Abstract
Growing evidence suggests that dysregulated microRNAs were critical in the development of tumors and the progression number of malignancies. This research aimed to check the effect of microRNA 320a-3p transfection on gastric cancer (GC) cell lines. Following transfection, the efficacy was determined by the RT-PCR method. After that, MTT, scratch assay, DAPI staining, RT-PCR, and flow cytometry were used respectively. The results demonstrated that the viability of GC cells considerably decreased following transfection. Moreover, microRNA 320a-3p transfection significantly suppressed cell migration and induced apoptosis in these cells. We found that transfection of microRNA 320a-3p remarkably decreased PD-L1 gene expression and influenced epithelial-mesenchymal transition (EMT)-related and apoptotic gene expressions. The findings propose that microRNA 320a-3p could decrease cell proliferation and migration and induce apoptosis by increasing TP53 and CASP3 expression levels in GC cells. Notably, microRNA 320a-3p might be a potential target in GC immunotherapy by suppressing the PD-L1 gene expression.
Similar content being viewed by others
Introduction
Gastric cancer (GC), is considered one of the most malignant human cancers, standing of the top five malignant tumors worldwide in terms of frequency and mortality1,2. For patients in the early stage of GC, timely radical surgery following diagnosis can serve as an effective method to control and, even cure the disease under certain conditions. Chemotherapy is also the standard therapy for advanced GC. The fluorouracil (5-Fu)/capecitabine, taxanes (paclitaxel or docetaxel), and platinum or a combinations of these drugs are commonly used as chemotherapeutic drugs. However, the clinical benefits of these treatments are minimal, and the prognosis for GC patients has not significantly improved due to the side effect and toxicity of chemotherapeutic agents and also the drug resistance development3,4.
Recently, microRNAs (miRNAs) have received much attention due to their significant regulatory roles in developing human cancers, including GC5. MicroRNAs are small, endogenous molecules that regulate crucial cellular processes such as proliferation, differentiation, invasion, and apoptosis6,7. Accumulated experimental evidence shows the deregulation of these non-coding RNAs in cancer tissues compared with normal tissues. MiRNAs with high expression in cancer cells act as oncogenes microRNAs (oncomiRs) by suppressing tumor suppressor genes. The tumor suppressor miRNAs (TS miRNAs), which could down-regulate oncogenes, on the contrary, are often under-expressed in tumor cells8,9. In this regard, microRNA restoration could be a promising approach in cancer treatment by adjusting the downregulated miRNAs in tumor tissues and decreasing tumor progression. MicroRNA 320a-3p, located at 8p21.3, is a significant member of the microRNA 320a family, and has been shown to be correlated with inhibiting epithelial-mesenchymal transition (EMT), cell proliferation, and apoptosis10. Moreover, low expression of miR-320-3p has been reported in colorectal, prostate, breast, and ovarian cancers, as well as GC10,11,12. Various studies exhibited that the downregulation of microRNA 320a-3p contributed to different pathological activities of tumor cells, suggesting its role as a TS miRNA. However, microRNA 320a-5p, another member of this family, play a paradoxical role in contrast to microRNA 320a-3p and upregulated in gastric, colorectal and pancreatic adenocarcinomas10,13. More recently, it was revealed that microRNA 320a could bind to the specific region in the 3′-UTR of programmed cell death ligand 1 (PD-L1) mRNA14. PD-L1, also known as CD274, is a transmembrane protein and the most crucial factor in regulating T cell functions and preventing immune response by binding to its receptor, programmed death 1 (PD-1). It has been discovered that PD-L1 is not only expressed in antigen-presenting cells but is also commonly expressed on the surface of tumor cells, thereby resulting in the escape of tumor cells from the immune system14,15,16. Moreover, upregulation of PD-L1 has been found in various human malignancies, including GC, and is associated with tumor progression17,18. In this investigation, we intended to explore the therapeutic effects and mechanism of microRNA 320a in GC. We first verified the low expression level of microRNA 320a-3p in GC cells and then transfected GC cells with microRNA 320a-3p mimics to find its effects on cell proliferation, migration, apoptosis, stemness, and cell cycle. We also utilized bioinformatics analyses to confirm PD-L1 as a potential target of microRNA 320a-3p. Numerous human cancers have been also linked to the dysregulation and mutation of genes producing cyclin-dependent kinases (CDKs)19. Cyclin dependent kinase 16 (CDK16) can boost cancer cell growth via phosphorylating the tumor-suppressor genes p27 and p53, thereby decreasing their expression through the ubiquitin-proteasome degradation pathway20,21,22. Several studies have detected that the expression of miR-320 family members was negatively associated with the EMT-related phenotype of cancer cells10. Hong et al. imply that hsa-miR-320a-3p plays a role in regulating EMT, by suppressing the level of E-cadherin and upregulating the expression of vimentin via directly targeting forkhead box M1 (FOXM1)23. Moreover, based on the Zhang et al.’s study, miR-320 induces apoptosis by activation of caspase-3 however, it inhibits cell proliferation, migration, and tumorigenesis in cervical cancer cells24. Besides, it was indicated that the inhibitory function of p53 on tumor escape involved the transactivation of hsa-miR-320a-3p. The PDL1 repression by hsa-miR-320a-3p was linked to p53-regulated tumor escape14. In this regard, we evaluated the expression levels of CDK16, as well as, E-cadherin (CDH1) and vimentin (VIM) for anti-migratory effect, and also, P53 (TP53), and Caspase-3 (CASP3) for anti-apoptotic activity in GC-transfected cells to understand better the molecular mechanism of microRNA 320a-3p in gastric carcinogenesis.
Results
Bioinformatic analysis of MicroRNA 320a-3p
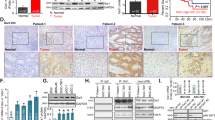
We used the mir-TV database to evaluate the expression level of microRNA 320a-3p in GC patients. The results showed that microRNA 320a-3p is significantly overexpressed in primary GC (n = 436) compared to normal tissues (n = 41) (Fig. 1A, P < 0.0001, logFC:0.45). We also investigated the prognostic characteristics of microRNA 320a-3p based on expression using the UALCAN database. We observed that microRNA 320a expression increased in GC stages 1–4 and grades 1–3 compared to normal samples. The highest expression was observed in stage 2 and grade 2 (Fig. 1B–D, P < 0.0001). In addition, this miRNA significantly increased in GC N0-N3 status (Fig. 1E). Survival analysis of microRNA 320a using the ENCORI database indicated no significant difference in overall survival (OS) between individuals with high expression levels and those with low or medium expression levels for microRNA (Fig. 1F). Target prediction of microRNA 320a-3p by the mirDB database revealed that PD-L1 has the highest score among the selected genes.
Differential expression and survival analysis of hsa-miR-320a-3p in STAD. Expression boxplot of hsa-miR-320a-3p comparing tumor and normal tissue by Mir-TV (A). Differentially expressed miR-320a-3p in STAD based on individual cancer stages (B), tumor histology (C), tumor grade (D), and nodal metastasis status (E) using UALCAL. Survival analysis of hsa-miR-320a in gastric patients (F) by ESOMIR (*P value < 0.05, **P value < 0.01, ***P value < 0.001, and ****P value < 0.0001).
Bioinformatic analysis of genes
We utilized the UALCAN database to investigate the expression patterns of target genes. Our analysis revealed that CDK16, TP53, PD-L1, CDH1, and CASP3 are overexpressed significantly in tumor tissues when compared to the control group (Fig. 2A–C, E,F). Conversely, VIM displays no significant differences between the groups (Fig. 2D). Furthermore, we observed that the upregulation of five out of the six genes, excluding VIM, is linked to higher grades of GC (Fig. 2A–D). Survival analysis using KM-plot showed that high expression levels of CDK16, CDH1, TP53, and VIM are linked to a worse prognosis (Fig. 2B–E). In contrast, the increased levels of CD274 and CASP3 correlate with a better prognosis for GC patients (Fig. 2A–F < 0.05).
Target gene expression patterns in STAD. Differentially expressed genes in STAD based on sample type, individual cancer stages, and patient survival using the UALCAN and KM-plot databases. CD274 (Row A), CDK16 (Row B), CDH1 (Row C), VIM (Row D), TP53 (Row E), CASP3 (Row F) (*P-value < 0.05, **P-value < 0.01, ***P-value < 0.001, and ****P-value < 0.0001).
Protein-protein interaction
We utilized the STRING database to obtain the Protein-Protein Interaction (PPI) network for six chosen genes. STRING analysis showed that the “microRNAs in cancer” pathways have the highest score in relation to these genes based on KEGG pathways. Cytoscape software was used to analyze this network based on the degree of connectivity. The PPI network of target genes consists of five nodes connected by 17 links. Cytoscape network analysis indicated that CASP3, CDH1, and TP53 were the top genes, each exhibiting four connections with other genes. Following this, CD274 and VIM displayed three connections, whereas CDK16 showed no interactions with other genes (Fig. 3).
Microarray analysis by R for genes
The GEO database was utilized to obtain the GSE6308 dataset, which consists of 45 pairs of GC and normal tissue samples. We employ the P-heatmap package from R software to analyze the expression patterns of these genes in GC versus the control group. In the heatmap, the vertical axis represents 90 samples from both normal and cancer tissues, while the horizontal axis lists the target genes. Higher red color saturation indicates gene overexpression, whereas blue signifies downregulation (Fig. 4).
MicroRNA 320a-3p mimic was upregulated in GC cell lines
To determine the microRNA 320a-3p expression level, microRNA 320a-3p mimic and negative control were transfected into AGS, MKN-45, and KATO III cells. RT-PCR confirmed the ectopic expression of microRNA 320a-3p. The results elucidated that the microRNA 320a-3p mimic was upregulated in GC cell lines (AGS, MKN-45, and KATO III) after transfection with microRNA 320a-3p, compared to control groups (NC, ELC, and control) (Fig. 5A–C).
mir320a-3p expression in GC cell lines. Relative expression of mir320a-3p in GC cells as a control group compared to electroporation group (ELC), Negative control (NC) which is transfected by non-target mimic, 20, 40, 60,80 pmol mir320a-3p mimic group in AGC (A), MKN45 (B), and KATO III (C) cell lines. (D) The effect of mir320a-3p restoration on cell viability of GC cell lines (AGS, MKN-45, and KATO III) after 24 and 48 h of restoration. (E) The IC50 concentrations in study groups. Statistical significance was assessed using an unpaired t-test (*P value < 0.05, **P value < 0.01, ***P value < 0.001, and ****P value < 0.0001).
MicroRNA 320a-3p overexpression inhibits GC cell proliferation
We set out a MTT assay to evaluate the effect of microRNA 320a-3p on GC cell line proliferation. This showed that transfection with 80 pmol of microRNA 320a-3p after 48 h incubation has a significant antiproliferative effect in a dose and time-dependent manner (Fig. 5D). The IC50 concentrations were obtained at 67.09 pmol and 39.15 pmol for AGS cells, 73.11 pmol and 53.03 pmol for MKN-45, and 58.37 pmol 33.78 pmol for KATO III cells for 24 and 48 h, respectively (Fig. 5E). These results propose that overexpression of microRNA 320a-3p is associated with anti-proliferative activity on GC cell lines.
MicroRNA 320a-3p promotes GC cells apoptosis
To investigate whether microRNA 320a-3p can induce apoptosis in GC cells, Annexin V/PI was used. The result of the apoptosis assay indicated that microRNA 320a-3p transfection induces apoptosis in all GC cell lines (Fig. 6A). We also applied DAPI staining to assess microRNA 320a-3p induced apoptosis in GC cells. DAPI staining results exhibited remarkably elevated induction of GC cell apoptosis in the transfected group compared to NC (Fig. 6B).
(A) The effect of mir320a-3p restoration on apoptosis of GC cell lines (AGS, MKN-45, and KATO III) after restoration. The cells were transfected with IC-50 concentration to characterize apoptosis rates by the Annexin-V/PI method. According to the graphs, the Q1 area displayed cells with necrosis, Q2, Q3, and Q4 areas represented the late, early apoptotic, and viable cells, respectively. As the figure shows, the percentage of apoptosis increased in transfected groups compared to control cells. (B) DAPI staining determined the induction of apoptosis in AGS, MKN-45, and KATO III cell lines, respectively, compared to control and negative control groups. Apoptotic cells (Arrows indicate apoptotic cells)were identified based on nuclear condensation, fragmentation, and morphological features such as cell shrinkage and membrane blebbing. Statistical significance was assessed using an unpaired t-test (**P value < 0.01 and ***P value < 0.001).
MicroRNA 320a-3p expression is inversely associated with PD-L1 levels in GC
To explore the correlation of microRNA 320a-3p with PD-L1 expression levels in GC cell lines, we performed the RT-PCR to evaluate the PD-L1 expression in transfected cells. Compared to the control, the expression level of PD-L1 significantly decreased in AGS (P < 0.0001), MKN-45 (P < 0.0001), and KATO III (P < 0.001) cell lines (Fig. 7A).
Expression in GC cell lines. Relative CD274 (A), CDK16 (B), CDH1 (C), VIM (D), TP53 (E), CASP3 (F) genes expression in GC cells as a control group compared to electroporation group (ELC), Negative control (NC), and mir320a-3p mimic transfected group in AGS, MKN-45, and KATO III cell lines. (G) Wound healing assay. The scratch assays showed up-regulation of mir320a-3p decreased migration in AGS, MKN-45, and KATO III cells compared to control and negative control cells after 24 and 48 h of transfection. Statistical significance was assessed using an unpaired t-test (*P value < 0.05, **P value < 0.01, ***P value < 0.001, and ****P value < 0.0001).
MicroRNA 320a-3p suppressed the CDK16 expression in GC
We also assayed the impact of microRNA 320a-3p on the CDK16 expression level in AGS, MKN-45, and KATO III cell lines. The finding indicated that microRNA 320a-3p regulated CDK16 expression negatively, and the level of CDK16 expression was decreased in GC cells, particularly in MKN-45 cells (P < 0.0001), compared to other cell lines (Fig. 7B).
MicroRNA 320a-3p inhibits EMT-related gene expression in GC cells
To further determine the association between microRNA 320a-3p and EMT-related genes, we investigated the effect of microRNA 320a-3p mimic on the vimentin and E-cadherin expression. According to the result, the levels of vimentin and E-cadherin were significant compared to control groups (NC, ELC, and Control) (AGS (P < 0.01), MKN-45 (P < 0.001), and KATO III (P < 0.01) Fig. 7C and AGS (P < 0.001), MKN-45 (P < 0.001), and KATO III (P < 0.001) Fig. 7D).
MicroRNA 320a-3p increases the expression level of apoptotic genes in GC cells
To evaluate apoptotic gene expression, we measured the CASP3 and TP53 levels. In all cell lines, TP53 (AGS (P < 0.01), MKN-45 (P < 0.001), and KATO III (P < 0.01) Fig. 7E) and CASP3 (AGS (P < 0.001), MKN-45 (P < 0.01), and KATO III (P < 0.0001) Fig. 7F) levels increased significantly.
Overexpression of MicroRNA 320a-3p reduces GC cell migration
A wound-healing assay was accomplished to investigate the effect of microRNA 320a-3p on cell migration. As indicated in (Fig. 7G), microRNA 320a-3p upregulation suppressed cell migration in microRNA 320a-3p transfected AGS cells compared with the NC group in a time-dependent manner. There were no significant differences between the cell migration of NC and the control group. The results showed that microRNA 320a-3p has a considerable inhibitory effect on GC cell invasion and migration.
Discussion
PD-L1, a co-inhibitory immune checkpoint, is crucial in creating an immunosuppressive microenvironment, promoting the invasion and progression of cancer25. Numerous malignant tumors exhibit PD-L1 expression related to a poor prognosis, such as GC18. According to studies, miRNAs play essential roles in cancer progression26, and tumor-suppressing miRNAs regulate immune checkpoint inhibitors to control anti-tumor immunity27. The benefits of miRNAs as a therapeutic option for cancer treatment are the following: First, regulating target gene expression by miRNAs instead of eradicating it prevents gene expression repression completely. Next, because a single miRNA can affect numerous genes, disrupting miRNA expression impacts a wide range of genes. Therefore, the target gene’s expression returns to normal when miRNA expression is altered by a treatment approach25. Several studies have reported that microRNA 320a-3p, essential for suppressing EMT, tumor growth, and metastasis, is downregulated throughout the development of tumors10. In this regard, studies showed that the expression levels of microRNA 320a-3p were relatively decreased in GC patient serum28 and cancer tissues29. Hence, this study explores the association between microRNA 320a-3p, its altered expression pattern, and its functional role in controlling GC by targeting PD-L1 expression. The first step in this research was to ascertain the expression level of microRNA 320a-3p after transfection in GC cells. It should be noted that the expression level of microRNA 320a-3p was drastically enhanced in AGS, MKN-45, and KATO III cell lines. In the present study, the cell viability results exhibit that the microRNA 320a-3p mimic reduces cell proliferation in AGS, MKN-45, and KATO III cell lines. Notably, the inhibition of growth in AGS and KATO III cell lines was higher than MKN-45. Also, our results showed that restoration of microRNA 320a-3p at a higher concentration and 48 h after transfection enhances the stimulation of cell death in each of the cell lines. Therefore, it can be concluded that microRNA 320a-3p induces cell death in AGS, MKN-45, and KATO III cell lines. Several studies have revealed the anti-proliferative microRNA 320a-3p effect on cancer cells. Lu et al. demonstrated that restoration of microRNA 320a-3p inhibits cell proliferation in multiple myeloma via targeting pre-B-cell leukemia transcription factor 3 (PBX3)30.
Further, another study indicated that microRNA 320a-3p exerts an anti-proliferation and anti-invasion effect in SGC-7901 human GC cells by reducing cyclin D1 and MMP9 expression at both mRNA and protein levels29. Moreover, another study showed that in comparison to their respective control cells, microRNA 320a-3p mimics considerably reduced the proliferation rate of SGC-7901 cells. In contrast, microRNA 320a-3p inhibitors dramatically increased the proliferation of BGC-823 human GC cells31. The results of the apoptosis assay display that restoration of microRNA 320a-3p can induce the apoptosis of AGS, MKN-45, and KATO III cell lines. A study report indicated that microRNA 320a-3p mimics transfection-repressed glioma cell proliferation and induced apoptosis32. In this regard, we also checked the TP53 and CASP3 expression levels. TP53 mutations are prevalent in GC and are considered an early event in the process of tumorigenesis33. In this context, these mutations were also detected in widely used GC cell lines nowadays including KATOIII (deleted for TP53), NUGC-3 (p53 Y220C), MKN28 (p53 I251L), and MKN1 (p53 V143A)33,34. These findings further imply that the inactivation of the p53 gene may play a vital role in the transformation of gastric cells into a malignant phenotype. KATO-III cells could serve as a good model for studying the effect of p53 gene loss in cellular transformation34. As expected, the levels of TP53 and CASP3 increased significantly in all cell lines, indicating that the ectopic expression of microRNA 320a-3p promoted apoptosis in GC cells. Similarly, the study conducted by Pan et al. explains that microRNA 320a-3p induced glioma cells apoptosis (U87 and U251 cells) via CASP3 enzyme-dependent32.
Further, Bozgeyik et al. proposed that microRNA 320a-3p induces TP53-dependent apoptosis of prostate cancer cells through negatively regulating TP73-AS1 long non-coding RNA35. However, the significant results observed in NC and ELC groups may be due to apoptosis caused by electroporation in these groups. Of note, the apoptosis observed in the microRNA 320a-3p groups is highly significant compared to the NC and ELC groups. According to the literature, electroporation is utilized for gene transfer and improved medication delivery but can cause cell death. Under specific circumstances, electric pulses inducing electroporation can trigger apoptosis, necrosis, necroptosis, and pyroptosis36. Hence, it could be explained that the induction of apoptosis in these cells is caused by electroporation.
One of the remarkable results of this study was that the restoration of microRNA 320a-3p suppresses PD-L1 expression. Moreover, the expression level of PD-L1 in the AGS cell line was reduced significantly, as well as in MKN-45 and KATO III cell lines. In this frame, Costa et al. discovered that microRNA 320a-3p binds the 3′-UTR of PD-L1 and, consistently, microRNA 320a modulation influences PD-L1 levels in malignant pleural mesothelioma (MPM) cells. Interestingly, their findings imply that PD-L1 expression may be brought on by an inadequate TP53-regulated miRNA response, which may aid MPM immune evasion or carcinogenesis via tumor-intrinsic functions37. In line with this, bioinformatics results showed that PD-L1 expression level is elevated in GC tissue, while restoration of microRNA 320a reduces PD-L1 expression. Therefore, it could be elucidated that restoration of microRNA 320a-3p may be associated with lowering PD-L1 expression. This data suggested that microRNA 320a-3p mimic may exert anti-tumor activity by inhibiting PD-L1 expression.
CDK16 has been related to the development of various malignancies, and tissues taken from prostate and breast tumors have been found to express it substantially more than normal38. In the current study, gene expression analysis indicated that the CDK16 expression level decreased significantly in GC cell lines following microRNA 320a-3p transfection. Also, the CDK16 level exhibited more reduction in the MKN-45 cell line than in AGS and KATO III cell lines. A study conducted by Jia et al. indicated that miR-485-5p decreased protein production from the longer CDK16 transcript by targeting the 3′UTR between the distal and proximal polyA (pA) site and caused senescence-associated phenotypes in lung cancer cells19. Interestingly, based on the bioinformatic analysis, the level of CDK16 in tumor tissue is high, whereas we observed that the expression of CDK16 decreased after microRNA 320a-3p transfection. In this context, it can be said that microRNA 320a-3p mimic reduces the expression of CDK16. Besides, it is revealed that CDK16 knockdown causes a reduction in the expression of PD-L1 molecules, which in turn increases the anti-tumor activity of senescent cancer cells19. Last but not least, functional research should be done on the target genes discovered in this work by bioinformatics analysis that may be crucial in GC.
Growing evidence elucidates that microRNA 320a-3p is linked to the EMT process since members of the microRNA 320a-3p family were found to be negatively regulated in tumor cell invasion and migration10. Moreover, the anti-migratory effect of microRNA 320a-3p was demonstrated previously in GC by targeting Forkhead box protein M1 (FOXM1). FOXM1 promotes disease development in GC by activating urokinase plasminogen activator (uPA) and facilitating cell migration through activating cathepsin39. In this context, we checked the Vimentin and E-cadherin expression levels as essential proteins in migration after transfection in GC cells. To study possible mechanisms whereby microRNA 320a-3p impacts GC tumorigenesis, we checked the predicted microRNA 320a-3p targets and ascertained E-cadherin is one of these targets. Therefore, the E-cadherin reduction might be correlated with this reason. However, more research is required on this topic.
This study provides valuable insights into the role of microRNA 320a-3p in GC; however, our study have some limitations. The study focused on a limited number of GC cell lines, possibly limiting the generalizability of the results. The lack of in vivo studies and clinical validation further highlights the need for future research to confirm the therapeutic potential of microRNA 320a-3p. Furthermore, while bioinformatic analyses were utilized to identify target genes and pathways, comprehensive experimental validation of all predicted targets remains necessary. Further analysis, such as investigating caspase 3 cleavage by western blot or using a specific caspase assay, would provide additional confirmation of apoptosis induction and strengthen the mechanistic understanding of microRNA 320a-3p’s role in gastric cancer cells. However, more research is required on this topic.
Conclusion
MicroRNA replacement therapy has been demonstrated to be a beneficial strategy in cancer research projects. Our study revealed that microRNA 320a-3p could restrain cell viability, proliferation, and migration and induce apoptosis of GC cells by decreasing the expression levels of CD274, CDK16, and VIM and increasing CASP3 and TP53 expression levels. These outcomes expressed that microRNA 320a-3p represented a tumor suppressor role in GC partly by regulating the genes mentioned above and their related signaling pathways. However, this study was limited to in vitro experiments, and further studies are needed to conduct in vivo and clinical experiments. These results may provide a reasonable strategy for using microRNA 320a-3p in GC treatment. Based on the obtained data, more investigations are necessary to identify the exact role of microRNA 320a-3p in GC.
Materials and methods
Bioinformatic analysis
Gene analysis by R software
The expression microarray dataset GSE63089 was downloaded from GEO (Gene Expression Omnibus). The GSE63089 dataset has 90 samples comprising 45 pairs of GC vs. normal tissues. These expression microarray data were analyzed by R software, and the heatmap of these 90 samples for genes CASP3, PD-L1(CD274), CDH1, CDK16, VIM, and TP53 was designed by the P-heatmap package of R.
MiRNA and gene expression analysis
We used the Mir-TV database to investigate the expression pattern of microRNA 320a-3p in GC compared to normal tissues. Furthermore, the UALCAN database was utilized to evaluate the prognostic characteristics of microRNA 320a in GC from various angles, such as cancer stages (stages 1–4), nodal metastasis status (N 0–4), and tumor grades (grade 1–3). ENCORI is also utilized to conduct survival analysis of miR320a in gastric patients. We also used the mirDB database to target the prediction of selected genes. The UALCAN database is also used to perform prognostic bioinformatic analysis on target genes CASP3, PDL1(CD274), CDH1, CDK16, VIM, and TP53, analyzing their expression levels and the correlations between gene expression and cancer stages. Additionally, we employed km-plot analysis to examine how gene expression patterns affect overall survival times (OS) in gastric patients.
PPI network
Predicted protein-protein interactions (PPI) were performed using STRING. The interactions comprise both direct (physical) and indirect (functional) associations; they are the result of computational prediction, knowledge transfer between organisms, and interactions aggregation of other (primary) databases. The selected genes were uploaded to the STRING database to generate the PPI network and then imported to Cytoscape software to analyze the network. The PPI pairs were extracted with a combined score of > 0.4.
Cell culture
The human GC cells (AGS, MKN-45, and KATO ш) were procured from the Iranian Institute Pasteur (Tehran, Iran). The Cells were cultivated in RPMI-1640 medium, which supplements with 1% penicillin-streptomycin antibiotic mixes plus 10% fetal bovine serum (FBS) (Gibco, NY). The cells were kept at 37 °C in a sterilized incubator with 5% CO2 and 95% humidity and periodically cultured when they reached almost 70% confluency.
MiRNA transfection
MicroRNA 320a-3p mimics with (5′-AAAAGCUGGGUUGAGAGGGCGA-3′) nucleotide sequence, obtained from http://www.mirbase.org, and miR negative control mimics were purchased from GenePharma Co, Shanghai. The electroporation technique was used for cell transfection following the manufacturer’s guidelines. For determining optimal time and dose, the GC cells were transfected with 20, 40, 60, and 80 picomoles (pmol) of microRNA 320a-3p mimics using Gene Pulser X cell Electroporation System (Bio-Rad) and then cultured for 24, 48, and 72 h. The efficacy of microRNA 320a-3p transfection was assessed by qRT-PCR.
MTT assay
MicroRNA 320a-3p mimics and miR negative control mimics were transfected into GC cells, and then the 1 × 104 cells were seeded into 96-well plates and incubated at 37 °C. At 24, 48, and 72 h post-seeding, cell viability was examined using MTT (3-(4, 5-dimethylthiazol-2-yl)- 2, 5-diphenyltetrazolium bromide) method as previously detailed40.
DAPI staining
The DAPI staining was performed to detect apoptotic morphology and determine apoptotic cells. DAPI staining was performed as previously described41.
Apoptosis assay
The transfected cells, with microRNA 320a-3p mimics and miR negative control mimics, were seeded into 6-well plates at a density of 4 × 105 cells/well for evaluating the effect of microRNA 320a-3p on cell apoptosis. After 24 h of incubation at 37 °C, the cultured cells were dissociated by trypsin (Sigma-Aldrich), washed by Phosphate buffered saline (PBS), and collected. Afterward, the cells were stained with annexin V- fluorescein isothiocyanate /propidium iodide (AnnexinV-FITC/PI) kit at 4 °C and darkness in line with the manufacturer’s instructions. The cellular apoptotic rate was determined using flow cytometry (Sysmex-Cyflow), and the results were analyzed by the FlowJo 10.0 software.
Scratch assay
The wound-healing assay (scratch assay) assessed cell migration capability. Briefly, 5 × 105 transfected cells were plated to reach 70–85% confluence in 24-well plates. A sterile pipette tip created an artificial wound gap in the cell monolayer. Afterward, the cells were washed twice with PBS to remove detached cells, and RPMI-1640 medium containing 1% FBS (low serum) was added to the wells to minimize the effects of proliferation on wound closure. The width of the migration area was evaluated at 0, 24, and 48 h under an inverted microscope (Optika, XDS-3, Italy) in the microRNA 320a-3p-transfected group compared to the NC group.
Real-time PCR (qRT-PCR)
The total cellular RNA was isolated from GC cells utilizing Trizol reagent (RiboEX-GeneAll) accordant with the manufacturer’s procedure. Reverse transcription of microRNA 320a-3p and target gene RNAs into complementary DNA (cDNA) was performed using BONmiR (BON-miR, Tehran, Iran) and Yekta Tajhiz (Yekta Tajhiz Azma, Tehran, Iran) cDNA synthesis kits, respectively. In addition, the qRT-PCR assay was carried out to assess the expression levels of microRNA 320a-3p, CD274, CDK16, CDH1, VIM, TP53, and CASP3 using SYBR Green Master Mix (Ampliqon, Denmark). The primer sequences are indicated in Table 1. In addition, SNORD-48 and GAPDH were utilized as internal controls for normalizing the expression ratio of microRNA 320a-3p and genes, respectively. The findings were analyzed by the 2 (−ΔΔCt) method.
Statistical analysis
Data are presented as mean ± standard deviation (SD). All statistical analyses were done by GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA, USA). Student’s t-test and ANOVA were utilized to determine the statistical differences between groups. A P-value less than 0.05 was determined statistically significant.
Data availability
The data that support the findings of this study are available within the paper and its Supplementary Information. The other ones that not openly available due to reasons of sensitivity are available from the corresponding author upon reasonable request.
Abbreviations
- GC:
-
Gastric cancer
- EMT:
-
Epithelial-mesenchymal transition
- PD-L1:
-
Programmed cell death-ligand 1
- miRNAs:
-
MicroRNAs
- OncomiRs:
-
Oncogenes microRNAs
- TS miRNAs:
-
Tumor suppressor miRNAs
- PD-1:
-
Programmed death 1
- CDKs:
-
Cyclin-dependent kinases
- CDK16:
-
Cyclin dependent kinase 16
- E-cadherin; CDH1:
-
Epithelial cadherin
- PPI:
-
Protein-protein interactions
- FBS:
-
Fetal bovine serum
- PBS:
-
Phosphate buffered saline
- Annexin V-FITC/PI:
-
Annexin V-fluorescein isothiocyanate/propidium iodide
- cDNA:
-
Complementary DNA
- SD:
-
Standard deviation
- OS:
-
Overall survival
- PBX3:
-
Pre-B-cell leukemia transcription factor 3
- MPM:
-
Malignant pleural mesothelioma
- FOXM1:
-
Forkhead box protein M1
- UPA:
-
Urokinase plasminogen activator
- pA:
-
PolyA
- 5-Fu:
-
Fluorouracil
- STAD:
-
Stomach adenocarcinoma
References
Chandra, R. et al. The changing face of gastric cancer: epidemiologic trends and advances in novel therapies. Cancer Gene Ther. 28, 390–399 (2021).
López, M. J. et al. Characteristics of gastric cancer around the world. Crit. Rev. Oncol. /Hematol, 103841 (2022).
Li, K., Zhang, A., Li, X., Zhang, H. & Zhao L. Advances in clinical immunotherapy for gastric cancer. Biochim. Biophys. Acta (BBA)-Rev Cancer. 1876, 188615 (2021).
Panahizadeh, R. et al. A literature review of recent advances in gastric cancer treatment: exploring the cross-talk between targeted therapies. Cancer Cell. Int. 25 https://doi.org/10.1186/s12935-025-03655-8 (2025).
Kipkeeva, F. et al. MicroRNA in gastric cancer development: mechanisms and biomarkers. Diagnostics 10, 891 (2020).
Kabekkodu, S. P. et al. Clustered MiRNAs and their role in biological functions and diseases. Biol. Rev. 93, 1955–1986 (2018).
Mirzaei, S. et al. MicroRNA-146 family: molecular insights into their role in regulation of signaling pathways in glioma progression. Pathol. Res. Pract., 155707 (2024).
Kim, T. & Croce, C. M. MicroRNA: trends in clinical trials of cancer diagnosis and therapy strategies. Exp. Mol. Med. 55, 1314–1321 (2023).
Asghariazar, V., Kadkhodayi, M., Sarailoo, M., Jolfayi, A. G. & Baradaran, B. MicroRNA-143 as a potential tumor suppressor in cancer: an insight into molecular targets and signaling pathways. Pathol. Res. Pract. 250, 154792. https://doi.org/10.1016/j.prp.2023.154792 (2023).
Liang, Y., Li, S. & Tang, L. MicroRNA 320, an anti-oncogene target MiRNA for cancer therapy. Biomedicines 9, 591 (2021).
Carvalho, T. M. et al. MicroRNAs miR-142-5p, miR-150-5p, miR-320a-3p, and miR-4433b-5p in serum and tissue: potential biomarkers in sporadic breast cancer. Front. Genet. 13, 865472 (2022).
Lieb, V. et al. Serum levels of miR-320 family members are associated with clinical parameters and diagnosis in prostate cancer patients. Oncotarget 9, 10402 (2018).
Khandelwal, A. et al. Circulating miR-320a acts as a tumor suppressor and prognostic factor in non-small cell lung cancer. Front. Oncol. 11, 645475 (2021).
Costa, C. et al. P53-regulated miR-320a targets PDL1 and is downregulated in malignant mesothelioma. Cell. Death Dis. 11, 748 (2020).
Yohei, M. et al. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut 66, 1463. https://doi.org/10.1136/gutjnl-2016-311421 (2017).
Chen, S. et al. Mechanisms regulating PD-L1 expression on tumor and immune cells. J. Immunother. Cancer 7, 1–12 (2019).
Liu, X. et al. High PD-L1 expression in gastric cancer (GC) patients and correlation with molecular features. Pathol. Res. Pract. 216, 152881 (2020).
Gu, L. et al. PD-L1 and gastric cancer prognosis: A systematic review and meta-analysis. PLoS One 12, e0182692 (2017).
Jia, Q. et al. Lung cancer cells expressing a shortened CDK16 3′ UTR escape senescence through impaired miR-485‐5p targeting. Mol. Oncol. 16, 1347–1364 (2022).
Xie, J. et al. CDK16 phosphorylates and degrades p53 to promote radioresistance and predicts prognosis in lung cancer. Theranostics 8, 650 (2018).
Wang, H. et al. CDK16 overexpressed in non-small cell lung cancer and regulates cancer cell growth and apoptosis via a p27-dependent mechanism. Biomed. Pharmacother. 103, 399–405 (2018).
Yanagi, T., Krajewska, M., Matsuzawa, S. & Reed, J. C. PCTAIRE1 phosphorylates p27 and regulates mitosis in cancer cells. Cancer Res. 74, 5795–5807 (2014).
Hong, H. et al. The novel circCLK3/miR-320a/FoxM1 axis promotes cervical cancer progression. Cell Death Dis. 10, 950 (2019).
Zhang, T. et al. Down-regulation of miR-320 associated with cancer progression and cell apoptosis via targeting Mcl-1 in cervical cancer. Tumor Biol. 37, 8931–8940 (2016).
Alemohammad, H. et al. The importance of immune checkpoints in immune monitoring: A future paradigm shift in the treatment of cancer. Biomed. Pharmacother. 146, 112516 (2022).
Zhao, L. et al. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget 7, 45370 (2016).
Skafi, N., Fayyad-Kazan, M. & Badran, B. Immunomodulatory role for MicroRNAs: regulation of PD-1/PD-L1 and CTLA-4 immune checkpoints expression. Gene 754, 144888 (2020).
Hao, N. B., He, Y. F., Li, X. Q., Wang, K. & Wang, R. L. The role of MiRNA and LncRNA in gastric cancer. Oncotarget 8, 81572 (2017).
Zhao, Y., Dong, Q. & Wang, E. MicroRNA-320 inhibits invasion and induces apoptosis by targeting CRKL and inhibiting ERK and AKT signaling in gastric cancer cells. OncoTargets Ther. 10, 1049 (2017).
Lu, Y. et al. miR-320a regulates cell proliferation and apoptosis in multiple myeloma by targeting pre-B-cell leukemia transcription factor 3. Biochem. Biophys. Res. Commun. 473, 1315–1320 (2016).
Ge, X. et al. miR-320a modulates cell growth and chemosensitivity via regulating ADAM10 in gastric cancer. Mol. Med. Rep. 16, 9664–9670 (2017).
Pan, C. et al. MiR-320 inhibits the growth of glioma cells through downregulating PBX3. Biol. Res. 50, 1–9 (2017).
Blanchet, A., Bourgmayer, A., Kurtz, J. E., Mellitzer, G. & Gaiddon, C. Isoforms of the p53 family and gastric cancer: a menage a Trois for an unfinished affair. Cancers 13, 916 (2021).
Matozaki, T. et al. Missense mutations and a deletion of the p53 gene in human gastric cancer. Biochem. Biophys. Res. Commun. 182, 215–223 (1992).
Bozgeyik, E. et al. miR-320a promotes p53-dependent apoptosis of prostate cancer cells by negatively regulating TP73-AS1 in vitro. Biochem. Biophys. Res. Commun. 619, 130–136 (2022).
Napotnik, T. B., Polajžer, T. & Miklavčič, D. Cell death due to electroporation–a review. Bioelectrochemistry 141, 107871 (2021).
Costa, C. et al. P53-regulated miR-320a targets PDL1 and is downregulated in malignant mesothelioma. Cell. Death Dis. 11, 1–15 (2020).
Dixon-Clarke, S. E. et al. Structure and inhibitor specificity of the PCTAIRE-family kinase CDK16. Biochem. J. 474, 699–713 (2017).
Weidle, U. H., Birzele, F., Auslaender, S. & Brinkmann, U. Down-regulated MicroRNAs in gastric carcinoma May be targets for therapeutic intervention and replacement therapy. Anticancer Res. 41, 4185–4202 (2021).
Asghariazar, V. et al. MicroRNA-143 act as a tumor suppressor MicroRNA in human lung cancer cells by inhibiting cell proliferation, invasion, and migration. Mol. Biol. Rep. 49, 7637–7647. https://doi.org/10.1007/s11033-022-07580-1 (2022).
Asghariazar, V., Kadkhodayi, M., Mansoori, B., Mohammadi, A. & Baradaran, B. Restoration of miR-143 reduces migration and proliferation of bladder cancer cells by regulating signaling pathways involved in EMT. Mol. Cell. Probes. 61, 101794. https://doi.org/10.1016/j.mcp.2022.101794 (2022).
Acknowledgements
The authors wish to acknowledge the assistance of Dr. Shahnaz Hosseinzadeh, Dr. Dariush Shanehbandi, and Prof. Mohsen Sagha for their advice and efforts.
Funding
This study was supported by the Ardabil University of Medical Sciences (finance code 1002769).
Author information
Authors and Affiliations
Contributions
“Vahid Asghariazar and Elham Safarzadeh conceived and planned the study; Vahid Asghariazar, Shima Makaremi, Negin Amani, and Erfan Zare carried out the experiment and collected available literature; Vahid Asghariazar, Mahtab Kadkhodayi, Majid Eterafi, Mohammad Ghasem Golmohammadi and Elham Safarzadeh prepared the manuscript, analyzed the statistical data and verified the accuracy of the tests.”
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Ardabil University of Medical Sciences, Ardabil, Iran (IR.ARUMS.REC.1398.569).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Asghariazar, V., Makaremi, S., Amani, N. et al. MicroRNA 320a-3p up-regulation reduces PD-L1 expression in gastric cancer cells: an experimental and bioinformatic study. Sci Rep 15, 8239 (2025). https://doi.org/10.1038/s41598-025-92537-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-92537-0