Abstract
During the winter 2023–2024, an upsurge of Mycoplasma pneumoniae (M.pneumoniae) was noted in the Netherlands. To investigate the distribution of M.pneumoniae sequence types from different patient populations and to explore genotypic macrolide resistance which is common in East Asia but not (yet) in Europe. M.pneumoniae positive throat/nasal samples from participatory respiratory surveillance, patients visiting general practitioners with an acute respiratory infection including community acquired pneumonia (CAP) and hospitalised patients with CAP were included, representing different disease severity. The M.pneumoniae were typed with multilocus sequence typing and the 23 S rRNA region was sequenced to determine macrolide resistance markers. In total, 153 M.pneumoniae were sequenced, six sequence types (STs) and only one bacterium with macrolide resistance marker were detected. No link between STs or bacterial load (PCR cycle threshold) and source population of M.pneumoniae was detected. In the Netherlands, the M.pneumoniae upsurge in 2023–2024 existed of multiple commonly found STs. No link between ST and severity of illness was detected. Macrolide resistance remained sporadic.
Similar content being viewed by others
Introduction
Mycoplasma pneumoniae (M. pneumoniae) infection is a common cause of community-acquired pneumonia (CAP), with regional outbreaks occurring every 2–7 years1,2.
In the Netherlands, prevalence of M. pneumoniae detections was relatively low since the last national outbreak in the 2011/2012 winter until the winter of 2023/2024 which showed a steep increase in detections by Dutch medical microbiology laboratories, based on the national laboratory-based reporting system “virologische weekstaten” and in a regional hospital3,4. However, at that time no M. pneumoniae were typed and no resistance for first choice treatment with macrolide antibiotics was determined.
In this retrospective sequence study, we therefore aimed to obtain a molecular epidemiological picture of M. pneumoniae from cases with respiratory tract symptoms from (1) those self-reporting with ARI from the community (Infectieradar), (2) those attending the general practitioner (GP) with community acquired pneumonia (CAP) or other acute respiratory infections (ARI) (Nivel) and (3) hospitalised patients with CAP during the winter of 2023/2024 in the Netherlands (Hospitals). In addition, we wanted to investigate macrolide resistance given that recent publications show high levels of resistance (88–90%)5,6,7.
Methods
Retrospective study
A retrospective study was undertaken on upper respiratory tract samples collected for either surveillance or clinical diagnosis that were tested positive for M. pneumoniae by PCR.
Sample origin
Only positive samples were included in this study. The positive samples originated from three sources that loosely represent markers for disease severity, namely, patients with mild symptoms (Infectieradar), those visiting a GP because of CAP or ARI symptoms (Nivel) and those hospitalized with ARI complaints (Hospitals).
Infectieradar
Infectieradar is a participatory syndromic and molecular surveillance system for ARI8 Samples (nose swab and throat swab combined in one tube virus transport medium (VTM)) are self-collected voluntarily by Infectieradar participants after completing an online questionnaire in which they reported symptoms of an ARI. Samples are submitted to RIVM where they are routinely tested for M. pneumoniae since the start of Infectieradar in 2022. M. pneumoniae has been detected for the first time in samples collected in week 26/2023. M. pneumoniae positive samples collected through week 9/2024 were included.
GPs of the Nivel Primary Care Database—GP sentinel surveillance
Nivel Primary Care Database – Sentinel Practice network performs sentinel clinical and virological surveillance for ARI9. Each GP collects a nose swab and throat swab combined in one tube VTM from up to five patients per week. M. pneumoniae positive samples from patients visiting a GP of the network and collected from week 40/2023 through week 5/2024 and analyzed at RIVM were included. In this period a targeted case-control study was performed for potential causes of an unusual upsurge of CAP among older children and young adults (manuscript in preparation). This case-control study included testing for bacteria, among which M. pneumoniae, whilst routinely GP sentinel surveillance samples are only tested for respiratory viruses. The M. pneumoniae positive samples originated from patients with an acute respiratory infection and diagnosed with CAP or with another acute respiratory infection but not diagnosed with CAP or influenza-like illness (ILI) (further referred to as OARI).
Hospitals
Since the COVID-19 pandemic, respiratory clinical samples at hospitals are collected by swabbing both the throat and nasal area with a single swab. Samples from hospitalised CAP patients in two Rotterdam regional hospitals (Maasstad Ziekenhuis and Franciscus Gasthuis) were included from week 16/2023 through week 9/2024. For diagnostic purposes, samples are routinely tested for M. pneumoniae on request of the treating doctor of the patients. As of week 16/2023, an upsurge in detections of M. pneumoniae in 2023 was observed in these hospitals. Therefore, samples were included since week 16/2023.
Laboratory methods
Respiratory clinical samples were either tested by commercial real-time PCR test (RespiFinder® 2Smart, PathoFinder, Netherlands; Infectieradar samples) which includes M. pneumoniae, in-house developed real-time PCR test targeting the multicopy P1 adhesive gene of M. pneumoniae (hospital samples) or determined by real-time BIOFIRE® FILMARRAY® Pneumonia Panel plus (GP sentinel surveillance and hospital samples) which includes M. pneumoniae. M. pneumoniae positive samples were further analyzed and sequenced at Maasstad Hospital. DNA was extracted using the Emag system (BioMerieux, France), Ct value determined by in-house developed real-time PCR test (p1 gene) and sequenced on Minion platform (Oxford Nanopore technologies, Oxford, UK). P1 gene primer sequences were as follows: Forward: GGAACGCGAACCACTTGTGT, Reverse: GATATAACCGCGCCTCAAAMC, probe: Yakima Yellow-ACTARTTCCGCTGGACRACCCCG.
The Multilocus Sequence Typing (MLST) scheme by PubMLST10,11adjusted with adk, gmk and atpA outer primer pairs from12were used to obtain longer PCR products to better suit the workflow with the Nanopore rapid barcoding kit (SQK-RBK114.96). PCRs consisted of an initial denaturation step of 3 min at 94 °C, followed by 35 cycles of 60 s at 94 °C, 60 s at 60 °C, and 60 s at 72 °C. A final extension step was maintained for 10 min at 72 °C. After 35 cycles, PCR products were quantified by Qubit fluorometer (ThermoFischer, USA), normalised and processed following the Rapid sequencing genomic DNA protocol before loading onto flow cells (R10.4.1) on a MinIon 1B device. Fastq files were generated using MinKNow 24.02.8 and analysed using CLC genomic Workbench 24.0.1 (Qiagen, Germany) using a custom workflow; reads were filtered on quality (Q > 12) and length (> 100 bp), map long reads to reference tool (minimap2) was used to map reads to the eight MLST reference genes, a consensus was generated per target (> 20x coverage) using the MLST typing workflow. In case 20x coverage was not obtained, alignment was manually curated to call the Sequence Type (ST) if possible. A minimum spanning tree (UPGMA) was created in BioNumerics 7.6.3 (AppliedMaths, Biomerieux, France). Potential macrolide resistance was called by sequencing a 654 bp region of the 23 S rRNA, targeting the hotspot region for macrolide resistance markers13, following the same workflow as for MLST5.
Results
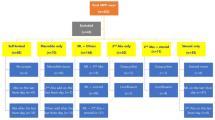
A total of 209 M. pneumoniae positive throat and/or nasal swabs were included in this study of which 158 (76%) were successfully sequenced. The samples that could not be sequenced contained low bacterial load (Cycle Threshold (Ct) value > 35). Out of the 158 successfully sequenced samples, 44 (28%) were collected from Infectieradar, 15 (9%) from patients attending the GP with an acute respiratory infection other than CAP or ILI (OARI), 43 (27%) from patients attending the GP with clinically diagnosed CAP, and 56 (35%) from hospitalized CAP patients (Table 1)0.154 (97%) were successfully typed by MLST and of 153 (96.8%) the genotypic macrolide resistance was determined. For four samples the MLST was not conclusive for determination of the sequence type and for five no 23 S rRNA sequence was obtained.
Sequence types
In total, 6 different sequence types (ST) were detected. No new ST was found in this study. Figure 1 shows the distribution of ST detected by time of sampling.
To investigate prevalence of specific ST in hospitalized patients vs. milder cases from the GP or community (Infectieradar) a minimum spanning tree was constructed to depict the ST in comparison to sampling source: Infectieradar, GP visitors (CAP or OARI), and hospitalised patients (Fig. 2). No obvious difference in distribution of ST by source of samples was detected. ST19 was only found in hospital samples, although rare (n = 2) and ST2 was only found in small numbers in hospital (n = 1) and Infectieradar (n = 2) samples. Supplementary Table 1 provides an overview of the prevalence of each ST stratified by age groups.
Macrolide resistance
Macrolides and doxycycline antibiotics are the first-line treatment of confirmed M. pneumoniae respiratory tract infections. Possible macrolide resistance was investigated by sequencing the 23 S rRNA region covering A2058G/C/T, A2059G/C, A2062G and C2611A/G (E. coli numbering) marker nucleotide mutations associated with macrolide resistance13. For 153 samples the 23 S rRNA region was fully or partially successfully sequenced. Only one sample showed a known resistance marker (A2058G, ST3, GP diagnosed CAP patient, no antibiotics prescribed at consultation) indicating genotypic macrolide resistance was minimally present in our populations. No other mutations were detected. Macrolide prescriptions was low for the GP population of M. pneumoniae patients with successful macrolide resistance marker analysis (Table 2) while macrolides (azithromycin or claritromycin) are the first choice of antibiotic for diagnosed M. pneumoniae CAP patients in the Netherlands (NHG).
Discussion and conclusion
In a response to the increase of M. pneumoniae positive hospitalized patients during the 2023/2024 season in the Netherlands and among GP diagnosed CAP patients (manuscript in preparation), this retrospective typing study was conducted. We investigated M. pneumoniae positive samples from a diverse range of clinical settings, to investigate possible over-representation of specific MLST sequence types in more severe cases and determine presence of genotypic indication for macrolide resistance. Clinical samples obtained from participatory surveillance (Infectieradar) and GP sentinel surveillance for acute respiratory infections or hospitalised patients with a respiratory infection were included, representing different stages of M. pneumoniae infection severity.
When looking at the typing data, diverse STs were detected over time and multiple MLST profiles were found at the three different settings, suggesting that the uprise of M. pneumoniae in the Dutch population was caused by a diverse set of STs. The same predominant STs in this study (ST3, ST7 and ST14) were also predominantly found in other countries5,12,14,15,16. Additionally, our data does not support a more virulent sequence type as five ST were found both among community and GP patients with mild respiratory disease as well as in hospitalised patients and GP patients with CAP. ST19 was only detected among hospitalised patients, but given there were only two samples with ST19 no conclusions can be drawn. Neither do we see evidence for a M. pneumoniae load difference (based on Ct value; Table 1) between the categorised samples. This is in-line with other studies that also found no evidence for a link between bacterial load in nasopharynx/throat samples and clinical severity of disease14. Many M. pneumoniae studies target children and adolescents because they suffer most of ‘walking pneumonia’, which is a mild case of CAP often caused by M. pneumoniae. Our study includes also a substantial proportion adults of higher age as we included also mild cases with M. pneumoniae infection found in ARI surveillance. No significant differences in ST distribution within each age group was found between age groups (Supplementary Table S1).
In the National Dutch GP guideline, amoxicillin is the first-choice therapeutic agent for empiric treatment of CAP patients, which is not appropriate for treatment of M. pneumoniae13,17. This policy is reflected by the number of GP patients that received amoxicillin when consulting their GP with CAP in our study. Only few got azithromycin or doxycycline prescribed. Azithromycin is second choice for young children and pregnant women, whereas doxycycline is second choice for older children (≥ 8 years) and adults. In the hospital setting the first-choice antibiotic to treat mild to moderate unknown cause of origin CAP is amoxicillin, more severe unknown cause of origin CAP patients is given cefuroxim or cefotaxim and ciprofloxacin18. The use of macrolides in outpatients and hospitals is relatively high in the Netherlands with ten years data 2014–2023 on macrolide use in the Netherlands for outpatients mean 1.17 (range 1.07–1.22) DDD/1,000 inhabitant-days and mean 2.82 (range 2.50–3.18) DDD/1,000 inhabitant-days for hospitals19, although not known how frequently used to treat a M. pneumoniae respiratory infection. Our study indicates that despite this common use, only one (ST3) isolate from GP surveillance with genotypic indication for macrolide resistance was found among the successfully sequenced isolates, which is in line with earlier Dutch data20 However, this seems surprising as macrolide resistance in China (88%) and Japan is widely detected among the same M. pneumoniae STs found in this study5,16,21(.These studies have found an association between the same STs found in our study and macrolide resistance, which could not be confirmed in our study. Likely the more frequent use of macrolide antibiotics to treat M. pneumoniae infection in those countries explains this observation7 rather than a certain ST has always high prevalence of macrolide resistance. In addition, spread of resistant M. pneumoniae of certain ST might have contributed to linkage of macrolide resistance with ST. In Europe, however, the rate of macrolide-resistant M. pneumoniae is currently still low, ranging from 0 to 19% by country22,23. Therefore, our data suggest that in the Netherlands, and likely in Europe as well23,24, macrolides are still a good second-choice antibiotic if treatment with amoxicillin fails in the absence of diagnostic testing of the possible cause of CAP, or a laboratory confirmed M. pneumoniae pneumonia has been determined. Although doxycycline is second choice in older children and adults in the Netherlands we have not looked into resistance, as doxycycline resistant M. pneumoniae from clinical cases have previously never been reported13.
One limitation of our study is that a more discriminatory typing method, e.g. culture combined with whole genome sequencing, could be applied to provide a more in-depth analysis. However, given the diversity of sequence types already discovered in our study, this would most likely not alter the overall outcome25,26. Another limitation is that samples in the hospital part of our study that were tested for M. pneumoniae by PCR were selected from patients with respiratory symptoms who had been tested first for influenza virus, Sars-CoV-2 and Respiratory Syncytial Virus infection with PCR. Furthermore, diagnostics in all populations included in this study relied on molecular detection by PCR which has a known limitation for its inability to differentiate between asymptomatic colonization or symptomatic infection27. Therefore, linking detection of M. pneumoniae in an upper respiratory tract sample to cause of CAP and severity of disease is challenging and requires other approaches. Thirdly, we analysed data of people with respiratory symptoms. It is important to emphasise that although the major clinical presentation is pulmonary infection, presentations with extrapulmonary manifestations can occur, such as erythema exsudativum multiforme and encephalitis28,29. However, there is no evidence that specific ST are linked to these other presentations or lead to increased incidence of resistance when treated with macrolides30. Despite these limitations, our approach is fit for determining the level of macrolide resistance and the distribution of ST in the upsurge of 2023/2024 in the Netherlands.
In conclusion, the upsurge of M. pneumoniae during the winter of 2023–2024 in the Netherlands, was caused by multiple sequence types detected in multiple patient populations and that carried only sporadic macrolide resistance.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Waites, K. B. & Talkington, D. F. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. https://doi.org/10.1128/CMR.17.4.697-728.2004 (2004).
Waites, K. B., Xiao, L., Liu, Y., Balish, M. F. & Atkinson, T. P. Mycoplasma pneumoniae from the respiratory tract and beyond. Clin. Microbiol. Rev. https://doi.org/10.1128/CMR.00114-16 (2017).
Bolluyt, D. C. et al. Increased incidence of Mycoplasma pneumoniae infections and hospital admissions in the Netherlands, November to December 2023. Eurosurveillance 29, 66 (2024).
RIVM. Virologische weekstaten. https://www.rivm.nl/virologische-weekstaten.
Chen, Y., Li, X., Fu, Y., Yu, Y. & Zhou, H. Whole-genome sequencing unveils the outbreak of Mycoplasma pneumoniae in mainland China. Lancet Microbe 6, 66 (2024).
Sun, H. & Xiao, L. Overview of the epidemic characteristics of Mycoplasma pneumoniae infection around COVID pandemic. iLABMED 1, 148–157 (2023).
Wang, G., Wu, P., Tang, R. & Zhang, W. Global prevalence of resistance to macrolides in Mycoplasma pneumoniae: A systematic review and meta-analysis. J. Antimicrob. Chemotherapy 77, 66 (2022).
Smit, T. et al. Flexible and scalable participatory syndromic and virological surveillance for respiratory infections: Our experiences in The Netherlands. medRxiv https://doi.org/10.1101/2024.04.24.24306278 (2024).
Donker, G. A. Nivel Primary Care Database—Sentinel Practices 2015. NIVEL 1–178 (2016).
Brown, R. J., Spiller, B. O. & Chalker, V. J. Molecular typing of Mycoplasma pneumoniae: Where do we stand?. Future Microbiol. 10, 1793–1795. https://doi.org/10.2217/fmb.15.96 (2015).
Jolley, K. A., Bray, J. E. & Maiden, M. C. J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3, 66 (2018).
Sohyeon, K., Donghyeok, K., Jeong-Hoon, C., Kyujam, H. & Sang, J. Multilocus sequence typing analysis of Mycoplasma pneumoniae strains isolates in Korea, 2006–2019. Public Health Wkly. Rep. 13, 66 (2020).
Pereyre, S., Goret, J. & Bébéar, C. Mycoplasma pneumoniae: Current knowledge on macrolide resistance and treatment. Front. Microbiol. https://doi.org/10.3389/fmicb.2016.00974 (2016).
Xu, M. et al. Molecular epidemiology of Mycoplasma pneumoniae pneumonia in children, Wuhan, 2020–2022. BMC Microbiol. 24, 66 (2024).
Kenri, T. et al. Periodic genotype shifts in clinically prevalent Mycoplasma pneumoniae strains in Japan. Front. Cell Infect. Microbiol. 10, 66 (2020).
Kenri, T. et al. Genotyping of Mycoplasma pneumoniae strains isolated in Japan during 2019 and 2020: spread of p1 gene type 2c and 2j variant strains. Front. Microbiol. 14, 66 (2023).
NHG Guideline Acute Cough, NHG Guideline M78, February 2024. (2011).
Stichting werkgroep antibiotica beleid. SWAB Guideline Community Acquired Pneumoniae. (2024).
NethMap 2023 Consumption of Antimicrobial Agents and Antimicrobial Resistance among Medically Important Bacteria in the Netherlands (2024).
Spuesens, E. B. M. et al. Macrolide resistance determination and molecular typing of Mycoplasma pneumoniae in respiratory specimens collected between 1997 and 2008 in the Netherlands. J. Clin. Microbiol. 50, 66 (2012).
Morozumi, M. et al. Sequence type changes associated with decreasing macrolide-resistant Mycoplasma pneumoniae, Japan. Emerg. Infect. Dis. 26, 66 (2020).
Loconsole, D. et al. Update on the epidemiology of macrolide-resistant Mycoplasma pneumoniae in Europe: A systematic review. Infect. Dis. Rep. 13, 811–820. https://doi.org/10.3390/idr13030073 (2021).
Nordholm, A. C. et al. Mycoplasma pneumoniae epidemic in Denmark, October to December, 2023. Eurosurveillance 29, 66 (2024).
Dumke, R. The high-incidence period of Mycoplasma pneumoniae infections 2023/2024 in southeast Germany was associated with a low level of macrolide resistance. Infection https://doi.org/10.1007/s15010-024-02336-4 (2024).
Dumke, R. Molecular tools for typing Mycoplasma pneumoniae and Mycoplasma genitalium. Front. Microbiol. https://doi.org/10.3389/fmicb.2022.904494 (2022).
Meyer Sauteur, P. M. et al. Mycoplasma pneumoniae genotypes and clinical outcome in children. J. Clin. Microbiol. 59, 66 (2021).
Spuesens, E. B. M. et al. Carriage of Mycoplasma pneumoniae in the upper respiratory tract of symptomatic and asymptomatic children: An observational study. PLoS Med. 10, 6 (2013).
Gandelman, J. S., Kim, E. Y., Grzegorczyk, A. M., Zejnullahu, K. & Edson, R. S. Mycoplasma pneumoniae induced rash and mucositis in a previously healthy man: A case report and brief review of the literature. Open Forum Infect. Dis. https://doi.org/10.1093/ofid/ofaa437 (2020).
Tong, C. W., Menson, E., Lin, J. P. & Lim, M. Prevalence of mycoplasma encephalitis. Lancet Infect. Dis. https://doi.org/10.1016/S1473-3099(11)70130-X (2011).
Diaz, M. H., Benitez, A. J. & Winchell, J. M. Investigations of Mycoplasma pneumoniae infections in the United States: Trends in molecular typing and macrolide resistance from 2006 to 2013. J. Clin. Microbiol. 53, 66 (2015).
Acknowledgements
We thank Gabriel Goderski and Michelle van den Oever, RIVM, for samples logistics and Biofire testing of the GP samples. We thank Albert Jan van Hoek, Wanda Han, Mansoer Elahi and Jordi de Bakker, RIVM, for Infectieradar project management, sample logistics and diagnostic testing. We thank Nivel Primary Care Database – Sentinel Practices team, participating practices and their patients and all Infectieradar participants. We thank Lieuwe Roorda, Hans Renkens, Ellen Reedijk and Joost de Ruiter for designing MSLT assay and implementing the M. pneumoniae analysis workflow at Maasstad Hospital.
Funding
Funding for the analysis of Infectieradar and GP sentinel surveillance samples was received from the Dutch Ministry of Health, Welfare and Sport. Maasstad Hospital provided funding to type the hospital samples. These funders had no influence on how the study was performed and writing the paper.
Author information
Authors and Affiliations
Contributions
PS, SL, SP, AM, DE initiated the study, arranged funding, samples, did analysis and wrote the article. PWS, SP, AM arranged ethical review at the various committees, DD performed all laboratory work on MLST and sequencing for resistance profiling and contributed in analysis and writing up. MG, MH aided in making the samples available for this study. MG, MH, SP, RvG and SL provided critical review of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical statement
Non-WMO approval (WMO = Wet Medisch-wetenschappelijk Onderzoek met mensen; the Dutch law regulating medical scientific research with humans) was obtained at Maasstad hospital (L2024027) and Franciscus Gasthuis and Vlietland, Rotterdam the Netherlands (2024-031). For research based on data and samples collected in the GP sentinel surveillance a waiver for ethical approval was obtained from the Clinical Expert Centre RIVM (IDS-688). For Infectieradar a waiver for ethical approval was obtained from the Medical Ethics Review Committee Utrecht (reference number: WAG/avd/20/008757; protocol 20–131) given the nature of data collection. Due to the retrospective nature of the study, the Medical Ethics Review Committee Utrecht, the institutional review boards at Expert Centre RIVM, Maasstad hospital, and the institutional review board at Franciscus Gasthuis and Vlietland waived the need of obtaining informed consent. Hereby authors confirm that all experiments were performed in accordance with relevant guidelines and regulations that apply for this retrospective study on residual clinical samples.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Smit, P.W., Eggink, D., Paltansing, S. et al. Mycoplasma pneumoniae MLST detected in the upsurge of pneumonia during the 2023 to 2024 winter season in the Netherlands. Sci Rep 15, 6985 (2025). https://doi.org/10.1038/s41598-025-88990-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-88990-6