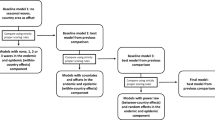
Abstract
Avian influenza poses substantial risks to animal welfare and public health. The recent surge in highly pathogenic avian influenza (HPAI) outbreaks has led to extensive poultry culling, highlighting the need for early warning systems. Using data on H5 HPAI virus (HPAIV) occurrence from the World Organization for Animal Health and the Food and Agriculture Organization, we employed a spatial time-series modelling framework to predict occurrences in Japan and South Korea, 2020–2024. This framework decomposes time-series data into endemic and epidemic components and has previously been used to model HPAIV in Europe. We identified 1,310 HPAIV detections from 2020 to 2024, the majority being H5N1 (55.3%) and H5N8 (35.0%). These data consisted of 827 and 483 detections in wild and domestic birds, respectively. The model included seasonality and covariates in both endemic and epidemic components and revealed consistent yearly seasonal patterns. This contrasts with previous modelling of European data where seasonality changed over time. The model predicted 81% of detections as epidemic, primarily due to within-region transmission (53%), whereas only 19% were endemic. This model effectively predicts weekly H5 HPAIV detections, aiding decision-makers in identifying high-risk periods. This study confirms the robustness and usefulness of endemic-epidemic modelling of HPAIV in different regions of the world.
Similar content being viewed by others
Introduction
Highly pathogenic avian influenza (HPAI) has caused outbreaks in wild birds and poultry worldwide for many years, with increasing numbers of outbreaks in Europe in 2020–20231. However, since early 2023, Europe has seen a decrease in HPAI virus (HPAIV) oubreaks2, and so far in 2024, the number of detections is the lowest since the 2019–2020 epidemiological year1. Studies have suggested that the European outbreaks of H5N1 were transmitted through the trans-Atlantic flyway to North America in 2021, from where the infection expanded southwards to South America, causing mass mortality events along this pathway3,4. Recently, a study found that H5N1 has reached as far south as Antarctica5. In Japan, HPAIV re-emerged in 2004; outbreaks with H5N1, H5N8 and H5N6 subtypes have been detected, and the most severe outbreaks here have been caused by H5 subtypes6. In the centre of the East Asian-Australasian flyway, South Korea and Japan are at risk of HPAIV introduction from a large number of migratory birds7. Both countries have reported a substantial number of outbreaks of avian influenza (AI) during the last decade, making it possible to study the spatiotemporal dynamics of HPAIV occurrence. Moreover, recent research has identified East Asia as a potential source of HPAI outbreaks in both Central Asia and Alaska; in addition the area is exposed to transmission from continental Europe, the Black Sea Mediterranean region and central Asia, connecting the two continents8. Thus, East Asia is well connected with the rest of the world in terms of HPAIV spread.
It is established that AI virus (AIV) can infect mammals, and in recent years, several mammalian species, including seals (Pinnipedia), foxes (Vulpes vulpes), tigers (Panthera tigris), cats (Felis catus) and mink (Neovison vison)9, have been found infected with HPAIV. The subtype overwhelmingly found in mammals is H5N19. Since early 2024, cattle have been infected and H5N1 has now been detected in many US states, likely facilitated by a single introduction10. Additionally, several human H5N1 infections in the U.S. have been linked to contact with infected poultry or dairy cows11. In late 2024, H5N1 was also confirmed in a pig in Oregon12. Since pigs can be co-infected with avian, human, and swine influenza strains, this raises concerns about genetic reassortment, which could lead to the emergence of novel strains with enhanced transmissibility or pathogenicity in humans12. Actions to prevent the spread of AIV to poultry include regulations to reduce contact with wild birds and spread via farm workers. Moreover, regular screening and disinfection of fomites such as farm equipment, clothing, and vehicles can help identify and mitigate contamination risks13,14. Additionally, vaccination can be utilized in some places15. To implement timely actions against the spread of avian influenza to poultry, and thereby also reducing the risk of spill over to humans, it is imperative to have prediction models in place16. Recent years have seen the development of many such models, ranging from syndromic surveillance models using google trends to detect outbreaks17, to spatiotemporal regression type models for international spread18, to high-resolution spatiotemporal risk models for outbreaks19. We recently utilized a previously developed endemic-epidemic model framework for occurrence of HPAIV to predict HPAIV detections and transmission between countries in Europe20. This framework utilises near-real-time register data to evaluate the risk of outbreaks in a specific area based on estimating the history of HPAIV detections of both local and external epidemic outbreaks, as well as contributions from an endemic component of transmission. The aim of this study was to investigate if this model framework could be utilized on registered HPAIV detections from East Asia to predict the risk of detections in domestic or wild birds.
Materials and methods
Data, spatial resolution, and landscape variables
Due to lack of sufficient data from multiple East Asian countries, we restricted our analyses to Japan and South Korea, as both countries consistently reported HPAIV detections throughout the study period, and being neighbours, they constitute a coherent study area. We obtained data on the occurrence of AIV from WOAH-WAHIS2 and EMPRES-i21 for all of East Asia from week 43 in 2020 until week 21 in 2024. The time period was restricted in order to reflect the recent HPAIV detections in the region. To identify HPAIV detections, for the WOAH-WAHIS data we used the definitions “High pathogenicity avian influenza viruses (poultry) (Inf. with)” and “Influenza A viruses of high pathogenicity (Inf. with) (non-poultry including wild birds)”, and for the EMPRES-i data we used the definition “Influenza—Avian “ and serotypes containing the word “HPAI". We excluded non-avian species from both data sets and defined each data entry as an HPAIV detection, disregarding the number of birds affected. We did not differentiate between domestic and wild birds in the data sets. We removed duplicate entries between the two datasets, if they overlapped in the date of detection, longitude and latitude, serotype and bird species. We then obtained shapefiles of administrative areas of Japan and South Korea from GADM22 and modified these to create 8 Japanese regions and 9 South Korean regions based on geography (Fig. 1A). For each of these regions (n = 17), we aggregated the data into weekly detections (n = 187). For each region, any week within the study period that had no reported detections in a region was recorded as a “zero detection” week.
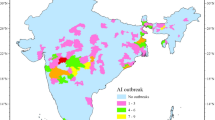
(A) Map of reported highly pathogenic avian influenza virus (H5 subtype) detections per 10,000 km2 between 2020 and 2024 (21 October 2020 to 22 May 2024) summed over 17 regions in Japan and South Korea. The dataset contained a total of 1,310 detections of varying sizes in both wild and domestic birds. (B) Waterway connections (dark blue) created for the spatial adjacency order of the regions (light blue) included in the endemic-epidemic time-series model in this study (remote islands belong to regions on the mainland and thus are considered part of the region with no need for connecting waterways). The maps were created using the package tmap23 in R 4.3.324.
We examined seasonality in the data using the spectrum function in R version 4.3.324, applied to the weekly aggregated detection counts across all regions. This approach identifies periodic patterns by estimating the power spectral density and presents the results as a periodogram. Peaks in the periodogram represent dominant frequencies in the time series, with their height indicating the strength of the periodicity. We explored landscape variables that might be associated with presence of wild birds, i.e., coastal areas, waterways, and wetlands, where migratory birds are known to aggregate in high numbers25. For each region, we calculated the length of coastline (km) using Natural Earth’s coastline raster26 (resolution of 10 × 10 m) and used Global Lakes and Wetlands Database rasters27, 1km2 resolution). We used R 4.3.324 package sp28,29 to calculate both the length of coastline and the area (km2) of waterways/wetlands within each region. We combined waterways and wetlands and used the following landcover definitions in the raster file: “Lake”, “Reservoir”, “River”, “Freshwater Marsh, Floodplain”, “Coastal Wetland (incl. Mangrove, Estuary, Delta, Lagoon)”, “Pan, Brackish/Saline Wetland”, “Bog, Fen, Mire (Peatland)”, and “Intermittent Wetland/Lake”.
Statistical analysis
As described in Kjær et al.20, we applied a multivariate time-series model (Meyer et al.30) to understand the spatiotemporal dynamics of HPAIV detections. This framework decomposes data into endemic and epidemic components. The endemic part represents the baseline rate of detections, while the epidemic part captures occasional spatiotemporal clusters. Our model accounts for within-region (λ) and between-region (ϕ) effects in the epidemic component. The main formula for the model’s mean count per region (i) and time (t) is:
here, eit represents the expected counts as an offset for the endemic predictor νit. The epidemic component includes within-region effects (λit) and between-region effects (ϕit) captured by a spatial weights matrix describing how close the regions are located (wji). We incorporated long-range transmission by adjusting spatial weights as a power-law model with distance-decay20,30. The model integrates endemic and epidemic transmission components, allowing for log-linear predictors within each region, such as landscape features. To capture seasonality in detections, we furthermore included 1, 2, or 3 seasonal waves in both the endemic and epidemic components. To minimize model complexity, seasonal waves were added only to the within-region effects in the epidemic components and not to the between-country effects. The model equations below illustrate the inclusion of 3 seasonal waves:
Lastly, the model allows for the inclusion of region-specific random effects (both correlated and uncorrelated) to capture any remaining heterogeneity not explained by the covariates, endemic and epidemic effects, or seasonal waves. For detailed equations, refer to Kjær et al.20, Meyer et al.30, Held et al.31, and Paul and Held32.
As in Kjær et al.20, we added water shapes to the spatial weights matrix calculated for the model, in order to account for birds being able to traverse waterways (Fig. 1B). In the original spatial weights matrix, if two regions are divided by waterways, the spatial weights calculated by the adjacency matrix will automatically produce a zero value; by including water as a shape between such regions these waterways in the adjacency matrix act as a connecting region (Fig. 1B).
We utilized the R package surveillance30,33 in R version 4.3.324 to fit various models to our data. The dataset was split into a training (from week 43 in 2020 to week 10 in 2024, and week 16 to week 21 in 2024) and test set (from week 11 to week 15 in 2024). We trained models on the training data and evaluated their performance on the test data.
We compared several models to identify the best fit for our data, including a basic model with simple between-region effects (only transmission from regions sharing a border), models with seasonal waves, and those incorporating long-range transmission and random effects. For all models, we added region area relative to all regions as an offset in the endemic component. We assessed model performance using ranked probability score (RPS) and logarithmic score (logS), selecting the best models based on calibration. Subsequently, we included the log-transformed variables area of waterways/wetlands and length of coastline as either covariates or offsets to the endemic and within-country epidemic components. After validating these models against the baseline without seasonality or covariates, we compared them to models with spatial weights as a power-law model with distance-decay and region-specific random effects. Calibration checks and scoring rules guided the selection of the final forecasting model. For models with random effects, we used Nelder-Mead penalization to maximize marginal likelihood. We conducted 10,000 simulations. Based on visual convergence, we assessed the long-term forecasting ability of the final model, covering the respective timeframes of the datasets. Forecasting involved sequential calls to negative binomial distributions, utilizing parameter estimates and simulated counts from previous time points.
Results
Data
The data obtained from WOAH-WAHIS contained 1,221 HPAIV detections for 187 weeks (from 21 October 2020 to 22 May 2024) in 17 regions in Japan and South Korea, whereas EMPRES-i contained 575 HPAIV detections (from 4 November 2020 to 29 April 2024). Of these detections, 486 were duplicates. Therefore, the final dataset included 1,310 HPAIV detections (see Table 1) between 21 October 2020 and 22 May 2024. Of these 1,310 detections, 827 were detections in wild birds and 483 were detections in domestic birds. During this time period, only HPAIV H5 subtypes were reported, the majority of which were H5N1 (55.3%) and H5N8 (35.0%, Table 1). These proportions are similar to what was previously reported from Europe (Kjær et al.20). Due to data constraints, we combined all reported HPAIV H5 subtypes in our analyses. In general, Japan reported more detections (n = 716) than South Korea (n = 594), however the South Korean regions Gyeonggi-do, Gangwon-do, Jeollabuk-do, and Jeollanam-do had higher number of detections relative to their areas than any other regions, including the regions in Japan (Fig. 1A). Detections were generally reported during the months October to May (Fig. 2), with wild and domestic bird detections showing similar seasonal patterns (Supplementary Figure S1). For details regarding HPAIV H5 detections within individual regions in each country, see Tables 1 and 2. Although the WOAH-WAHIS data included the number of affected birds for each entry, we opted to model only outbreak detections. This decision accounts for potential bias: some countries, such as Denmark, only sample one or a few initially infected birds in an area and therefore do not sample or test all wild birds within a known outbreak area. Additionally, domestic birds are typically affected in much larger numbers than wild birds, potentially distorting patterns of outbreak severity if both groups are modelled together. Therefore, we here focus on occurrence from a surveillance perspective.
Analysing the detection data for seasonality revealed a dominant period of 48 weeks (Supplementary Figure S2). This indicates a repeating cycle every 48 weeks, suggesting a near-annual pattern slightly shorter than a full 52-week year. Other smaller peaks in the periodogram reveals further potential seasonal cycles and these findings confirm the presence of seasonality in the detection data, supporting the inclusion of seasonal components in our endemic-epidemic time-series model.
Endemic-epidemic time-series model
To assess seasonality, we compared the baseline model without seasonality to models with one, two, and three seasonal waves in both endemic and epidemic (within-region effects) components. The optimal model, with the lowest RPS and logS scores, featured one seasonal wave in both components and was selected as the new baseline (Table 3). Most models passed calibration tests (p ≥ 0.05). Adding covariates for waterways/wetlands and coastline length improved the model, with the best-calibrated version including these as offsets in the endemic component and as covariates in the epidemic component (Table 3). Adding spatial weights between regions wji as a power-law model with distance-decay to all other regions and region-specific random effects further enhanced the model’s performance (Table 3).
The final model thus included one seasonal wave in both the endemic component and the epidemic within-region component, covariates as offset in the endemic component and as covariates in the epidemic within-region component, power law to account for long-range transmission and correlated random effects. Correlated random effects allow unobserved variables that may explain the remaining heterogeneity in the model to be correlated with all observed variables. The endemic component showed peaks during late autumn/winter (Supplementary Figure S3), whereas the within-region epidemic seasonality showed peaks during late summer/early autumn. Epidemic between-region transmission was mostly seen in Tohoku, Chubo, Chungcheongbuk-do and Jeju (Figs. 3 and 4). The model showed a negative association between coastline length and detections, and a positive association with wetland area, though these were not statistically significant (Table 4). Based on the final model results, most reported HPAIV detections in Japan and South Korea were epidemic (80.8%), with 53.2% within-region and 27.6% between-region transmission; 19.1% were endemic (Fig. 5). Comparing the final model to the original baseline model shows that the final model’s overall predictions encompass slightly more of the actual observed detections, particularly for the years 2022/2023 and 2023/2024 (Fig. 5). Whereas the baseline model overestimated total number of predictions (observed n = 1,310) by 2.06% (n = 1,342), the final model only overestimated number of predictions by 1.3% (n = 1,327). The logS scoring rule is calculated as the logarithm of the probability estimate for the actual outcome and based on these logS values; the baseline model predicted true compared to actual detections on average 83% for the test set (logS = 0.19), whereas the final model predicted true on average 85% for the test set (logS = 0.16). Thus, the final model was only slightly better than the baseline model. The prediction plots showed a slight tendency for our final models to underestimate the number of detections during periods of high detection (Figs. 3, 4, and 5).
Model fit for the 8 regions of Japan illustrates the relative contributions of model components for the final multivariate time-series model, with grey showing the endemic component, blue the within-region epidemic component, and orange the between-region epidemic component. The dots represent the actual counts of reported highly pathogenic avian influenza virus (H5 subtype) detections in domestic and wild birds. While the depicted actual counts from week 11–15 in 2024 are shown, they were not included in the model’s training set and therefore are not part of the model fit.
Model fit for the 9 regions of South Korea illustrates the relative contributions of model components for the final multivariate time-series model, with grey showing the endemic component, blue the within-region epidemic component, and orange the between-region epidemic component. The dots represent the actual counts of reported highly pathogenic avian influenza virus (H5 subtype) detections in domestic and wild birds. While the depicted actual counts from week 11–15 in 2024 are shown, they were not included in the model’s training set and therefore are not part of the model fit.
The aggregated model fit across all regions of Japan and South Korea illustrates the relative contributions of model components for both the original baseline model and the final multivariate time-series model. The dots represent the actual counts of reported highly pathogenic avian influenza virus (H5 subtype) detections in domestic and wild birds. While the depicted actual counts from week 11–15 in 2024 are shown, they were not included in the model’s training set and therefore are not part of the model fit.
The exponentially transformed deviations of the random intercepts revealed that Hokkaido, Kyushu, Shikoku, Jeollanam-do, and Gangwon-do had more within-country cases than accounted for by the model (Supplementary Figure S4, pink colour). Maps of the random intercepts for epidemic between-region transmission indicated that regions such as Kyushu, Gyeonggi-do, and Jeollanam-do, followed by Hokkaido, received more cases from other regions than explained by the model (Supplementary Figure S4, pink colour). Maps of the endemic random intercepts highlighted regions with lower endemic incidence than predicted by the model (Supplementary Figure S4, shown as cyan). Fan plots showing one-week-ahead predictions indicated the final model predicted detections reasonably well, with a lag of one week for some peaks (Supplementary Figures S5-S6).
Long-term forecasts from 2020 to 2024 using the final model predicted numerous detections annually during the autumn/spring periods (Fig. 6). The median number of simulated detections (n = 1,295) was slightly lower than the observed detections (n = 1,310). This pattern was consistent across individual regions (Supplementary Figures S7 and S8).
The simulation-based long-term forecast for the final model starts from week 43 in 2020 (left-hand dot) and extends through week 21 in 2024. The plots display the weekly number of predicted and observed detections of highly pathogenic avian influenza virus (H5 subtype) aggregated across all regions. The fan charts represent the 1% and 99% quantiles of the simulations (n = 10,000) each week, with the mean depicted as a white line. Actual reported detections are shown with open circles. Data from weeks 11 to 15 in 2024 were not used to train the model.
Discussion
In this study, we used the same modelling framework for HPAIV in Japan and South Korea as we have previously used for Europe20. The European model detected a change in the seasonality of detections in recent years, with detections changing from being exclusively during spring/autumn/winter to also occurring during the summer period, although at a lower level (and the pattern may have changed again in 2024). However, the pattern for Japan and South Korea was found to be consistently seasonal with most detections during the autumn/spring periods. This highlights that there are regional differences between occurrence patterns in Europe and East Asia, which must be considered when predicting outbreaks and designing surveillance systems.
In contrast to the European model for the years 2021–2022, the final model for Japan and South Korea used for predictions included a seasonal wave in the endemic component. The within-region effects of the epidemic component also included one seasonal wave. The model predicted higher probability of detections during the autumn-early spring months, and overall, the model fit showed good predictive power for all regions over time (Figs. 3 and 4). The model tended to underestimate detections during high-incidence periods, though it predicted the timing of detection peaks well (Figs. 3–5). While most detections were predicted as epidemic, including long-distance transmission, the model responded to reported data and could miss initial surges, as seen in Hokkaido and Chugoku. However, predictions adjusted to new levels based on prior data, typical of one-step-ahead forecasting (Supplementary Figures S5 and S6). This highlights the need for careful consideration in forecasting HPAI risk periods and decision-making, with greater uncertainty in regions with fewer observations. Despite a good fit to observed detections, the model over- and underestimated detections within regions (Supplementary Figures S3 and S4). Underestimation (RI > 1) occurred in regions like Hokkaido, Kyushu, Gangwon-do, and Jeollanam-do, while overestimation (RI < 1) was seen in Chubo, Chukogu, Kansai, Kanto, and Tohoku. The model also underestimated between-region transmission in regions with extensive coastlines, such as Hokkaido and Kyushu, where migratory birds from beyond neighbouring regions may introduce variability. The endemic component, accounting for unreported outbreaks or transmission from areas not included in the study or transmission from migratory birds coming from areas and countries not included in the study, showed underestimation (R > 1) in several regions, possibly due to underreporting or the model attributing more detections to surrounding regions, thus reducing the estimated effect of migratory bird transmission. Incorporating covariates such as coastline length and area of waterways/wetlands improved the model, indicating a link between these factors and HPAIV detection. However, as with the European models, the effect of these covariates in the epidemic component was statistically non-significant. Other studies suggest landscape variables influence AIV detections25,34,35, but seasonality, like bird migrations, may be more crucial drivers of epidemic HPAI. Fujimoro and Haga36 found weather associations with HPAI outbreaks in Japan, suggesting that including weather variables could enhance model prediction.
Since early 2023, Europe has seen a decline in HPAIV detections2, with 2024 showing the lowest number since the 2019–2020 epidemiological year1. However, HPAIV now circulates year-round in wild birds, albeit at low levels, complicating model fitting15. This shift could be due to factors such as flock immunity, depletion of wild bird populations, changes in migration, reduced viral contamination, and decreased reporting in wild birds1. In contrast, HPAIV in Japan and South Korea shows consistent seasonality. This may be due to the virus never having had the opportunity to spread quickly within dense colonies, as it has done in tern (Sternidae spp.), gull (Larinae spp.), cormorant (Phalacrocoracidae spp.) and other seabird colonies in Europe37,38,39,40. Also, the large goose species and swans that overwinter in Japan and South Korea breed in the Arctic regions of Russia, and never come into direct contact with those from Europe41,42,43. In contrast, some Chinese overwintering tundra swans (Cygnus columbianus columbianus) breed as far west as the Yamal Peninsula in the western Russian Arctic, where they mix with swans, white-fronted geese (Anser albifrons), and many other ducks and swans that will overwinter in western Europe, creating a transmission point for their wintering grounds along the Yangtze River41,42,43. In their wintering grounds these birds tend not to mix with those overwintering in Japan and South Korea, which breed further to the east41,42,43. These differences may explain the contrasting occurrence patterns in Europe and East Asia. According to the logS for the European model (logS = 1.091) for the years 2021–2022, the model only predicted true on average 33.6% for the test set. With the Japan/South Korea model predicting true on average 85% for the test set, this latter model seems to be a better predictive model, which could be explained by the more consistent patterns over time seen for Japan and South Korea but also having to fit a smaller number of regions (countries for the European model) in the Japan/South Korea model.
The final model estimated 80.8% of HPAIV detections in Japan and South Korea as epidemic, driven by within- and between-region transmission. In contrast to the European model, which found 12.2% of detections endemic, this model identified 19.1% as endemic, suggesting unreported local HPAIV circulation and input from migratory birds outside the study area. The higher endemic proportion and consistent seasonality imply that HPAIV occurrence in Japan and South Korea remains heavily dependent on migratory birds.
The modelling framework used here has some caveats as was also explained in Kjær et al.20 First, pooling the different H5 subtypes might cause the predictive ability of our model to be lower, as different HPAIV strains may have different transmission potentials44,45. Most of the subtypes reported for our study regions were H5N1 and H5N8, and in a review paper on AIV parameter estimates by Kirkeby and Ward45, the median basic reproduction number (R0, based on reviewing published articles) was found to be 2.18 for H5N1 and 0.94 for H5N8. Even though there might be a high uncertainty surrounding these values, this clearly shows that there is a large difference between the two subtypes and thus any future models would benefit from running separate models for different subtypes (albeit data constraints, as in our case, may be a limiting factor). Second, similarly to our European models, we combined HPAIV detections in both domestic and wild birds due to data constraints. Although the assumption that HPAIV can be transmitted from wild to domestic birds and back may not hold, the seasonal patterns for each dataset exhibit agreement (Supplementary Material Figure S1), which show us that combining the data is warranted. Lastly, the WOAH and EMPRES-i data do not report sampling scheme or prevention measures, and we do not have a measure of surveillance or detection effort for each region in our model. Differences in surveillance efforts can bias model results, overrepresenting areas with intensive monitoring while underestimating outbreak levels in poorly monitored regions, distorting spatial dynamics and reducing accuracy. Incomplete and delayed reporting in WOAH-WAHIS and EMPRES-i data further skews analyses, with underreporting in wild birds particularly problematic, especially if the analysis is used to estimate risk continuously. These limitations underscore the need for better reporting mechanisms and complementary data sources. The model’s reliance on data quality constrained its scope to parts of East Asia, highlighting the importance of consistent international data collection to improve predictive accuracy and expand coverage. In the face of these caveats, the model for Japan and South Korea still fitted well to observed HPAIV reports from WOAH-WAHIS and EMPRES-i. The results show the utility and practicality of using this type of modelling framework together with publicly available data, where some information is unavailable. The model presented here may be of use to policy makers in identifying the spatio-temporal risk of HPAI within the Japan and South Korea regions.
Conclusion
We here used a spatio-temporal model framework on 1,310 HPAIV detections from wild birds and poultry in Japan and South Korea in 2020–2024. The model predicts risks of detections at the regional level within both countries, and the final model included seasonality and covariates to predict detections one week ahead. This model can be continuously used as an indicator for the regional risk of HPAIV detections, taking detections in neighbouring regions into account. Thus, it can be used for policy makers to timely implement actions to prevent spread to poultry farms.
Data availability
Data retrieved from the World Organisation for Animal Health (WOAH) (2024) – Periodical Data Extraction WAHIS SharePoint. Retrieved on 2024–06–21 from https://oieoffice365.sharepoint.com/sites/PeriodicaldataextractionsOIE-WAHIS. Reproduced with permission. WOAH bears no responsibility for the integrity or accuracy of the data contained herein, but not limited to, any deletion, manipulation, or reformatting of data that may have occurred beyond its control. Data from EMPRES-i were downloaded from https://data.apps.fao.org/empres-i/ with the following disclaimer: The designations employed and the presentation of material in this information product are not warranted to be error free and do not imply the expression of any opinion whatsoever on the part of FAO concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. FAO makes every effort to ensure, but does not guarantee, the accuracy, completeness or authenticity of the information in this information product. Data and model code of this study are available on figshare: https://doi.org/10.6084/m9.figshare.27127134, and can also be provided by the corresponding author by request.
References
European Food Safety Authority et al. Avian influenza overview March – June 2024. EFSA J. 22, e8930 (2024).
WAHIS: World Animal Health Information System (of the OIE). wahis.woah.org/#/home (2024).
Harvey, J. A., Mullinax, J. M., Runge, M. C. & Prosser, D. J. The changing dynamics of highly pathogenic avian influenza H5N1: Next steps for management & science in North America. Biol. Conserv. 282, 110041 (2023).
Caliendo, V. et al. Transatlantic spread of highly pathogenic avian influenza H5N1 by wild birds from Europe to North America in 2021. bioRxiv 2022.01.13.476155 (2022) https://doi.org/10.1101/2022.01.13.476155.
Bennison, A. et al. Detection and spread of high pathogenicity avian influenza virus H5N1 in the Antarctic Region. bioRxiv 2023.11.23.568045 (2023) https://doi.org/10.1101/2023.11.23.568045.
Moriguchi, S. et al. Surveillance system for avian influenza in wild birds and implications of its improvement with insights into the highly pathogenic avian influenza outbreaks in Japan. Prev. Vet. Med. 187, 105234 (2021).
Nam, J. H. et al. Surveillance of avian influenza viruses from 2009 to 2013 in South Korea. Sci. Rep. 2021(11), 1–7 (2021).
Hill, N. J. et al. Ecological divergence of wild birds drives avian influenza spillover and global spread. PLOS Pathog. 18, e1010062 (2022).
ENETWILD Consortium et al. The role of mammals in Avian Influenza: a review. EFSA Support. Publ. 21, (2024).
Burrough, E. R. et al. Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4b Virus Infection in Domestic Dairy Cattle and Cats, United States, 2024 - Volume 30, Number 7—July 2024 - Emerging Infectious Diseases journal - CDC. Emerg. Infect. Dis. 30, (2024).
Garg, S. et al. Highly pathogenic avian influenza A(H5N1) virus infections in humans. N. Engl. J. Med. https://doi.org/10.1056/NEJMOA2414610/SUPPL_FILE/NEJMOA2414610_DATA-SHARING.PDF (2024).
Centers for Disease Control and Prevention (CDC). CDC A(H5N1) Bird Flu Response Update November 4, 2024. https://www.cdc.gov/bird-flu/spotlights/h5n1-response-11012024.html (2024).
Tiwari, A., Patnayak, D. P., Chander, Y., Parsad, M. & Goyal, S. M. Survival of two avian respiratory viruses on porous and nonporous surfaces. Avian Dis. 50, 284–287 (2006).
United States Department of Agriculture (USDA). Biosecurity Factors and the Introduction and Spread of HPAI : Findings from Epidemiological Studies. (2015).
Guinat, C. et al. Promising effects of duck vaccination against highly pathogenic avian influenza, France 2023–24. bioRxiv 2024.08.28.609837 (2024) https://doi.org/10.1101/2024.08.28.609837.
Lambert, S. et al. A systematic review of mechanistic models used to study avian influenza virus transmission and control. Vet. Res. 54, 96 (2023).
Lu, Y. et al. An epidemic avian influenza prediction model based on google trends. Lett. Org. Chem. 16, 303–310 (2018).
Azat, C., Alvarado-Rybak, M., Aguilera, J. F. & Benavides, J. A. Spatio-temporal dynamics and drivers of highly pathogenic avian influenza H5N1 in Chile. Front. Vet. Sci. 11, 1387040 (2024).
Prosser, D. J. et al. Using an adaptive modeling framework to identify avian influenza spillover risk at the wild-domestic interface. Sci. Rep. 2024(14), 1–13 (2024).
Kjær, L. J. et al. Using surveillance data for early warning modelling of highly pathogenic avian influenza in Europe reveals a seasonal shift in transmission, 2016–2022. Sci. Rep. 13, 1–16 (2023).
Food and Agriculture Organization of the United Nations. EMPRES-i. https://empres-i.apps.fao.org/ (2024).
GADM. GADM, GADM, the Database of Global Administrative Areas. https://gadm.org (2023).
Tennekes, M. tmap: Thematic Maps in R. J. Stat. Softw. 84, 1–39 (2018).
R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing http://www.r-project.org (2024).
Belkhiria, J., Hijmans, R. J., Boyce, W., Crossley, B. M. & Martínez-López, B. Identification of high risk areas for avian influenza outbreaks in California using disease distribution models. PLoS One 13, e0190824 (2018).
Natural Earth. Coastline - Free vector and raster map data at 1:10m, 1:50m, and 1:110m scales. https://www.naturalearthdata.com/downloads/10m-physical-vectors/10m-coastline/ (2022).
Global Lakes and Wetlands raster data. World Wildlife Fund https://www.worldwildlife.org/pages/global-lakes-and-wetlands-database (2022).
Pebesma, E. J. & Bivand, R. S. Classes and methods for spatial data in R. R News 5, (2005).
Bivand, R. S., Pebesma, E. & Gómez-Rubio, V. Applied Spatial Data Analysis with R: Second Edition. Appl. Spat. Data Anal. with R Second Ed. 1–405 (2013) https://doi.org/10.1007/978-1-4614-7618-4/COVER.
Meyer, S., Held, L. & Höhle, M. Spatio-Temporal Analysis of Epidemic Phenomena Using the R Package surveillance. J. Stat. Softw. 77, (2014).
Held, L., Höhle, M. & Hofmann, M. A statistical framework for the analysis of multivariate infectious disease surveillance counts: Stat. Modelling 5, 187–199 (2005).
Paul, M. & Held, L. Predictive assessment of a non-linear random effects model for multivariate time series of infectious disease counts. Stat. Med. 30, 1118–1136 (2011).
Salmon, M., Schumacher, D. & Höhle, M. Monitoring Count Time Series in R: Aberration Detection in Public Health Surveillance. J. Stat. Softw. 70, 1–35 (2016).
Kjær, L. J. et al. Landscape effects and spatial patterns of avian influenza virus in Danish wild birds, 2006–2020. Transbound. Emerg. Dis. https://doi.org/10.1111/tbed.14040 (2021).
Ward, M. P., Maftei, D., Apostu, C. & Suru, A. Association between outbreaks of highly pathogenic avian influenza subtype H5N1 and migratory waterfowl (Family Anatidae ) populations. Zoonoses Public Health 56, 1–9 (2009).
Fujimoto, Y. & Haga, T. Association between highly pathogenic avian influenza outbreaks and weather conditions in Japan. J. Vet. Med. Sci. https://doi.org/10.1292/JVMS.23-0521 (2024).
Falchieri, M. et al. Shift in HPAI infection dynamics causes significant losses in seabird populations across Great Britain. Vet. Rec. 191, 294–296 (2022).
Knief, U. et al. Highly pathogenic avian influenza causes mass mortality in Sandwich Tern Thalasseus sandvicensis breeding colonies across north-western Europe. Bird Conserv. Int. 34, e6 (2024).
Lean, F. Z. X. et al. Highly pathogenic avian influenza virus H5N1 infection in skua and gulls in the United Kingdom, 2022. Vet. Pathol. 61, 421–431 (2024).
Bregnballe, T. et al. Outbreaks of highly pathogenic avian influenza (HPAI) epidemics in Baltic Great Cormorant Phalacrocorax carbo colonies in 2021 and 2022. Bird Study https://doi.org/10.1080/000636572399168 (2024).
Fang, L. et al. Two distinct flyways with different population trends of Bewick’s Swan Cygnus columbianus bewickii in East Asia. Wildfowl 6, 13–42 (2020).
Deng, X. et al. Contrasting trends in two East Asian populations of the Greater White-fronted Goose Anser albifrons. Wildfowl 6, 181–205 (2020).
Li, C. et al. Population trends and migration routes of the East Asian Bean Goose Anser fabalis middendorffii and A. f. serrirostris. Wildfowl 6, 124–156 (2020).
Niqueux, É. et al. Quantitative transmission characteristics of different H5 low pathogenic avian influenza viruses in Muscovy ducks. Vet. Microbiol. 168, 78–87 (2014).
Kirkeby, C. & Ward, M. P. A review of estimated transmission parameters for the spread of avian influenza viruses. Transbound. Emerg. Dis. 69, 3238–3246 (2022).
Acknowledgements
Work by LJK was partly funded by Erasmus+ Traineeship grant 2023. Participation of CTK, AEB, CKH, and LJK was funded by the ENIGMA project, a veterinary contingency project funded by the Danish Veterinary and Food Administration.
Author information
Authors and Affiliations
Contributions
LJK analysed the data and drafted the manuscript. CTK, AEB, and MPW contributed to analysis and drafting the manuscript. CKH and ADF contributed to drafting the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kjær, L.J., Kirkeby, C.T., Boklund, A.E. et al. Prediction models show differences in highly pathogenic avian influenza outbreaks in Japan and South Korea compared to Europe. Sci Rep 15, 6783 (2025). https://doi.org/10.1038/s41598-025-91384-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-91384-3