Abstract
Salmonella Typhimurium is an invasive intracellular pathogen that employs various factors for its survival within host cells. To mitigate S. Typhimurium survival, it is crucial to identify factors that influence bacterial survival and to develop drugs that inhibit these factors. In this study, we investigated the effects of nafcillin and diosmin, both of which have been identified as inhibitors of Lon protease, on the intracellular survival of S. Typhimurium and its survival under various stress conditions. Additionally, we examined the expression of genes associated with the type II toxin-antitoxin system to enhance our understanding of the impact of these systems on the bacterium’s survival. Our findings indicate that while nafcillin and diosmin did not affect S. Typhimurium attachment, they significantly reduced bacterial intracellular survival, particularly in Hep2 cells after 16 h. These inhibitors were also effective in decreasing bacterial survival under oxidative and acidic stress conditions. Furthermore, gene expression analysis revealed that although there were variations in the expression of TA system genes in S. Typhimurium across different cell lines, the relEB system emerged as the most effective among those studied, exhibiting the highest increase in expression. This study highlights the efficacy of nafcillin and diosmin in reducing the intracellular survival of S. Typhimurium as well as its survival under stress conditions. These findings suggest potential new strategies for developing therapies aimed at preventing S. Typhimurium infections.
Similar content being viewed by others
Introduction
Salmonella enterica serovar Typhimurium is a Gram-negative, facultative intracellular bacterium that serves as a significant enteric pathogen affecting both humans and animals1. It poses a substantial threat to food-producing animals, including cattle, pigs, and poultry, and is primarily transmitted through the consumption of contaminated food or water2.
After being ingested, S. Typhimurium attaches to the epithelial cells of the ileum and colon, initiating gastrointestinal infection3. The bacterium targets specialized epithelial cells known as M cells in the small intestine, prompting intracellular changes that facilitate their entry via the secretion of effector proteins3,4. The Type III secretion system (T3SS-1) plays a pivotal role in the invasion of intestinal epithelial cells by enabling bacterial entry and survival within the host4. S. Typhimurium uses two main strategies for cell entry: the “trigger” mechanism, which involves manipulation of the host’s cytoskeleton, and the receptor-mediated “zipper” mechanism5. Successful adhesion and invasion are critical for establishing infection. Once inside, Salmonella resides within specialized vacuoles that shield it from the host’s immune defenses6. Thus, blocking bacterial adhesion, invasion, and survival is crucial for managing infections, but delivering effective antimicrobial agents to target cells presents significant challenges7.
In both human and animal health settings, various antimicrobial agents, including fluoroquinolones, are used to treat intracellular infections like those caused by S. Typhimurium8,9. However, increasing antibiotic resistance among these bacteria has become a pressing concern8. Moreover, some antibiotics fail to eradicate intracellular bacteria even at inhibitory concentrations10. Therefore, it is imperative to identify the factors influencing bacterial adhesion and invasion while exploring new compounds that can effectively inhibit these processes as well as intracellular survival in S. Typhimurium. Such efforts are essential for developing innovative strategies to combat infections caused by this significant pathogen11.
The coordinated expression of genes within the Salmonella Pathogenicity Islands (SPI-1 and SPI-2) is crucial for the initial invasion and subsequent establishment of infection in host organisms12. SPI-1 encodes proteins that include a Salmonella type III secretion system (TTSS), which plays a key role in injecting effector molecules that manipulate the host’s cytoskeletal structure, facilitating bacterial internalization. Once inside the host cell, genes from SPI-2 are activated, encoding another TTSS that secretes effectors aimed at redirecting endocytic trafficking13. This process prevents the recruitment and fusion of lysosomes with Salmonella-containing vacuoles, allowing the bacteria to survive within infected cells14.
In its natural life cycle, Salmonella faces numerous environmental challenges, including increased temperatures, acidic gastric conditions, elevated salt levels from bile, and oxidative stress stemming from the host’s immune response15,16. Within this framework, Lon protease—a well-conserved ATP-dependent serine protease—has a crucial role in the physiology of bacteria17. It contributes to the degradation of misfolded proteins, the regulation of gene expression, and the modulation of stress responses18. Furthermore, Lon protease is capable of degrading antitoxins within toxin-antitoxin systems, which leads to the activation of these systems19.
By selectively degrading damaged or regulatory proteins, Lon protease helps maintain cellular homeostasis and enables adaptation to fluctuating environmental stressors19. Previous research has indicated the involvement of Lon in the expression of SPI-1 and SPI-2 genes in S. Typhimurium20, making it essential to further investigate its role in modulating virulence during S. Typhimurium attachment and invasion.
Type II toxin-antitoxin systems add another layer of regulatory complexity to bacterial survival and virulence21. These systems consist of a stable toxin paired with a labile antitoxin, which together govern cell fate under stress conditions22,23. When faced with environmental challenges—such as nutrient deprivation or oxidative stress—the antitoxin may be degraded by Lon protease, allowing the toxin to exert its effects23. This can result in growth arrest or programmed cell death, potentially serving as a survival strategy for the bacterial population under stressful conditions24,25.
In a prior investigation, we examined strategies for drug repositioning aimed at Lon protease in S. Typhimurium, successfully identifying diosmin and nafcillin as potent inhibitors of this enzyme26. This study focuses on evaluating how these Lon protease inhibitors, diosmin and nafcillin, affect the attachment, invasion, and survival of S. Typhimurium when exposed to different stress conditions.
Materials and methods
Bacterial strains and cell culturing
This research utilized the bacterial strain S. Typhimurium ATCC 14028, which was preserved in Brain Heart Infusion (BHI) broth (Merck, Germany) with 20% glycerol at -80 °C. The strain was subsequently grown on Luria-Bertani (LB) agar plates at 37 °C.
Three cell lines were employed: the human colorectal cancer SW480 cell line, the human laryngeal Hep2 cell line, and mouse macrophage leukemia RAW264.7 cell. These cell lines were obtained from the Cell Bank Department of the Pasteur Institute of Iran.
The SW480 line was maintained in Roswell Park Memorial Institute (RPMI)-1640 medium, while the Hep2 line was cultured in Dulbecco’s Modified Eagle Medium (DMEM)-F12. RAW264.7 cells were grown in DMEM. All media contained 10% fetal bovine serum (FBS) and 100 U/ml of penicillin-streptomycin, and the cells were incubated at 37 °C in a humidified environment with 5% CO2.
Cytotoxicity assays
The impact of nafcillin and diosmin on the viability of SW480, Hep2, and RAW264.7 cells was evaluated using the MTT assay. The cell lines were seeded in 96-well plates at a density of 1.0 × 10^4 cells per well and incubated for 24 h at 37 °C in a 5% CO2 atmosphere. Once a monolayer was established, the cells were washed three times with phosphate-buffered saline (PBS). Following this, 200 µL of nafcillin or diosmin at various concentrations was added to each well for an additional 24-hour incubation. After this period, 10 µL of MTT solution dissolved in PBS was introduced, and the cells were incubated for another 4 h. Subsequently, the media were removed, and 100 µL of dimethyl sulfoxide (DMSO) was added to each well before measuring the absorbance at 570 nm using a microplate reader (Thermo Fisher Scientific, USA). All experiments were conducted in triplicate. Cell viability was calculated using the following formula27:
Cell adhesion and invasion assays
Based on the MTT test results, concentrations of 256 µg/mL and 128 µg/mL of diosmin were selected for further experiments, as these levels did not exhibit cytotoxicity to the cell lines. Furthermore, although none of the assessed nafcillin concentrations showed cytotoxic effects on the cells, our previous study established the minimum inhibitory concentration (MIC) of this antibiotic against S. Typhimurium at 16 µg/mL26. Consequently, sub-MIC concentrations of 8 µg/mL and 4 µg/mL of nafcillin were chosen for additional investigation.
Adhesion and invasion assays were performed according to previously described methods, with slight modifications28,29. The SW480, Hep2, and RAW264.7 cell lines were cultured in 24-well plates at a density of 1 × 10^5 cells per well for 24 h. Following this initial incubation, the monolayers were infected with S. Typhimurium at a multiplicity of infection (MOI) of 100 and incubated for 1–4 h at 37 °C in a 5% CO₂ atmosphere. This was done for both untreated and treated groups, which included diosmin at concentrations of 256 and 128 µg/mL, and nafcillin at concentrations of 4 and 8 µg/mL. After incubation, the medium was discarded, and the cells were washed twice with 1× PBS to eliminate non-adherent bacteria. The cell lines were then lysed using 1% Triton X-100, and the lysates were serially diluted and plated on LB agar for viable bacterial counts after overnight incubation.
For the invasion assay, SW480, Hep2, and RAW264.7 cell lines were also plated in 24-well plates at a density of 1 × 10^5 cells per well for 24 h. Following this initial incubation, the monolayers were infected with S. Typhimurium at a multiplicity of infection (MOI) of 100. In addition to the control group (cells infected with bacteria alone), diosmin at doses of 256 and 128 µg/mL and nafcillin at doses of 4 and 8 µg/mL were added to the treatment group. Infected cells from the untreated and treated groups were washed twice with PBS after 1 and 16 h of incubation. Gentamicin (100 µg/mL) was added to prevent the growth of extracellular bacteria, and the cells were incubated with 1% Triton X-100 for 1 h before lysis. The resulting suspensions were serially diluted and plated on LB agar plates for counting viable bacteria after overnight incubation.
Microscopy
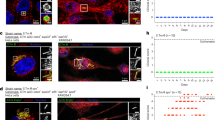
Giemsa staining was performed to visualize the adherence of S. Typhimurium to SW480, Hep2, and RAW264.7 cells in both control and treated groups with some modifications30. The cell lines were seeded in 6-well plates with glass cover slips at a density of 1 × 10^5 cells per well and incubated for 24 h at 37 °C with 5% CO2. Then, they were infected with a 2 mL bacterial suspension (1 × 10^7) in cell culture medium, either alone or with 8 µg/mL nafcillin or 256 µg/mL diosmin along with 10% FBS for 3 h. After washing with PBS, the cells were fixed with 100% methanol for 20 min and stained with 10% Giemsa for 30 min, followed by three rinses with PBS. Adherence of S. Typhimurium to the cells was assessed using a light microscope (Olympus, Japan) at a magnification of 1000x.
Bacterial growth under stress conditions
The bacterial cultures were exposed to different stress conditions with minor modifications20. The S. Typhimurium strain was grown on LB agar and incubated for 24 h at 37 °C. A single colony from this culture was inoculated into 5 mL of LB broth, followed by another 24-hour incubation at the same temperature. Next, a second subculture was created by diluting this culture 100-fold in 50 mL of LB broth, which was then incubated at 37 °C while shaking at 200 rpm until an optical density of 0.4 at 600 nm (OD600) was reached. The bacterial cells were harvested using centrifugation at 4000 × g for 15 min. The cells were then re-suspended in pre-warmed LB broth as a control and in various conditions: LB broth adjusted to pH 3.0 (acid stress using hydrochloric acid, pre-chilled LB broth at 4 °C (cold stress), pre-warmed LB broth at 42 °C (heat stress), LB broth with 5 mM H2O2 (oxidative stress), and LB broth with 5% NaCl (osmotic stress). The bacteria were exposed to these stressors for 4 h, either in LB broth alone or supplemented with diosmin at concentrations of 256 and 128 µg/mL, and nafcillin at 4 and 8 µg/mL. Following the exposures, the cultures were washed twice with sterile normal saline (0.85%) before undergoing serial dilutions and plating on LB agar, with colony counts taken after a 24-hour incubation at 37 °C.
Quantitative real-time PCR (qRT-PCR)
The expression of genes related to Lon protease and type II toxin-antitoxin (TA) systems (gnat/rhh, vapC/vapB, parB/parE, phd/doc, relE/relB) in S. Typhimurium was examined using qRT-PCR under cell line experiments and different stress conditions. For the cell line studies, about 10^6 SW480, Hep2, and RAW264.7 cells were seeded in 6-well plates and incubated at 37 °C in a 5% CO2 for 24 h. These cells were then infected with 2 mL of a S. Typhimurium suspension (1 × 10^8 CFU/mL) in either cell culture medium or that supplemented with 8 µg/mL nafcillin or 256 µg/mL diosmin, plus 10% FBS for 3 h. RNA extraction was performed using a TRIzol-based method31. For stress conditions, total RNA from the bacteria was extracted via the TRIzol method after a 3-hour exposure to stressors during logarithmic-phase growth in LB broth alone or with nafcillin or diosmin.
RNA yield and purity were evaluated with a spectrophotometer at 260/280 nm and 260/230 nm (NanoDrop, Thermo Fisher Scientific, USA). DNase I treatment was done per the manufacturer’s protocol (Thermo Fisher). RNA was reverse-transcribed using the AddScript cDNA synthesis kit (AddBio, South Korea) according to manufacturer instructions. Primer sequences used in the study are listed in Table 1.
qRT-PCR was conducted using a Rotor-Gene thermal cycler with the SYBR Green method (Ampliqon Co, Denmark). The total volume for reactions was 20 µL, containing 1 µL of cDNA, 10 µL of SYBR Green master mix, 7 µL of nuclease-free water, and 1 µL of each primer. The cycling protocol included an initial denaturation step at 95 °C for 12 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 20 s, and extension at 72 °C for 25 s. The invA gene was used as a reference for normalizing gene expression levels, with relative fold changes calculated using the 2−ΔΔCt method32.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA, USA). A one-way analysis of variance (ANOVA) was utilized, followed by Tukey’s range test, with a significance level set at p < 0.05.
Results
Cytotoxicity of nafcillin and diosmin
The MTT cytotoxicity assay was conducted on SW480, Hep2, and RAW264.7 cell lines to evaluate the cytotoxic concentrations of nafcillin and diosmin. For all tested concentrations of nafcillin across the three cell lines, cell viability exceeded 70%, indicating non-cytotoxic effects according to international standard protocols33. In contrast, while most concentrations of diosmin were also non-cytotoxic, notable exceptions included 1024 µg/mL (64.74% viability) and 512 µg/mL (64.66% viability) for SW480 cells; 1024 µg/mL (65.31% viability) for Hep2 cells; and 1024 µg/mL (48.97% viability) for RAW264.7 cells (Fig. 1).
Impact of nafcillin and diosmin on S. Typhimurium adhesion and invasion
The effects of nafcillin and diosmin on the adhesion, invasion, and intracellular survival of S. Typhimurium in SW480, Hep2, and RAW264.7 cells were examined. Results indicated that none of the tested concentrations of either drug produced a statistically significant reduction in bacterial adhesion after 1 and 4 h of incubation in the respective cell lines (Fig. 2).
In terms of bacterial invasion, assessed via the gentamicin method, after 2 h of incubation with nafcillin and diosmin, there was no significant decrease in invasion observed in SW480 and Hep2 cells. However, in RAW264.7 cells treated with 4 µg/mL nafcillin, a significant reduction of 1.56 logarithms in the S. Typhimurium count was recorded (p = 0.0016).
After 16 h of incubation, there was no significant reduction in bacterial invasion for RAW264.7 cells. Nevertheless, in SW480 cells, treatment with 8 µg/mL nafcillin resulted in a notable 2.08 log reduction in bacterial count (p = 0.0014). Additionally, in Hep2 cells, a significant reduction in bacterial count was observed at 256 µg/mL diosmin, with a 1.25 log reduction (p = 0.0075). Further reductions were seen with nafcillin, including a 3.95 log reduction at 8 µg/mL (p < 0.0001) and a 2.95 log reduction at 4 µg/mL (p < 0.0001), (Fig. 3).
Giemsa staining
To assess the attachment of S. Typhimurium to the tested cells in the presence of 8 µg/mL nafcillin or 256 µg/mL diosmin, a comparison was made to the control group. Following three hours of bacterial infection, the cells were washed, fixed, and stained using Giemsa stain. Our observations indicated that the number of bacteria adhering to SW480, Hep2, and RAW264.7 cells in the treated group did not remarkably differ from that of the control group (Fig. 4).
Survival of S. Typhimurium under stress conditions
To investigate the impact of various stressors on the survival of S. Typhimurium in the presence of different concentrations of nafcillin and diosmin, colony count assays were conducted. The results indicated that treatment with diosmin and nafcillin had no significant effect on bacterial survival under cold stress. At 42 °C, the administration of 8 and 4 µg/mL nafcillin resulted in a 3.08 (p = 0.0003) and a 2.3 (p = 0.0030) log reduction in bacterial counts, respectively, when compared to the control group.
Under osmotic stress, a 2.74 (p = 0.0002) log reduction was observed in the presence of 8 µg/mL nafcillin compared to the control group. In conditions of acidic stress, a 2.88 (p < 0.0001) and a 1.7 (p = 0.0047) log reduction was noted for the 8 and 4 µg/mL nafcillin treatments, respectively, while a 2.36 (p = 0.0004) and a 1.4 (p = 0.0167) log reduction was observed for 256 and 128 µg/mL diosmin, respectively. Additionally, during oxidative stress, a 2.03 (p = 0.0003) and a 1.82 (p = 0.0007) log reduction was recorded with 8 and 4 µg/mL nafcillin, respectively, and a 0.96 (p = 0.0488) and a 1.38 (p = 0.0052) log reduction was noted with 256 and 128 µg/mL diosmin, respectively, in comparison to the control group (Fig. 5).
Quantitative real-time PCR analysis
This study aimed to evaluate the expression levels of the lon protease, type II toxin-antitoxin (TA) system, ssrA, and hilA in S. Typhimurium infecting SW480, Hep2, and RAW264.7 cell lines. Additionally, we investigated the expression of lon protease and the type II TA system under various stress conditions through real-time PCR.
Our results indicated that among the systems examined in S. Typhimurium infecting Hep2 cells, the relE/relB system exhibited the most significant increase in expression, with values of 21.4 and 43.4, followed by the gnat/rhh system at 5.8 and 6.63 (p < 0.0001). For S. Typhimurium infecting SW480 cells, the relE/relB system again showed the highest expression increase, recorded at 18.64 and 27.85, along with the phd/doc system at 9.49 and 19.49 (p < 0.0001). In S. Typhimurium infecting RAW264.7 cells, there was an elevation in expression for most systems analyzed, with the relE/relB system showing a peak increase of 30.46 and 41.43 (p < 0.0001). In groups treated with diosmin and nafcillin, a decrease in expression was noted across all studied genes compared to the untreated group; however, the presence of diosmin and nafcillin increased the expression of the hilA gene (Fig. 6).
Analysis of the relative expression levels of Type II TA System genes in S. Typhimurium ATCC 14028 infecting SW480, Hep2, and RAW264.7 cell lines. For each group of genes, three bars are presented: the first bar indicates gene expression levels in infecting cell, the second represents expression in infecting cell exposed to nafcillin (8 µg/mL), and the third shows expression in infecting cell treated with Diosmin (256 µg /mL). Relative expression normalization was conducted using the reference gene invA and error bars reflect the standard deviations derived from three biological replicates (P < 0.0001, determined by one-way ANOVA).
Regarding gene expression under different stress conditions, we observed an increase in lon protease expression across all four stress types: heat, oxidative, osmotic, and acidic. Notably, the gnat/rhh genes experienced the highest increase in expression during the heat stress conditions, with increases of 3.21 and 8.1 (p < 0.0001). Under osmotic stress, most genes studied demonstrated elevated expression, with the gnat/rhh genes showing increases of 4.02 and 1.88 (p < 0.0001). In acidic stress, only the parB/parE system exhibited a significant increase, recorded at 2.18 and 4.14 (p < 0.0001). Conversely, during oxidative stress, none of the systems analyzed displayed a considerable increase (p < 0.0001). Furthermore, treatment with diosmin and nafcillin resulted in reduced expression of the type II toxin-antitoxin systems (Fig. 7).
Analysis of the relative expression levels of Type II TA System genes in S. Typhimurium ATCC 14028 in different stress conditions. For each group of genes, three bars are presented: the first bar indicates gene expression levels in stress condition, the second represents expression in stress condition exposed to Nafcillin (8 µg/mL), and the third shows expression in stress condition treated with Diosmin (256 µg /mL). Relative expression normalization was conducted using the reference gene invA and error bars reflect the standard deviations derived from three biological replicates (P < 0.0001, determined by one-way ANOVA).
Discussion
S. Typhimurium is a foodborne pathogen responsible for diseases in humans and animals. Its ability to attach to and invade host cells while responding to various environmental stresses is crucial for its virulence34. The ability of S. Typhimurium to endure within macrophages and other host cells, as well as in various stressful conditions, is closely associated with its pathogenicity15,35. This bacterium adapts to challenging environments while maintaining its ability to cause disease36. When S. Typhimurium successfully evades the host’s immune response and persists within cells, it can result in prolonged infections, heightened disease severity, and complications like systemic infections37. By reducing its intracellular survival, we may mitigate these adverse outcomes, leading to improved infection management37. Furthermore, intracellular bacteria often evade the effects of antibiotics because their location within host cells limits antibiotic penetration. By targeting the mechanisms that enable S. Typhimurium to thrive inside cells, we could enhance the efficacy of existing antibiotics or develop new therapeutic approaches38. Among the various molecular mechanisms that regulate cell invasion and responses to stress, the Lon protease and type II antitoxin (TA) systems have been implicated in bacterial survival under these conditions, according to the literature39. This study investigates the role of nafcillin and diosmin as previously identified as inhibitors of Lon protease26 in S. Typhimurium intracellular survival, and resilience against various stresses.
Adhesion and invasion assays were conducted using SW480, Hep2, and Raw 264.7 cells. The adhesion of S. Typhimurium to these cells remained relatively unchanged at different concentrations of nafcillin and diosmin during the study period. However, nafcillin and diosmin, as inhibitors of Lon protease, effectively reduced the survival of S. Typhimurium in Hep2 cells after 16 h. In the case of SW480 cells, an 8 µg/ml concentration of nafcillin significantly reduced the survival of S. Typhimurium after 16 h, while a decrease in survival was also observed with 4 µg/ml nafcillin and 256 µg/ml diosmin; however, these reductions were not statistically significant. In the macrophage cell line Raw 264.7, bacterial invasion was also reduced in the presence of nafcillin and diosmin during the initial two hours. Notably, 8 µg/ml nafcillin significantly decreased S. Typhimurium survival during this period, with a subsequent decrease in relative survival observed at 16 h, although this reduction was not statistically significant. Kirthika et al.20 previously investigated the adhesion and invasion of S. Typhimurium strains with the Lon gene deleted in HeLa, HepG2, and Caco-2 cells, comparing them to wild-type strains. Their findings indicate that the deletion of the lon gene significantly enhanced both adhesion and invasion into host cells. However, the intracellular survival of the Lon mutant strains was notably impaired compared to that of the wild-type strains. In our study, we observed reduced intracellular survival in epithelial cells at 16 h, while in macrophages, survival was diminished during the first two hours. These findings suggest that the effects of Lon protease may vary depending on the cell type.
The expression of type II toxin-antitoxin (TA) systems, the hilA gene as an SPI-1 activating factor40, and ssrA as one of the SPI-2 activating factors in S. Typhimurium41 infecting different cell lines was investigated. Although the expression levels of type II TA system genes varied across different cell lines—likely due to the distinct cellular environments affecting gene activity—the relEB system exhibited a significant increase in bacteria isolated from all three cell types. This finding underscores the importance of the relEB system in the intracellular adaptation of S. Typhimurium.
Upon addition of nafcillin and diosmin to the cell cultures, expression levels of these TA systems decreased, indicating that these compounds inhibit Lon protease activity, thereby affecting TA system function. Interestingly, while the presence of nafcillin and diosmin did not significantly impact the expression of ssrA, an increase in hilA gene expression was observed under these conditions. Kirthika et al.20 also reported that deletion of Lon protease resulted in a substantial increase in the expression of candidate SPI-1 genes such as invF and hilC, while candidate SPI-2 genes like sifA and sseJ showed no such increase. This suggests a specific relationship between Lon protease and SPI-1 rather than SPI-2. Other studies have implicated type II toxin-antitoxin systems in bacterial intracellular survival. For instance, Paul et al. found that, while adhesion rates were not significantly different between wild-type (WT) and isogenic mutants, invasion rates were impaired in both Δhha and ΔtomB mutants42. Tiwari et al. examined the growth of wild-type and mazF-mutated strains of the type II toxin system in THP-1 macrophages, revealing that the survival of the mazF mutant strain was fourfold impaired compared to the wild-type strain at day 6 post-infection43.
We subjected S. Typhimurium to various stress conditions, including temperature, osmotic, oxidative, and acidic stress, in the presence of nafcillin and diosmin as Lon protease inhibitors, and assessed the expression of lon protease genes and type II TA systems. Our results indicated that bacterial survival was significantly reduced under oxidative and acidic stress when nafcillin and diosmin were present. In contrast, only a significant reduction in survival was observed under heat and osmotic stress at an 8 µg/ml concentration of nafcillin, likely attributable to its antibiotic properties in addition to its role as a Lon protease inhibitor.
Regarding gene expression profiles, lon protease gene expression was significantly elevated across all stress conditions examined. According to the literature, Lon protease is essential for regulating oxidative and acid stress in S. Typhimurium20. Additionally, the deletion of this protease in Actinobacillus pleuropneumoniae leads to inhibited growth when exposed to thermal, osmotic, and oxidative stresses44. Similarly, the deletion of Lon protease in Dickeya solani results in increased sensitivity to specific stress conditions, particularly osmotic and high-temperature stresses45.
However, the role of toxin-antitoxin (TA) systems in response to different stresses appears to vary across studies. For instance, a study of six type II TA systems in Klebsiella pneumoniae found no significant changes in their expression levels following exposure to oxidative stress, indicating that these TA systems may not be involved in this particular response46. In contrast, it has been clearly demonstrated that the two type II TA systems, yefM-yoeB and relBE, play a role in the oxidative stress response in Streptococcus pneumoniae47. Deletion of either or both TA systems significantly reduced the survival of S. pneumoniae after exposure to hydrogen peroxide (H2O2)47. Furthermore, the ParDE TA system has been shown to enhance cell survival in Escherichia coli at 42 °C48. The expression of relE/RHH-like TA systems, fic/phd, and brnTA was increased following exposure of Brucella species to acidic conditions (pH 5.5), suggesting a potential role for these TA systems under acidic stress49. In our study, no significant increase in expression was observed for any of the systems examined under oxidative stress. Conversely, most systems exhibited increased expression under osmotic stress. Overall, the expression levels of type II TA systems varied depending on the specific type of stress, suggesting that each system may respond differently to distinct stress conditions.
Conclusion
The results presented in this study demonstrate that Lon protease is crucial for the survival of S. Typhimurium within various host cells and in response to oxidative, acidic, osmotic, and thermal stresses. Our findings also highlight the effectiveness of nafcillin and diosmin as Lon protease inhibitors in reducing intracellular survival and viability, particularly under acidic and oxidative stress conditions. This suggests their potential utility in mitigating the pathogenesis and systemic infection associated with S. Typhimurium.
Data availability
The datasets were analyzed during the current study available from the corresponding author on reasonable request.
References
Mkangara, M. Prevention and control of human Salmonella enterica infections: An implication in food safety. Int. J. Food Sci. 2023(1), 8899596 (2023).
Teklemariam, A. D. et al. Human salmonellosis: A continuous global threat in the farm-to-fork food safety continuum. Foods 12(9), 1756 (2023).
Ménard, S. et al. Cross-talk between the intestinal epithelium and Salmonella Typhimurium. Front. Microbiol. 13, 906238 (2022).
Deng, L. & Wang, S. Colonization resistance: the role of gut microbiota in preventing Salmonella invasion and infection. Gut Microbes 16(1), 2424914 (2024).
Focarelli, F. Role of Salmonella enterica Sv Typhimurium Copper Homeostasis Proteins at the host-pathogen Interface (Newcastle University, 2023).
Petit, T. J. & Lebreton, A. Adaptations of intracellular bacteria to vacuolar or cytosolic niches. Trends Microbiol. 30(8), 736–748 (2022).
Ye, J. & Chen, X. Current promising strategies against antibiotic-resistant bacterial infections. Antibiotics 12(1), 67 (2022).
Narimisa, N., Razavi, S. & Masjedian Jazi, F. Prevalence of antibiotic resistance in Salmonella Typhimurium isolates originating from Iran: A systematic review and meta-analysis. Front. Veterinary Sci. 11, 1388790 (2024).
Lamichhane, B. et al. Salmonellosis: An overview of epidemiology, pathogenesis, and innovative approaches to mitigate the antimicrobial resistant infections. Antibiotics 13(1), 76 (2024).
Wang, C. et al. Nanocarriers for the delivery of antibiotics into cells against intracellular bacterial infection. Biomaterials Sci. 11(2), 432–444 (2023).
Ding, D. et al. The spread of antibiotic resistance to humans and potential protection strategies. Ecotoxicol. Environ. Saf. 254, 114734 (2023).
Liu, X. et al. Research progress of Salmonella Pathogenicity Island. Int. J. Biology Life Sci. 2(3), 7–11 (2023).
Lawrence, A-L-E. et al. Salmonella enterica serovar typhimurium SPI-1 and SPI-2 shape the global transcriptional landscape in a human intestinal organoid model system. MBio 12 (3). https://doi.org/10.1128/mbio.00399-21 (2021). 00399 – 21.
Thurston, T. L. & Holden, D. W. The Salmonella Typhi SPI-2 injectisome enigma. Microbiology 169(10), 001405 (2023).
Pradhan, D. & Negi, V. D. Stress-induced adaptations in Salmonella: a ground for shaping its pathogenesis. Microbiol. Res. 229, 126311 (2019).
Khan, C. A. The dynamic interactions between Salmonella and the microbiota, within the challenging niche of the gastrointestinal tract. Int. Sch. Res. Notices 2014(1), 846049 (2014).
Liu, Y. et al. RNA-seq reveals the critical role of Lon protease in stress response and Brucella virulence. Microb. Pathog. 130, 112–119 (2019).
Kirthika, P., Lloren, K. K. S., Jawalagatti, V. & Lee, J. H. Structure substrate specificity and role of Lon protease in bacterial pathogenesis and survival. Int. J. Mol. Sci. 24(4), 3422 (2023).
Singh, G., Yadav, M., Ghosh, C. & Rathore, J. S. Bacterial toxin-antitoxin modules: classification, functions, and association with persistence. Curr. Res. Microb. Sci. 2, 100047 (2021).
Kirthika, P., Senevirathne, A., Jawalagatti, V., Park, S. & Lee, J. H. Deletion of the lon gene augments expression of Salmonella Pathogenicity Island (SPI)-1 and metal ion uptake genes leading to the accumulation of bactericidal hydroxyl radicals and host pro-inflammatory cytokine-mediated rapid intracellular clearance. Gut Microbes. 11(6), 1695–1712 (2020).
Narimisa, N., Amraei, F., Kalani, B. S. & Jazi, F. M. Evaluation of gene expression and protein structural modeling involved in persister cell formation in Salmonella Typhimurium. Brazilian J. Microbiol. 52, 207–217 (2021).
Fernández-García, L. et al. Toxin-antitoxin systems in clinical pathogens. Toxins 8(7), 227 (2016).
Boss, L. & Kędzierska, B. Bacterial toxin-antitoxin systems’ cross-interactions—implications for practical use in medicine and biotechnology. Toxins 15(6), 380 (2023).
Bhowmick, A. et al. Role of VapBC4 toxin-antitoxin system of Sulfolobus acidocaldarius in heat stress adaptation. mBio 15(12), e02753–e02724 (2024).
Narimisa, N., Khoshbayan, A., Gharaghani, S., Razavi, S. & Jazi, F. M. Inhibitory effects of nafcillin and diosmin on biofilm formation by Salmonella Typhimurium. BMC Microbiol. 24(1), 1–11 (2024).
Narimisa, N., Razavi, S., Khoshbayan, A., Gharaghani, S. & Jazi, F. M. Targeting lon protease to inhibit persister cell formation in Salmonella Typhimurium: A drug repositioning approach. Front. Cell. Infect. Microbiol. 14, 1427312 (2024).
Gómara-Lomero, M., López-Calleja, A. I., Rezusta, A., Aínsa, J. A. & Ramón-García, S. In vitro synergy screens of FDA-approved drugs reveal novel zidovudine-and azithromycin-based combinations with last-line antibiotics against Klebsiella pneumoniae. Sci. Rep. 13(1), 14429 (2023).
Birhanu, B. T., Lee, E-B., Lee, S-J. & Park, S-C. Targeting Salmonella Typhimurium invasion and intracellular survival using pyrogallol. Front. Microbiol. 12, 631426 (2021).
Song, X. et al. Transcriptome analysis of virulence gene regulation by the ATP-dependent lon protease in Salmonella Typhimurium. Future Microbiol. 14(13), 1109–1122 (2019).
Kaur, D. & Mukhopadhaya, A. Outer membrane protein OmpV mediates Salmonella enterica serovar typhimurium adhesion to intestinal epithelial cells via fibronectin and α1β1 integrin. Cell. Microbiol. 22(5), e13172 (2020).
Rio, D. C., Ares, M., Hannon, G. J. & Nilsen, T. W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harbor Protocols 2010(6), pdb. prot5439. (2010).
Rao, X., Huang, X., Zhou, Z. & Lin, X. An improvement of the 2ˆ (–delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat.Bioinf. Biomath. 3(3), 71 (2013).
Jones, B. & Stacey, G. Safety considerations for in vitro toxicology testing. Cell. Cult. Methods Vitro Toxicol. 43–66. (2001).
Anderson, C. J. & Kendall, M. M. Salmonella enterica serovar typhimurium strategies for host adaptation. Front. Microbiol. 8, 1983 (2017).
Luk, C. H. et al. Salmonella enters a dormant state within human epithelial cells for persistent infection. PLoS Pathog. 17(4), e1009550 (2021).
Marmion, M., Macori, G., Ferone, M., Whyte, P. & Scannell, A. Survive and thrive: Control mechanisms that facilitate bacterial adaptation to survive manufacturing-related stress. Int. J. Food Microbiol. 368, 109612 (2022).
Sutar, A. A., Dashpute, R. S., Shinde, Y. D., Mukherjee, S. & Chowdhury, C. A systemic review on fitness and survival of Salmonella in dynamic environment and conceivable ways of its mitigation. Indian J. Microbiol. 64(2), 267–286 (2024).
Carryn, S. et al. Intracellular pharmacodynamics of antibiotics. Infect. Disease Clin. 17(3), 615–634 (2003).
Kamruzzaman, M., Wu, A. Y. & Iredell, J. R. Biological functions of type II toxin-antitoxin systems in bacteria. Microorganisms 9(6), 1276 (2021).
Lostroh, C. P., Bajaj, V. & Lee, C. A. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI-1. Mol. Microbiol. 37(2), 300–315 (2000).
Garmendia, J., Beuzon, C. R., Ruiz-Albert, J. & Holden, D. W. The roles of SsrA–SsrB and OmpR–EnvZ in the regulation of genes encoding the Salmonella Typhimurium SPI-2 type III secretion system. Microbiology 149 (9), 2385–2396 (2003).
Paul, P. et al. The Hha–TomB toxin–antitoxin module in Salmonella enterica Serovar Typhimurium limits its intracellular survival profile and regulates host immune response. Cell Biol. Toxicol. 1–17. (2022).
Tiwari, P. et al. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat. Commun. 6(1), 6059 (2015).
Xie, F. et al. The lon protease homologue LonA, not LonC, contributes to the stress tolerance and biofilm formation of Actinobacillus pleuropneumoniae. Microb. Pathog. 93, 38–43 (2016).
Figaj, D. et al. Lon protease is important for growth under stressful conditions and pathogenicity of the phytopathogen, bacterium Dickeya Solani. Int. J. Mol. Sci. 21(10), 3687 (2020).
Narimisa, N., Amraei, F., Kalani, B. S., Mohammadzadeh, R. & Jazi, F. M. Effects of sub-inhibitory concentrations of antibiotics and oxidative stress on the expression of type II toxin-antitoxin system genes in Klebsiella pneumoniae. J. Global Antimicrob. Resist. 21, 51–56 (2020).
Chan, W. T. et al. The Streptococcus pneumoniae yefm-yoeb and relBE toxin-antitoxin operons participate in oxidative stress and biofilm formation. Toxins 10(9), 378 (2018).
Kamruzzaman, M. & Iredell, J. A ParDE-family toxin antitoxin system in major resistance plasmids of Enterobacteriaceae confers antibiotic and heat tolerance. Sci. Rep. 9(1), 9872 (2019).
Amraei, F., Narimisa, N., Mohammadzadeh, R. & Lohrasbi, V. The expression of type II TA system genes following exposure to the sub-inhibitory concentration of gentamicin and acid stress in Brucella spp. Microb. Pathog. 144, 104194 (2020).
Narimisa, N., Amraei, F., Kalani, B. S. & Jazi, F. M. Evaluation of gene expression and protein structural modeling involved in persister cell formation in Salmonella Typhimurium. Braz J. Microbiol. 52(1), 207–217 (2021).
Kasturi, K. N., Drgon, T. & Real-Time, P. C. R. Method for detection of Salmonella spp. Environ. Samples Appl. Environ. Microbiol. 83(14). (2017).
Acknowledgements
This study was financially supported for a PhD thesis in Iran University of Medical Sciences (Tehran, Iran), for which we are very grateful.
Funding
This study was financially supported by a research grant (No.24791) in Iran University of Medical Sciences (Tehran, Iran).
Author information
Authors and Affiliations
Contributions
N.N. contributed to the conception, methodology, design, and writing—original draft. N.N. and N.B. and A.K. performed data collection and analysis. F.M.J. and S.R. and S.G. were involved interpretation of the data, review & editing of the study. F.M.J. and S.R. contributed to study supervision. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was approved by the Ethics Committee of Iran University of Medical Sciences by Ethics No. (IR.IUMS.FMD.REC.1402.262).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Narimisa, N., Bostanghadiri, N., Khoshbayan, A. et al. Impact of nafcillin and diosmin on the attachment, invasion, and stress survival of Salmonella Typhimurium. Sci Rep 15, 6308 (2025). https://doi.org/10.1038/s41598-025-90808-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-90808-4