 6, 961–967. https://doi.org/10.1038/nrc2011 (2006).
Vadevoo, S. M. P. et al. Peptides as multifunctional players in cancer therapy. Exp. Mol. Med. 55, 1099–1109. https://doi.org/10.1038/s12276-023-01016-x (2023).
Sun, J. et al. Evaluation of prostate cancer based on MALDI-TOF MS fingerprinting of nanoparticle-treated serum proteins/peptides. Talanta 220, 121331. https://doi.org/10.1016/j.talanta.2020.121331 (2020).
Kim, S. S. et al. Quantifiable peptide library bridges the gap for proteomics based biomarker discovery and validation on breast cancer. Sci. Rep. 13, 8991. https://doi.org/10.1038/s41598-023-36159-4 (2023).
Rungkamoltip, P., Roytrakul, S. & Navakanitworakul, R. MALDI-TOF MS analysis of serum peptidome patterns in Cervical Cancer. Biomedicines 11 https://doi.org/10.3390/biomedicines11082327 (2023).
Karp, N. A. & Lilley, K. S. Design and analysis issues in quantitative proteomics studies. Proteomics 7(Suppl 1), 42–50. https://doi.org/10.1002/pmic.200700683 (2007).
Li, X. et al. SP140 inhibitor suppressing TRIM22 expression regulates glioma progress through PI3K/AKT signaling pathway. Brain Behav. 14, e3465. https://doi.org/10.1002/brb3.3465 (2024).
Li, Q. et al. Aberrant expression of BLM correlates with malignant progression and Immune Infiltration in pancreatic adenocarcinoma. 2, 14–29, (2023). https://doi.org/10.14218/csp.2022.00015
Buchynska, L., Gordiienko, I., Glushchenko, N. & Iurchenko, N. The KRAS, ATR and CHEK1 expression levels in endometrial cancer are the risk factors predicting recurrence. PLoS ONE. 19, e0302075. https://doi.org/10.1371/journal.pone.0302075 (2024).
Fan, G. et al. The ATM and ATR kinases regulate centrosome clustering and tumor recurrence by targeting KIFC1 phosphorylation. Nat. Commun. 12, 20. https://doi.org/10.1038/s41467-020-20208-x (2021).
Hambarde, S. et al. EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart. Mol. Cell. 81, 2989–3006e2989. https://doi.org/10.1016/j.molcel.2021.05.027 (2021).
Gonzalez-Gonzalez, L., Alonso, J. & Periostin A matricellular protein with multiple functions in Cancer Development and Progression. Front. Oncol. 8, 225. https://doi.org/10.3389/fonc.2018.00225 (2018).
Li, L., Wu, J., Lyon, C. J., Jiang, L. & Hu, T. Y. Clinical peptidomics: Advances in instrumentation, analyses, and applications. BME Front. 4, 0019. https://doi.org/10.34133/bmef.0019 (2023).
Sandanayake, N. S. et al. Identification of potential serum peptide biomarkers of biliary tract cancer using MALDI MS profiling. BMC Clin. Pathol. 14 https://doi.org/10.1186/1472-6890-14-7 (2014).
Chaiyarit, P., Taweechaisupapong, S., Jaresitthikunchai, J., Phaonakrop, N. & Roytrakul, S. Comparative evaluation of 5-15-kDa salivary proteins from patients with different oral diseases by MALDI-TOF/TOF mass spectrometry. Clin. Oral Investig.. 19, 729–737. https://doi.org/10.1007/s00784-014-1293-3 (2015).
Yigitbasi, T. et al. An efficient biomarker panel for diagnosis of breast cancer using surface-enhanced laser desorption ionization time-of-flight mass spectrometry. Biomed. Rep. 8, 269–274. https://doi.org/10.3892/br.2018.1042 (2018).
Ploypetch, S. et al. Utilizing MALDI-TOF MS and LC–MS/MS to access serum peptidome-based biomarkers in canine oral tumors. Sci. Rep. 12, 21641. https://doi.org/10.1038/s41598-022-26132-y (2022).
Everley, R. A., Mott, T. M., Wyatt, S. A., Toney, D. M. & Croley, T. R. Liquid chromatography/mass spectrometry characterization of Escherichia coli and Shigella species. J. Am. Soc. Mass. Spectrom. 19, 1621–1628. https://doi.org/10.1016/j.jasms.2008.07.003 (2008).
Qi, H. et al. KNTC1 functions as a potential biomarker and oncogene regulating proliferation, migration and apoptosis in gastric cancer. Int. Immunopharmacol. 143, 113257. https://doi.org/10.1016/j.intimp.2024.113257 (2024).
Dogra, S. et al. Targeting doublecortin-like kinase 1 reveals a novel strategy to circumvent chemoresistance and metastasis in ovarian cancer. Cancer Lett. 578, 216437. https://doi.org/10.1016/j.canlet.2023.216437 (2023).
Mavroeidi, D., Georganta, A., Panagiotou, E., Syrigos, K. & Souliotis, V. L. Targeting ATR Pathway in solid tumors: Evidence of improving therapeutic outcomes. Int. J. Mol. Sci. 25 https://doi.org/10.3390/ijms25052767 (2024).
Kidiyoor, G. R. et al. ATR is essential for preservation of cell mechanics and nuclear integrity during interstitial migration. Nat. Commun. 11, 4828. https://doi.org/10.1038/s41467-020-18580-9 (2020).
Derby, S. J. et al. Inhibition of ATR opposes glioblastoma invasion through disruption of cytoskeletal networks and integrin internalization via macropinocytosis. Neuro Oncol. 26, 625–639. https://doi.org/10.1093/neuonc/noad210 (2024).
Gralewska, P., Gajek, A., Marczak, A. & Rogalska, A. Participation of the ATR/CHK1 pathway in replicative stress targeted therapy of high-grade ovarian cancer. J. Hematol. Oncol. 13, 39. https://doi.org/10.1186/s13045-020-00874-6 (2020).
Schneider, H. E. et al. Synthetic lethality between ATR and POLA1 reveals a potential new target for individualized cancer therapy. Neoplasia 57, 101038. https://doi.org/10.1016/j.neo.2024.101038 (2024).
Sivasubramaniam, S., Sun, X., Pan, Y. R., Wang, S. & Lee, E. Y. Cep164 is a mediator protein required for the maintenance of genomic stability through modulation of MDC1, RPA, and CHK1. Genes Dev. 22, 587–600. https://doi.org/10.1101/gad.1627708 (2008).
Diesch, J., Hannan, R. D. & Sanij, E. Genome wide mapping of UBF binding-sites in mouse and human cell lines. Genom. Data. 3, 103–105. https://doi.org/10.1016/j.gdata.2014.12.005 (2015).
Zhang, J. et al. UBTF facilitates melanoma progression via modulating MEK1/2-ERK1/2 signalling pathways by promoting GIT1 transcription. Cancer Cell. Int. 21, 543. https://doi.org/10.1186/s12935-021-02237-8 (2021).
Kaya, M., Eroz, R. & Kabakliogli, M. Expression of nucleolin, nucleophosmin, upstream binding transcription factor genes and propolis in wound models. J. Wound Care. 31, S28–S40. https://doi.org/10.12968/jowc.2022.31.Sup10.S28 (2022).
Kaur, E., Agrawal, R. & Sengupta, S. Functions of BLM helicase in cells: Is it acting like a double-edged Sword? Front. Genet. 12, 634789. https://doi.org/10.3389/fgene.2021.634789 (2021).
Wang, H. et al. BLM prevents instability of structure-forming DNA sequences at common fragile sites. PLoS Genet. 14, e1007816. https://doi.org/10.1371/journal.pgen.1007816 (2018).
Cohen, S. et al. A POLD3/BLM dependent pathway handles DSBs in transcribed chromatin upon excessive RNA:DNA hybrid accumulation. Nat. Commun. 13, (2012). https://doi.org/10.1038/s41467-022-29629-2 (2022).
Gatti, V., De Domenico, S., Melino, G. & Peschiaroli, A. Senataxin and R-loops homeostasis: Multifaced implications in carcinogenesis. Cell. Death Discov.. 9, 145. https://doi.org/10.1038/s41420-023-01441-x (2023).
Duan, Y., Du, Y., Mu, Y., Gu, Z. & Wang, C. Expression, prognostic value and mechanism of SP100 family in pancreatic adenocarcinoma. Aging (Albany NY). 15, 5569–5591. https://doi.org/10.18632/aging.204811 (2023).
Liu, X. et al. PPP1R15A-expressing monocytic MDSCs promote immunosuppressive liver microenvironment in fibrosis-associated hepatocellular carcinoma. JHEP Rep. 6, 101087. https://doi.org/10.1016/j.jhepr.2024.101087 (2024).
Ma, Y. et al. CEBPB-mediated upregulation of SERPINA1 promotes colorectal cancer progression by enhancing STAT3 signaling. Cell. Death Discov.. 10, 219. https://doi.org/10.1038/s41420-024-01990-9 (2024).
Cornillet, M. et al. Increased prevalence of Alpha-1-antitrypsin deficiency in patients with biliary tract cancer and its associated clinicopathological features. Cells 12 https://doi.org/10.3390/cells12121663 (2023).
Ponard, D. et al. SERPING1 mutation update: Mutation spectrum and C1 inhibitor phenotypes. Hum. Mutat. 41, 38–57. https://doi.org/10.1002/humu.23917 (2020).
Li, X. C., Wang, S., Zhu, J. R., Wang, Y. P. & Zhou, Y. N. J. T. C. R. Nomograms combined with SERPINE1-related module genes predict overall and recurrence-free survival after curative resection of gastric cancer: A study based on TCGA and GEO data. 9, 4393–4412 (2020).
Yang, B. et al. TGFB2 serves as a link between epithelial-mesenchymal transition and tumor mutation burden in gastric cancer. Int. Immunopharmacol. 84, 106532. https://doi.org/10.1016/j.intimp.2020.106532 (2020).
Zhou, C. et al. TGFB2-AS1 inhibits triple-negative breast cancer progression via interaction with SMARCA4 and regulating its targets TGFB2 and SOX2. Proc. Natl. Acad. Sci. U.S.A. 119, e2117988119. https://doi.org/10.1073/pnas.2117988119 (2022).
Wang, X. et al. Oxymatrine inhibits the migration of human colorectal carcinoma RKO cells via inhibition of PAI-1 and the TGF-beta1/Smad signaling pathway. Oncol. Rep. 37, 747–753. https://doi.org/10.3892/or.2016.5292 (2017).
Chen, Y. H. et al. The induction of Alpha-1 antitrypsin by Vitamin D in human T cells is TGF-β Dependent: A proposed anti-inflammatory role in Airway Disease. 8, (2021). https://doi.org/10.3389/fnut.2021.667203
Evans, D. R. & Guy, H. I. Mammalian pyrimidine biosynthesis: Fresh insights into an ancient Pathway*. J. Biol. Chem. 279, 33035–33038. https://doi.org/10.1074/jbc.R400007200 (2004).
Li, G., Li, D., Wang, T., He, S. & Pyrimidine Biosynthetic Enzyme, C. A. D. Its function, regulation, and diagnostic potential. Int. J. Mol. Sci. 22 https://doi.org/10.3390/ijms221910253 (2021).
Ridder, D. et al. Key enzymes in pyrimidine synthesis, CAD and CPS1, predict prognosis in hepatocellular carcinoma. Cancers 13, 744. https://doi.org/10.3390/cancers13040744 (2021).
Wang, H., Wang, X., Xu, L., Zhang, J. & Cao, H. High expression levels of pyrimidine metabolic rate–limiting enzymes are adverse prognostic factors in lung adenocarcinoma: A study based on the Cancer Genome Atlas and Gene expression Omnibus datasets. Purinergic Signal. 16, 347–366. https://doi.org/10.1007/s11302-020-09711-4 (2020).
Acknowledgements
All authors are truly thankful for helpful discussions with the late Prof. Narong Khuntikeo at Department of Surgery, Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand, Cholangiocarcinoma Research Institute (CARI), Khon Kaen University, Khon Kaen, Thailand and Cholangiocarcinoma Screening and Care Program (KKU), Khon Kaen University, Khon Kaen, Thailand. We are also indebted to all members of CASCAP, particularly the cohort members, and researchers at CARI, Faculty of Medicine, Khon Kaen University for collecting and proofing of CCA patient data. We also acknowledge Professor Ross H. Andrews for editing the MS via the Publication Clinic KKU, Thailand.
Funding
This work was supported by the National Science Research and Innovation Fund (NSRF) through Khon Kaen University to WL.
Author information
Authors and Affiliations
Contributions
Conceptualization— W.L., and A.T; Methodology—P.P., N.M., W.L., N.N., P.K., A.W. S.C. and J.J.; Formal analysis— V.T. and P.P.; Investigation— P.P., S.C., J.J. and S.R.; Resources— W.L., S.R. and A.T.; Data curation— V.T. and P.P.; Visualization—V.T., P.P., W.L., S.R. and A.T.; Supervision— W.L., S.R. and A.T.; Project administration— W.L. and A.T.; Funding acquisition— W.L.; Original draft preparation— V.T. and P.P.; Draft review and editing—all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Thanasukarn, V., Prajumwongs, P., Muangritdech, N. et al. Discovery of novel serum peptide biomarkers for cholangiocarcinoma recurrence through MALDI-TOF MS and LC–MS/MS peptidome analysis. Sci Rep 15, 2582 (2025). https://doi.org/10.1038/s41598-025-87124-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-87124-2
Keywords
This article is cited by
-
From detection to drug delivery: innovations in cancer biosensors from concept to clinical impact
Biotechnology for Sustainable Materials (2025)
-
Identification of serum peptide biomarkers for cholangiocarcinoma diagnosis and staging via MALDI-TOF MS and LC-MS/MS
British Journal of Cancer (2025)
-
Mass spectrometry-based serum peptidomic profiling reveals potential biomarker for canine hepatozoonosis
Scientific Reports (2025)
-
Serum peptide biomarkers by MALDI-TOF MS coupled with machine learning for diagnosis and classification of hepato-pancreato-biliary cancers
Scientific Reports (2025)