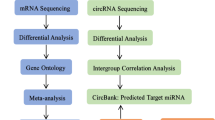
Abstract
This study aimed to evaluate the diagnostic value of miR-193b-5p and miR-511-5p in sepsis and septic shock from the perspective of immune regulation. Initially, serum exosomal miRNA sequencing was conducted on patients with sepsis (n = 6), septic shock (n = 6), and healthy controls (n = 3). Using bioinformatics analysis, miR-193b-5p and miR-511-5p were identified as immune-related miRNAs differentiating sepsis from septic shock. Subsequently, quantitative real-time polymerase chain reaction (qRT-PCR) was performed to validate the identified miRNAs in a cohort of 90 participants, including 30 patients with sepsis, 30 with septic shock, and 30 general surgery patients serving as controls. The results indicated that miR-193b-5p expression level was significantly reduced in septic patients and septic shock patients compared with healthy controls. Both miR-193b-5p and miR-511-5p exhibited diagnostic potential, distinguishing sepsis from the control group, with area under the curve values of 0.797 and 0.795, respectively (P < 0.05). Furthermore, miR-193b-5p expression level was inversely correlated with C-reactive protein level (r = -0.40, P < 0.001), procalcitonin level (r = -0.31, P = 0.003), and interleukin-6 level (r = -0.39, P < 0.001). A positive correlation was identified between miR-193b-5p and lymphocyte count (r = 0.26, P = 0.014). In conclusion, serum miR-193b-5p demonstrated potential as a diagnostic biomarker for sepsis and was associated with inflammation and immune regulation in sepsis.
Similar content being viewed by others
Introduction
Sepsis was defined as “life-threatening organ dysfunction caused by a dysregulated host response to infection” by the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis 3.0), and it is one of the most life-threatening critical infectious diseases. Septic shock is a subset of sepsis where particularly profound circulatory, cellular, and metabolic abnormalities substantially increase mortality1. There are approximately 48.9 million septic patients, and 11 million septic patients die worldwide annually, accounting for 19.7% of all the global deaths2. The morbidity and mortality rates of sepsis are extremely high, causing a huge financial burden and consumption of medical resources on countries around the world, as well as imposing heavy burden to families. The 2021 international guidelines for the management of sepsis and septic shock emphasize that early recognition and timely, appropriate treatment of these conditions can significantly reduce morbidity and mortality. Additionally, such measures improve both short- and long-term patient prognoses, decrease hospital stay, and alleviate the economic burden on individuals and the healthcare system3. However, at present, blood cultures and inflammatory markers, such as C-reactive protein (CRP), procalcitonin (PCT), interleukin (IL)-6, and white blood cell (WBC) count, fail to diagnose sepsis with sufficient speed and accuracy. According to the Sepsis 3.0 criteria, further investigation into the pathogenesis of sepsis and the identification of highly specific and sensitive biomarkers are necessary to establish a robust theoretical basis for clinical diagnosis and treatment.
Sepsis is characterized by a complex pathophysiology involving pro-inflammatory responses, immune suppression, and endothelial dysfunction. The immune inflammatory response plays a pivotal role in the development of sepsis4. In the early stages of the disease, various immune cells and their released inflammatory mediators and cytokines can trigger an exaggerated inflammatory response. Conversely, the later stages are marked by prolonged and severe immunosuppression. The immune-mediated damage, contributing to organ dysfunction, is regulated by a complex molecular network throughout the body. Investigating the role of the immune response in the development and progression of sepsis, identifying key regulatory points, and implementing interventions at these points can significantly improve the effectiveness of sepsis management in clinical practice5.
Over the past fifteen years, microRNAs (miRNAs) have emerged as promising biomarkers due to their sensitivity, specificity, and accessibility6. Meanwhile, miRNAs play a central role in sepsis pathogenesis7. MiRNAs are endogenous non-coding RNA molecules of approximately 21 nucleotides in length. They do not encode proteins, whereas regulate their expression by binding to target genes. Partial binding to the target gene can inhibit their expression, while complete binding can lead to the loss of function. MiRNAs account for only 1% of the human genome, while regulate up to 60% of genes and play a pivotal role in regulating gene expression networks8. They play critical roles in a wide range of cellular and organismal functions9, they have been identified as potential biomarkers for numerous diseases and viable targets for therapeutic interventions10. Although some miRNAs have exhibited to be associated with the development of sepsis11, sepsis is a heterogeneous, dynamic syndrome caused by imbalance in the inflammatory network. However, a single indicator or a small set of indicators is insufficient to comprehensively reflect the dysregulation of immune and inflammatory responses in sepsis. The present study therefore utilized high-throughput sequencing technology to obtain all miRNAs in serum exosomes from patients with sepsis and septic shock, and potential biomarkers were screened and verified for the early sepsis diagnosis from the perspective of immune function.
Results
Exosomal miRNA profiling
Exosomal miRNA profiling was conducted to isolate miRNAs from serum-derived exosomes of patients with sepsis and septic shock, as well as healthy controls (HCs). Through making comparison, differentially expressed miRNAs (DEMs) were identified. Using a cut-off of |log2-fold change| > 1 and a P-value < 0.05, 19 miRNAs exhibited differential expression between sepsis and septic shock, involving 9 upregulated and 10 downregulated miRNAs. Between septic patients and HCs, 124 DEMs were identified, including 108 upregulated and 16 downregulated DEMs. Additionally, 212 DEMs were detected between septic shock patients and HCs, consisting of 160 upregulated and 52 downregulated DEMs. Heatmaps and three volcano plots (Fig. 1a–d) present a visual representation of DEMs across patients with sepsis and septic shock, and HCs. Table 1 lists the top 10 upregulated and downregulated DEMs for each comparison group, ranked according to fold change.
Prediction of target genes
The target genes of the top 10 upregulated and downregulated DEMs were identified based on making comparisons between the groups. These target genes were acquired from TargetScan Human 8.0, miRWalk, and miRDB 6.0 databases. Using the Comparative Toxicogenomics Database (CTD), 21,628 sepsis-associated genes, 10,484 septic shock-associated genes, and 21,944 genes linked to both sepsis and septic shock were extracted after excluding those related to other infectious diseases. Genes overlapping across the four databases were considered as target genes. Furthermore, 356 upregulated and 614 downregulated target genes were predicted between sepsis and septic shock. Additionally, 670 upregulated and 327 downregulated target genes were identified between septic patients and HCs. In addition, 378 upregulated and 204 downregulated target genes were detected between septic shock patients and HCs. These target genes were visually represented by Venn diagrams (Fig. 1e-g).
Analysis of differentially expressed microRNAs and prediction of target genes. (a) The heatmap of upregulated and downregulated DEMs among sepsis, septic shock, and healthy controls. (b) The volcano plot of upregulated and downregulated DEMs between sepsis and septic shock. (c) The volcano plot of upregulated and downregulated DEMs between sepsis and healthy controls. (d) The volcano plot of upregulated and downregulated DEMs between septic shock and healthy controls. The red dot indicates the upregulated DEMs, and the green dot indicates the downregulated DEMs. (e) The Venn diagram of upregulated and downregulated target genes between sepsis and septic shock. (f) The Venn diagram of upregulated and downregulated target genes between sepsis and healthy controls. (g) The Venn diagram of upregulated and downregulated target genes between septic shock and healthy controls. S: Sepsis, SS: Septic shock, HCs: Healthy controls, CTD: Comparative Toxicogenomics Database.
Pathway Enrichment Analysis of genes related to Sepsis and Septic Shock
To investigate the pathway information of the target genes, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was conducted, and it was indicated that target genes were linked to immune regulation. In septic patients compared with those with septic shock, the downregulated target genes were significantly enriched in leukocyte transendothelial migration (hsa04670) and the T cell receptor (TCR) signaling pathway (hsa04660). The upregulated target genes between septic patients and HCs were enriched in Fc epsilon RI signaling pathway (hsa04664), Fc gamma R-mediated phagocytosis (hsa04666), and TCR signaling pathway (hsa04660). The KEGG pathway enrichment analysis revealed that upregulated target genes between septic shock and HCs were mainly involved in T helper (Th) 17 cell differentiation (hsa04659), TCR signaling pathway (hsa04660), Fc epsilon RI signaling pathway (hsa04664), leukocyte transendothelial migration (hsa04670), IL-17 signaling pathway (hsa04657), Th1 and Th2 cell differentiation (hsa04658), and NOD-like receptor signaling pathway (hsa04621). Bubble graphs were generated to visually represent the pathway enrichment analysis (Fig. 2a-c).
Analysis of target genes in KEGG pathways. (a) KEGG pathway analysis results for downregulated target genes between sepsis and septic shock. (b) KEGG pathway analysis results for upregulated target genes between sepsis and healthy controls. (c) KEGG pathway analysis results for upregulated target genes between septic shock and healthy controls. The x-axis represents the proportion of genes enriched in each pathway relative to the total number of genes in the group, along with the number of genes in each pathway. The circle size corresponds to the number of genes involved. The y-axis indicates the KEGG pathways, and the color scale represents -log10(Q value). KEGG: Kyoto Encyclopedia of Genes and Genomes.
Protein-protein Interaction (PPI) networks and extraction of hub genes
The study identified target genes enriched in immune regulation-associated signaling pathways, as listed in Table 2. A subsequent reciprocal analysis of these target genes led to the construction of a PPI network (Fig. 3a-c). The genes were thereafter ranked based on their degree, and hub genes were identified and listed in Supplementary Table S1. After removing isolated nodes, the PPI network of down-regulated target genes between sepsis and septic shock consisted of 12 nodes and 22 edges, involving five hub genes (MAPK3, PIK3CA, MAP2K7, CD8A, and CXCL12). On the other hand, the PPI network of upregulated target genes between septic patients and HCs included 13 nodes and 34 edges, comprising hub genes, such as PIK3CA, KRAS, PIK3CD, CRK, and CBL. Similarly, the PPI network of upregulated target genes between septic shock patients and HCs comprised 22 nodes and 67 edges. Key hub genes, including KRAS, MAPK14, PIK3CA, SMAD2, and TRAF6, were prominently involved in septic shock.
Construction of DEMs-messenger RNAs (mRNAs) networks
DEMs-mRNAs networks were constructed by incorporating immune-related target genes and DEMs that could predict these genes (Fig. 3d-f). According to the number of associations between DEMs and mRNAs, the analysis revealed that miR-511-5p and miR-193b-5p were linked to multiple genes in the network, exhibiting downregulation in the sepsis group compared with the septic shock group. On the other hand, miR-4433b-3p and miR-3158-5p were identified as critical miRNAs in the network, demonstrating upregulation in the sepsis group compared with the HC group. Noteworthy, compared with HCs, upregulated miR-3158-5p, miR-3158-3p, miR-584-5p, miR-342-5p, and miR-548o-3p were highlighted for their crucial roles in the septic shock.
Results of PPI network analysis of target genes related to immune and DEMs-mRNAs networks. (a) The downregulated hub genes between sepsis and septic shock. (b) The upregulated hub genes between sepsis and healthy controls. (c) The upregulated hub genes between septic shock and healthy controls. (d) The networks for downregulated DEMs and target genes between sepsis and septic shock. (e) The networks for upregulated DEMs and target genes between sepsis and healthy controls. (f) The upregulated DEMs and target genes between septic shock and healthy controls. DEMs: Differentially expressed miRNAs.
miR-193b-5p and mir-511-5p expression levels in patients with sepsis and septic shock
Analysis of the DEMs led to the selection of miR-193b-5p and miR-511-5p for validation of expression levels among septic patients (n = 30), septic shock patients (n = 30), and HCs (n = 30). Moreover, miR-193b-5p expression level was significantly lower in septic patients and septic shock patients compared with HCs. However, no significant difference was identified in miR-193b-5p expression level between septic patients and septic shock patients. Additionally, miR-511-5p did not exhibit significant difference in expression level among the three groups (Fig. 4a-b).
The potential of miR-193b-5p and mir-511-5p in the diagnosis of sepsis and distinguishing the severity of sepsis
In this study, the sepsis and septic shock groups were integrated into a single disease group and compared with the control group. Binary logistic regression analysis was employed to identify potential risk factors for sepsis. The results indicated that miRNA-193b-5p could serve as a protective factor against sepsis and septic shock after adjusting for age and sex [odds ratio (OR) = 0.267, 95% confidence interval (CI): 0.081–0.877] (Table 3). To further evaluate the diagnostic value of miRNA-193b-5p, receiver operating characteristic (ROC) curve analysis was conducted, and the area under the curve (AUC) was calculated. The AUC for miRNA-193b-5p was 0.797, indicating a promising predictive ability, while the AUC for miRNA-511-5p was 0.530, suggesting poor predictive performance. The combination of the two miRNAs yielded an AUC of 0.795. These findings suggest that miRNA-193b-5p could effectively distinguish septic patients from HCs. The optimal cut-off value for miRNA-193b-5p in predicting sepsis was 2.08, with a sensitivity of 85.0% and a specificity of 73.3% (Fig. 4c). The ROC curve analysis was also performed to assess the ability of these miRNAs to differentiate septic shock patients from septic patients (septic shock = 1, sepsis = 0). The AUC values for miRNA-193b-5p, miRNA-511-5p, and their combination in distinguishing septic shock from sepsis were 0.587, 0.520, and 0.590, respectively, indicating no significant differences in predictive performance among these markers (Fig. 4d). All participants, including those in the sepsis, septic shock, and HC groups, were divided into two subgroups based on miRNA-193b-5p levels: ≤2.08 and > 2.08. Clinical indicators were compared between these two subgroups. Significant differences were identified in several parameters, including the sequential organ failure assessment (SOFA) score, maximum temperature within the first 24 h after intensive care unit (ICU) admission, hypersensitive CRP level, PCT level, IL-6 level, lymphocyte percentage, lymphocyte count (LYC), and the prevalence of sepsis (P < 0.05) (Table 4). Other clinical parameters exhibited no significant differences, as detailed in Supplementary Table S2).
Correlation between miRNA-193b-5p, miRNA-511-5p, and immunity and inflammation indicators
In this study, Spearman’s correlation analysis was performed to evaluate the relationships between miRNA-193b-5p, miRNA-511-5p, and various immunity and inflammation indicators. The results revealed that the correlation coefficients between miRNA-193b-5p and complement 3 (C3), complement 4 (C4), immunoglobulin A (IgA), immunoglobulin G (IgG), immunoglobulin M (IgM), CRP, white blood cell (WBC) count, PCT, IL-6, LYC, CD3 T lymphocytes (CD3), CD4 T lymphocytes (CD4), CD8 T lymphocytes (CD8), CD4/CD8 T lymphocyte ratio (CD4/CD8), B cells, and natural killer cells (NK cells) were 0.17, 0.05, 0.13, -0.01, − 0.03, -0.40, -0.03, -0.31, -0.39, 0.26, -0.01, 0.02, -0.05, -0.04, 0.05, and − 0.01, respectively. The correlation coefficients between miRNA-193b-5p and CRP (r = -0.40, P< 0.001), PCT (r = -0.31, P = 0.003), IL-6 (r = -0.39, P < 0.001), and LYC (r = 0.26, P = 0.014) were statistically significant. The correlation coefficients between miRNA-511-5p and C3, C4, IgA, IgG, IgM, CRP, WBC count, PCT, IL-6, LYC, CD3, CD4, CD8, CD4/CD8, B cells, and NK cells were − 0.07, -0.11, -0.10, 0.04, -0.13, 0.01, -0.16, 0.02, -0.02, 0.02, 0.13, 0.05, 0.07, -0.10, 0.03, and 0.06, respectively, indicating non-significant correlations (all P > 0.05) (Fig. 4e).
The verification of the diagnosis value of miR-193b-5p and miR-511-5p in sepsis and septic shock groups. (a) Expression of miR-193b-5p in sepsis, septic shock, and control groups. (b) Expression of miR-511-5p in sepsis, septic shock, and control groups. (c) ROC curves illustrating the sensitivity and specificity of plasma miR-511-5p and miR-193b-5p, either individually or in combination, for sepsis diagnosis. (d) ROC curves depicting the sensitivity and specificity of plasma miR-511-5p and miR-193b-5p, either individually or in combination, for distinguishing the severity of sepsis. (e) Correlation between miRNAs and immunity and inflammatory indicators. C3: Complement 3, C4: Complement 4, IgA: Immunoglobulin A, IgG: Immunoglobulin G, IgM: Immunoglobulin M, hs-CRP: High-sensitivity C-reactive protein, WBC: White blood cell, PCT: Procalcitonin, IL-6: Interleukin-6, LYC: Lymphocyte count, CD3: T lymphocytes (CD3), CD4: T lymphocytes (CD4), CD8: T lymphocytes (CD8), CD4/CD8: T lymphocyte ratio (CD4/CD8), NK cells: Natural killer cells.
Discussion
Sepsis is characterized by systemic inflammation and a dysregulated immune response to invasive pathogens, leading to multiple organ dysfunction. The pathogenesis of sepsis is still unclear, and diagnostic markers are lacking. Dysregulated immune system function is a critical factor, triggering the onset and progression of sepsis12. Biomarkers reflecting immune function can promote more accurate diagnosis and staging of sepsis, enabling timely and effective treatment13. Moreover, miRNAs have emerged as promising biomarker candidates for sepsis14. Recent studies have reported varying findings for different miRNAs. For instance, Liang et al.15 demonstrated that serum miR-206 expression could be correlated positively with sepsis severity and prognosis. Plasma miR-147b has exhibited to differentiate bacterial sepsis from non-sepsis and septic shock16, while Zhu et al.17 suggested that miR-150-5p may alleviate apoptosis and serve as a therapeutic target for sepsis-induced myocardial depression. However, the impact of immune-related biomarkers in sepsis is unclear. The purpose of our study was to identify and verified diagnostic biomarkers of sepsis from the perspective of immune function regulation. In the present research, small RNA sequencing technology was employed to identify DEMs among septic patients, septic shock patients, and HCs, concentrating on miRNA profiles derived from serum exosomes. Target genes of the identified DEMs were predicted using public databases, and pathway enrichment analysis was conducted to identify immune-related pathways. By filtering the target genes enriched in immune-related pathways, DEMs-mRNAs interaction networks were constructed, enabling the selection of miRNAs associated with immune function as potential biomarkers for the early diagnosis of sepsis and septic shock. Further validation was carried out using quantitative real-time polymerase chain reaction (qRT-PCR), which confirmed that miR-193b-5p plays a role in sepsis by modulating the immune response.
In this study, public databases were employed to predict target genes associated with the identified DEMs. The KEGG pathway analysis of upregulated target genes in sepsis and septic shock groups, compared with HC group, revealed significant enrichment in multiple immune pathways and functions. Additionally, the KEGG pathway analysis of upregulated target genes in the septic shock group, compared with the sepsis group, demonstrated significant enrichment in the leukocyte transendothelial migration pathway (hsa04670) and the T cell receptor (TCR) signaling pathway (hsa04660). These findings indicate that dysregulation of innate and adaptive immune responses plays a critical role in the pathogenesis and progression of sepsis. To accurately identify the occurrence of septic shock in patients with sepsis, key genes enriched in immune pathways were screened, and a DEMs-mRNAs regulatory network was constructed to identify critical miRNAs. In the septic shock group, the upregulated target genes were MAPK3, PIK3CA, MAP2K7, CD8A, and CXCL12 as hub genes. In DEMs-miRNAs network, miRNAs with the most connections to these genes were the primary target miRNAs, and we screened out miR-193b-5p and miR-511-5p as the target miRNAs. MAP2K7 was associated with miR-193b-5p, while PIK3CA and CD8A were linked to miR-511-5p.
In our research, the analysis demonstrated that miR-193b-5p has significant clinical relevance in sepsis and septic shock. Its level was markedly reduced in these conditions. ROC curve analysis revealed an AUC of 0.797 for miR-193b-5p in assessing sepsis, with an optimal cutoff value of 2.08, yielding a sensitivity of 85.0% and a specificity of 73.3%. Patients were divided into two groups based on miR-193b-5p levels (≤ 2.08 and > 2.08). Lower levels (≤ 2.08) were associated with more severe inflammatory responses, remarkable clinical symptoms, and reduced LYC, indicating more severe condition. At the same time, miR-193b-5p was negatively correlated with PCT, CRP and IL-6 and positively correlated with LYC. These findings suggest that miR-193b-5p may serve as a potential biomarker for diagnosing and assessing the severity of sepsis. Increased expression level of miR-193b-5p may inhibit the progression of sepsis. Previous research18 reported that in vivo inhibition of miR-193b-5p plays a protective role in sepsis-induced lung damage. However, this study found that miR-193b-5p level was reduced in patients with sepsis and septic shock compared with HCs, suggesting that miR-193b-5p expression level may vary between systems affected by sepsis. In contrast, no significant difference in miR-511-5p expression level was identified between patients with sepsis or septic shock and HCs. Furthermore, miR-511-5p expression level had no correlation with immune-related indicators, suggesting limited utility for diagnosing sepsis through serum analysis.
However, the mechanisms by which miR-193b-5p influences downstream genes to affect sepsis onset and progression require further investigation. In DEMs-mRNAs network we previous constructed, miR-193b-5p associated with NFATC2, MAP2K7, CLDN2. MAP2K7 was hub genes in PPI networks of down-regulated target genes between sepsis and septic shock. MAP2K7 is a part of the MAP kinase family and have been implicated in proinflammatory cytokine responses. MAP2K7 exhibits significantly elevated expression level in the liver and kidney following lipopolysaccharide (LPS) injection19. In KEGG pathway enrichment analysis, MAP2K7 enriched in TCR signaling pathway. T-cell-mediated cellular immunity is also central to the pathogenesis of sepsis. Dysregulated TCR signaling can disrupt various T-cell biological functions, leading to immune dysfunction. Overactivation or suppression of TCR signals may result in a range of immune-related disorders20. Previous studies, such as those conducted by Kim et al.21, have demonstrated that downregulation of the TCR pathway in sepsis datasets is associated with poor prognosis. Notably, upregulation of the TCR signaling pathway was found in the septic shock group compared with the sepsis group, potentially reflecting early immune overactivation during septic shock. This early activation of adaptive immune pathways may amplify the hyper-inflammatory response, driving the transition from sepsis to septic shock. However, in the later stages of sepsis, T cell depletion becomes apparent, signifying immunosuppression and increased susceptibility to secondary infections. This biphasic immune response highlights the complex regulation of immunity in sepsis and its progression to septic shock.
This study utilized bioinformatics technology to identify sepsis biomarkers from the perspective of immune function regulation. We identified that miR-193b-5p may influence the development of sepsis by regulating immune-related pathways and validated this finding using qRT-PCR technology in a new patient cohort, thereby enhancing the reliability of the results. Furthermore, we explored the potential role of miR-193b-5p in sepsis progression, providing a theoretical basis for the accurate diagnosis and targeted treatment of sepsis. However, there were limitations in this research. The small sample size used for microRNA sequencing and qRT-PCR validation might affect the generalizability of the results. Future studies should verify the identified DEMs at the cellular and animal levels and incorporate larger sample sizes. Furthermore, this study only assessed the expression levels of miR-193b-5p and miR-511-5p at the time of patient inclusion, without dynamically monitoring their expression throughout disease progression. This limitation highlights the need for further research to better elucidate the clinical significance of these immune function-related miRNAs in sepsis. In conclusion, the findings indicate that serum miR-193b-5p can serve as a diagnostic marker for sepsis. Additionally, miR-193b-5p is associated with inflammation and immune regulation, highlighting its potential role in sepsis pathophysiology.
Materials and methods
Study population
Serum samples were collected from six patients with sepsis and six patients with septic shock who were admitted to the ICU of the General Hospital of Ningxia Medical University (Yinchuan, China) between August 2020 and August 2021. Additionally, three healthy volunteers were recruited as controls. Notably, miRNA sequencing was performed on these samples, and bioinformatics techniques were applied to identify miRNAs of interest.
A prospective observational study was subsequently conducted from January 1, 2023, to December 31, 2023. This study included patients with sepsis, septic shock, and general surgery patients (control group) who were admitted to the ICU of the same hospital. Blood samples were obtained within 24 h of ICU admission for qRT-PCR to validate the key miRNAs identified through sequencing. All study participants were adults (≥ 18 years old). Exclusion criteria included a history of oncological disease, immunosuppressive therapy, radiotherapy, or long-term use of chemotherapeutic agents. There were no significant differences in baseline clinical characteristics or SOFA scores among the three groups (P > 0.05). However, the septic shock group exhibited significantly higher Acute Physiology and Chronic Health Evaluation II (APACHE II) scores and maximum body temperatures within the first 24 h after ICU admission compared with the sepsis and control groups (P < 0.05). Additionally, the sepsis group exhibited a higher maximum body temperature during the same period compared with the control group (P < 0.05). Participants’ general and clinical characteristics are presented in Supplementary Tables S3 and S4. This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of General Hospital of Ningxia Medical University, China (Ethics Approval Number: 2020 − 722). All patients’ legal representatives and healthy volunteers have been informed and provided written informed consent forms in accordance with the Declaration of Helsinki.
Exosome extraction
Venous blood samples (20 mL) were collected and left at room temperature for 30 min before being placed at 4 °C for 3–4 h. The samples were centrifuged at 4000 × g for 20 min at 4 °C, and the supernatant was carefully extracted. This supernatant was centrifuged again at 1000 × g for 10 min at 4 °C. The resulting serum was transferred to RNase-free lyophilization tubes and stored at -80 °C. Exosome extraction was performed at 37 °C using ultracentrifugation.
DEMs screening
Following qualification of the exosome assay, RNA was measured using the highly sensitive Agilent 2100 pic600 to determine total quantity and fragment distribution. A cDNA library was constructed using the Small RNA (sRNA) Sample Pre kit. After library inspection, libraries were pooled based on effective concentration and target offline data requirements, followed by sequencing using Illumina SE50 technology. The raw sequencing data underwent quality control, length filtering, and reference sequence alignment to annotate and quantify sRNA and known miRNAs (Supplementary Table S5). DEMs were identified using DESeq2 package in R software, applying a negative binomial distribution with criteria of P < 0.05 and |log2-fold change| > 122. The top 10 upregulated and downregulated DEMs were selected for each comparison.
Prediction of target genes
To identify miRNA–mRNA interaction pairs, DEMs were queried across three publicly available databases: TargetScan Human23,24 (Version 8.0; http://www.targetscan.org/), miRWalk25 (Version 3.0; http://mirwalk.umm.uni-heidelberg.de/), and miRDB26,27 (Version 6.0; http://mirdb.org/). Sepsis- and septic shock-related genes were obtained from the CTD database28 (Update 2021; http://ctd.mdibl.org/) after excluding genes related to other infectious diseases. The final target genes were identified by intersecting results from the four databases using VENNY 2.1(https://bioinfogp.cnb.csic.es/tools/venny/).
Pathway Enrichment Analysis
KEGG enrichment analysis was conducted to explore the biological functions of genes, utilizing the Metascape database29 (Version3.5;https://metascape.org/) and the KEGG database30,31,32 (https://www.kegg.jp). These tools provided insights into the high-level functional roles and utilities of biological systems. Statistically significant entries were identified using screening criteria of P-value < 0.05 and Q-value < 0.05. Visualization of the KEGG enrichment results was performed using ChiPlot (https://www.chiplot.online/).
PPI networks and extraction of hub genes
Pathways associated with immune function were selected for further analysis. Genes enriched in these pathways were subjected to PPI network analysis using the STRING database (Version 11.5; https://cn.string-db.org/). The confidence score threshold was set at 0.40, with P < 0.05. Visualization of the PPI network was performed using Cytoscape software (Version 3.9.0, supported by Java 11.0.6). Hub genes were identified using the Maximal Clique Centrality (MCC) algorithm in the CytoHubba plugin. The top five genes in each analysis were designated as hub genes.
Construction of DEMs-mRNAs networks
The DEMs-mRNAs network was constructed using the DEMs identified and their target genes enriched in immune-relevant pathways. This network was visualized using Cytoscape software (Version 3.9.0) to facilitate the identification of key DEMs.
Verification of Key miRNAs
To validate the key identified miRNAs, a prospective study was conducted involving 90 participants: 30 patients with septic shock, 30 with sepsis, and 30 general surgery patients serving as controls. Venous blood samples (1 mL) were collected into sterilized, enzyme-free centrifuge tubes containing 5 mL RNA SolidTM reagent. The samples were stored at -30 °C until RNA extraction. The expression levels of miR-193b-5p and miR-511-5p were quantified using qRT-PCR with 2× SYBR Green qPCR Master Mix (None ROX) (Servicebio Technology Co., Ltd., Wuhan, China). The relative expression levels were calculated using the 2−ΔΔCt method.
Statistical analysis
Data were analyzed utilizing SPSS 26.0 software (IBM, Armonk, NY, USA). Hospitalization data for all participants were recorded using EpiData 3.0 software. Continuous variables following a normal distribution were reported as mean ± standard deviation (SD) and analyzed using Student’s t-test. Continuous variables with a skewed distribution were presented as median (interquartile range, IQR) and analyzed using the Mann-Whitney U test. Categorical variables, expressed as frequency and percentage, were compared using the Chi-square test. Differences among multiple groups were assessed through one-way analysis of variance (ANOVA) or the Kruskal-Wallis H-test. ROC curve analysis was performed using MedCalc 19.7.2 software to assess the diagnostic accuracy of miR-193b-5p and miR-511-5p for sepsis and septic shock. Spearman correlation analysis was used to examine the relationships between miR-193b-5p, miR-511-5p, and inflammation or immunity-related indicators. A significance level of α = 0.05 was applied, and P < 0.05 was considered statistically significant.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information.
References
Singer, M. et al. The Third International Consensus definitions for Sepsis and septic shock (Sepsis-3). Jama 315, 801–810 (2016).
Rudd, K. E. et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the global burden of Disease Study. Lancet 395, 200–211 (2020).
Evans, L. et al. Executive Summary: surviving Sepsis Campaign: International guidelines for the management of Sepsis and Septic Shock 2021. Crit. Care Med. 49, 1974–1982 (2021).
Arina, P. & Singer, M. Pathophysiology of sepsis. Curr. Opin. Anaesthesiol. 34, 77–84 (2021).
Hotchkiss, R. S., Monneret, G. & Payen, D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 13, 862–874 (2013).
Wu, X., Yang, J., Yu, L. & Long, D. Plasma miRNA-223 correlates with risk, inflammatory markers as well as prognosis in sepsis patients. Med. (Baltim). 97, e11352 (2018).
Antonakos, N., Gilbert, C., Théroude, C., Schrijver, I. T. & Roger, T. Modes of action and diagnostic value of miRNAs in sepsis. Front. Immunol. 13, 951798 (2022).
Hirschberger, S., Hinske, L. C. & Kreth, S. MiRNAs: dynamic regulators of immune cell functions in inflammation and cancer. Cancer Lett. 431, 11–21 (2018).
Ivey, K. N. & Srivastava, D. microRNAs as developmental regulators. Cold Spring Harb Perspect. Biol. 7, a008144 (2015).
Rupaimoole, R. & Slack, F. J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16, 203–222 (2017).
Vasilescu, C. et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS One. 4, e7405 (2009).
Hotchkiss, R. S. et al. Sepsis and septic shock. Nat. Rev. Dis. Primers. 2, 16045 (2016).
Kingsley, S. M. K. & Bhat, B. V. Role of microRNAs in sepsis. Inflamm. Res. 66, 553–569 (2017).
Shen, X. et al. Accuracy of circulating microRNAs in diagnosis of sepsis: a systematic review and meta-analysis. J. Intensive Care. 8, 84 (2020).
Liang, G. et al. The correlations between the serum expression of miR-206 and the severity and prognosis of sepsis. Ann. Palliat. Med. 9, 3222–3234 (2020).
Trung, N. T. et al. Circulating miR-147b as a diagnostic marker for patients with bacterial sepsis and septic shock. PLoS One. 16, e0261228 (2021).
Zhu, X. G., Zhang, T. N., Wen, R. & Liu, C. F. Overexpression of miR-150-5p Alleviates Apoptosis in Sepsis-Induced Myocardial Depression. Biomed Res Int. 3023186 (2020). (2020).
Dos Santos, C. C. et al. Mesenchymal stromal (stem) cell therapy modulates miR-193b-5p expression to attenuate sepsis-induced acute lung injury. Eur. Respir J. 59, 2004216 (2022).
Alsharif, K. F. et al. Oleuropein protects against lipopolysaccharide-induced sepsis and alleviates inflammatory responses in mice. IUBMB Life. 72, 2121–2132 (2020).
Shah, K., Al-Haidari, A., Sun, J. & Kazi, J. U. T cell receptor (TCR) signaling in health and disease. Signal. Transduct. Target. Ther. 6, 412 (2021).
Kim, K. S. et al. Immune gene expression networks in sepsis: a network biology approach. PLoS One. 16, e0247669 (2021).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Agarwal, V., Bell, G. W., Nam, J. W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4, e05005 (2015).
McGeary, S. E. et al. The biochemical basis of microRNA targeting efficacy. Science 366, eaav1741 (2019).
Sticht, C., De La Torre, C., Parveen, A. & Gretz, N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One. 13, e0206239 (2018).
Chen, Y. & Wang, X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 48, D127–d131 (2020).
Liu, W. & Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 20, 18 (2019).
Davis, A. P. et al. Comparative toxicogenomics database (CTD): update 2021. Nucleic Acids Res. 49, D1138–d1143 (2021).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–d592 (2023).
Funding
This work was supported by the Key Research and Development Program of Ningxia (2021BEG03064).
This work was supported by Ningxia Natural Science Foundation Project (2022A1782).
Author information
Authors and Affiliations
Contributions
C. L., X. S. and X. W. conceived the study, elaborated the study protocol, and interpreted the data; C. L., X. S. and X. Y. enrolled patients and collected the data; C. L., X. S. and R. Z. performed the statistical analysis; C. L. and X. S. drafted the article; J. C. and X. W. revised the article; X. W. supervised the whole study. All the authors approved the final version of the article to be submitted. C. L., X. S. and X. W. are the guarantors of the data. All authors commented on previous versions of the manuscript and all authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of General Hospital of Ningxia Medical University, China (Ethics Approval Number: 2020 − 722). All patients’ legal representatives and healthy volunteers have been informed and provided written informed consent forms in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
† Can Li and Xinxing Sun contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, C., Sun, X., Yang, X. et al. miRNA sequencing identifies immune-associated miRNAs and highlights the role of miR-193b-5p in sepsis and septic shock progression. Sci Rep 15, 5323 (2025). https://doi.org/10.1038/s41598-025-89946-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-89946-6