Abstract
The prevalence of nonalcoholic steatohepatitis (NASH) is rising annually, posing health and economic challenges, with limited treatments available. Diosgenin, a natural steroidal compound found in various plants, holds potential as a therapeutic candidate. Recent studies have confirmed diosgenin’s anti-inflammatory and metabolism-modulating properties. However, its therapeutic effects on NASH and the underlying mechanisms are still unclear. This study aims to explore diosgenin’s protective effects and pharmacological mechanisms against NASH using network pharmacology, molecular docking, and experimental validation. We gathered potential targets of diosgenin and NASH from various databases to generate protein-protein interaction (PPI) networks. GO and KEGG pathway enrichment analyses identified key targets and mechanisms. Molecular docking confirmed the binding capacity between diosgenin and core target proteins. Additionally, a NASH cell model was developed to validate the pharmacological effects of diosgenin. Our investigation identified nine key targets (ALB, AKT1, TP53, VEGFA, MAPK3, EGFR, STAT3, CASP3, IGF1) that interact with diosgenin. Molecular docking indicated potential bindings interactions, while enrichment analyses revealed that diosgenin may enhance fatty acid metabolism via the PI3K-Akt pathway. Cellular experiments confirmed that diosgenin activates this pathway, reduces SCD1 expression, and decreases triglyceride and IL-6 levels. Our study elucidates that diosgenin may ameliorate triglyceride deposition and inflammation through the PI3K-Akt pathway.
Similar content being viewed by others
Introduction
With the global increase in the prevalence of metabolic syndrome, diabetesand obesity, the incidence of nonalcoholic fatty liver disease (NAFLD) has also increased dramatically, and NAFLD has become the most common type of chronic liver disease worldwide. Nonalcoholic steatohepatitis (NASH) is an aggressive inflammatory subtype of NAFLD associated with 5% or more hepatic steatosis, hepatocellular damage (ballooning) and inflammation, with or without fibrosis1,2. 20% of NASH can progress to cirrhosis3 or even liver cancer4, and ultimately become the leading cause of liver transplantation3,5. Hepatic steatosis, characterized by the accumulation of triglyceride vesicles in hepatocytes2, arises from an imbalance between triglyceride supply and demand in the liver. This imbalance is attributed to heightened hepatic de novo lipogenesis, augmented fatty acid uptake, and decreased fatty acid beta-oxidation and triglyceride output6,7. The hepatic inflammatory response promotes liver fibrosis progression and is a key driver of cirrhosis8. Despite the dramatic public health concern that NASH poses, no effective therapeutic drugs have been approved for clinical use9,10. Consequently, there is a pressing need to explore novel and effective therapeutic strategies and drugs to address this growing health concern. In recent years, significant progress has been made in the study of traditional Chinese medicine (TCM) for the treatment of NASH, providing valuable insights for the development of NASH therapeutics. The active component vitexin from Shan Zha ameliorates NASH by significantly reducing hepatic macrophage infiltration and downregulating the expression of molecules associated with triglyceride synthesis11. Alisol A from Ze Xie alleviates steatohepatitis by inhibiting oxidative stress and stimulating autophagy via the AMPK/mTOR pathway12, and Alisol B has been reported to reduce ROS levels and suppress inflammatory cytokine expression13. These findings highlight the potential of TCM in NASH treatment.
Diosgenin is a steroidal compound found naturally in the matrix of plants such as fenugreek and wild yam14 that has anti-inflammatory, immunomodulatory, hypolipidemic, antiviral, antifungal, and anti-allergic effects15,16. In recent years, diosgenin has drawn increasing amounts of attention in the treatment of various metabolic diseases, including diabetes17, osteoporosis18, and hyperlipidemia19. Recently, diosgenin was shown to improve hepatic lipid metabolism20 by interfering with cholesterol absorption and transport21, inhibiting triglyceride synthesis, accelerating the breakdown of free fatty acids22 and affecting liver-gut circulation23. Diosgenin has also been confirmed to exhibit anti-inflammatory effects by inhibiting inflammatory signals from macrophages24 and significantly attenuating the inflammatory response in obese adipose tissue25. However, whether diosgenin has a therapeutic effect on NASH, an inflammatory subtype of NAFLD, remains unclear.
Network pharmacology, an emerging discipline based on systems biology theory, integrates various disciplines, such as polypharmacology, bioinformatics, and network analysis. In recent years, network pharmacology has been used to analyze the molecular associations between drugs and therapeutic objects from the overall perspective of the system level and biological network and to reveal the systemic pharmacological mechanism of drugs, serving as a guide for the development of new medicines and clinical therapy26,27. Therefore, this study aimed to screen the potential targets and signaling pathways of diosgenin in NASH using network pharmacology. Further validation through cytological experiments was conducted to elucidate diosgenin’s effects on NASH and its pharmacological mechanisms.
Materials and methods
Network pharmacology analysis
Prediction of the action targets of diosgenin
We first used “diosgenin” as the search term to obtain the active target in the HERB database28 (http://herb.ac.cn/). The 2D molecular structure formula of diosgenin was downloaded from the PubChem database29 (https://pubchem.ncbi.nlm.nih.gov/) and retrieved from the PharmMapper database30,31 (http://www.lilab-ecust.cn/pharmmapper/). Furthermore, potential targets were predicted from the SwissTargetPrediction database32 (http://www.swisstargetprediction.ch/). All the obtained targets were normalized for information using the UniProt database33 (https://www.uniprot.org/), which was subsequently subjected to gene name analysis. Subsequently, all the targets retrieved from the above databases were merged, and duplicate values were removed to obtain the active targets.
Identification of NASH-related targets
NASH-related targets were comprehensively gathered using the search terms “non-alcoholic steatohepatitis” and “nonalcoholic steatohepatitis”. We searched the GeneCards database34 (https://www.genecards.org), DisGeNET database35 (http://www.disgenet.org), OMIM database36 (http://www.omim.org), and TTD database37 (http://db.idrblab.net) to identify putative targets. Finally, all the above targets were combined, and duplicate values were removed to obtain NASH-related targets.
Analysis of overlapping targets of diosgenin in NASH target mapping
The targets of the above two steps were imported into Venn mapping software38 (http://www.bioinformatics.com.cn/static/others/jvenn/example.html) for visualization and mapping, after which the common targets were obtained. These targets are potential targets of diosgenin for the treatment of NASH.
GO and KEGG pathway enrichment analysis
The overlapping targets of diosgenin and NASH were imported into the DAVID database 6.839 (https://david.ncifcrf.gov/) for enrichment analysis of three aspects of GO enrichment: biological process (BP), molecular function (MF), and cellular component (CC) enrichment, as well as KEGG pathway enrichment40. The results were screened according to the criteria of a false discovery rate (FDR) < 0.05 and P < 0.05, and the top 10 pathways with the most highly enriched targets were selected for visualization via GO enrichment analysis and KEGG signaling pathway analysis via the “Microbiology Letter” tool38 (http://www.bioinformatics.com.cn/).
PPI network construction
We submitted the overlapping targets to the STRING database41 (https://string-db.org/) to predict functional protein association networks. For the protein–protein interaction (PPI) network constructed using the STRING database, the minimum interaction threshold was set to 0.4, with other parameters at default values. This generated a highly interconnected PPI network and derived combined PPI score data42. The composite score reflects protein interaction strength: higher scores indicate stronger interactions. The obtained network data were further imported into Cytoscape 3.7.243 (http://cytoscape.org/) software for the analysis of topological properties. The degree value indicates the number of direct interactions for a target, with higher values suggesting greater biological relevance and functional importance. Therefore, we used degree value analysis and ranking to screen the key targets of diosgenin in NASH.
Molecular docking validation of the binding capacity between diosgenin and targets
Protein structures were acquired from the RCSB Protein Data Bank (PDB) (Table 7). Use Pymol 2.3.0 to remove protein crystallization water and original ligands. Subsequently, import the protein structures into Autodock Tools (version 1.5.6) to add hydrogens, calculate charges, assign charges, specify atom types, and save the processed structures in the pdbqt format. The three-dimensional structure of diosgenin was retrieved from the PubChem database. This structure was subsequently imported into ChemBio3D Ultra 14.0 for energy minimization and saved in the mol2 format. The optimized diosgenin was then imported into Autodock Tools (version 1.5.6) to add hydrogens, calculate and assign charges, define rotatable bonds, and save in the pdbqt format. The molecular docking parameters of diosgenin with target proteins are shown in Table 7. Finally, Autodock Tools (version 1.5.6) was employed to compute the docking score, thereby assessing the degree of matching and docking activity between the target and its ligand. Based on prior studies, a binding affinity of less than − 4.25 kcal/mol is indicative of binding activity; a score below − 5.0 kcal/mol denotes enhanced binding activity; and a value lower than − 7 kcal/mol implies robust docking interactions between the ligand and the target44. The binding model was visualized by PyMol 2.3.0.
Cell experiments
Construction and processing of cellular models
HepG2 cells (National Model and Characteristic Experimental Cell Resource Bank/Chinese Academy of Sciences Typical Culture Repository Committee Cell Bank, China), a human hepatocellular carcinoma cell line, were cultured in MEM containing 10% fetal bovine serum (FND500; Shanghai Excell Biological Technology Co., Ltd., China). To construct the NASH cell model, HepG2 cells were treated with free fatty acids (FFAs; containing a palmitic acid to oleic acid molar ratio of 1:2; palmitic acid, P5585; Sigma, USA; oleic acid, O1383; Sigma, USA). The experimental groups were as follows: (i) normal control group: treated with the solvent BSA only; (ii) NASH model group: treated with FFA only; (iii) NASH + diosgenin group: Diosgenin (D1634, Sigma, USA) was added after FFA stimulation; and (iv) BSA + diosgenin group: Diosgenin was added in addition to the BSA treatment.
Cell activity assessment
The cytotoxic effects of FFA and diosgenin on HepG2 cells were detected by a CCK-8 assay (CK12, Beiren Chemical Technology Co., Ltd., China). HepG2 cells were seeded in 96-well plates at a density of 1 × 104/well, and media containing different concentrations of FFA (0, 0.1, 0.2, 0.3, 0.4, 0.5 mM) or diosgenin (0, 5, 10, 25, 50, 100 µM) were added for 24 h. CCK8 reagent was subsequently added, and the mixture was incubated at 37 °C for 1–2 h. The absorbance at a wavelength of 450 nm was subsequently determined. Cell viability was calculated as a percentage relative to untreated controls.
Oil Red O staining
The cells were cultured in 6-well plates (3 × 105 cells/well), treated with FFA and diosgenin, rinsed with PBS and then fixed in paraformaldehyde for 30 min. The triglycerides were stained with Oil Red O staining solution (O0625, Sigma‒Aldrich, USA), and the nuclei were stained with hematoxylin. After the slices were sealed with glycerol gelatin, the cells were observed under a microscope.
Measurement of cellular triglyceride (TG), total cholesterol (TC), and free cholesterol (FC) levels
The intracellular contents of TG, TC, and FC were determined according to the instructions in the kit (E1013-50/E1015-50/E1016-50; Beijing Pulley Gene Technology Co., Ltd., China).
Supernatant IL-6 determination
Enzyme-linked immunosorbent assays (ELISAs) were utilized to assess IL-6 levels in HepG2 cell culture supernatants following the instructions of an ELISA kit (EK106, UNIQUE Biologicals, China).
Western blot analysis
Cells were lysed using RIPA buffer, and total cellular proteins were extracted. Protein concentrations were determined with a BCA protein analysis kit. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were subsequently transferred to PVDF membranes, blocked with 5% milk, and incubated with primary antibodies overnight at 4 °C. p-AKT (Ser473; #12694), AKT (#9272), FASN (#3180), ACC (#3676), and p-ACC (#11818) antibodies from CST; PI3K (PTM-635) from PTM Bio; CPT1α (Ab128569); SCD1 (Ab1986) from Abcam; and GAPDH (#66000) from Proteintech were used. The blots were incubated with secondary antibodies for 1 h at room temperature. An Alpha Q detection system (GE Healthcare) was used for visualization.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8.0 software. The data are expressed as the mean ± standard deviation (x ± sd), and one-way ANOVA was used for multiple comparisons between groups. A P value < 0.05 was considered statistically significant.
Results
Network of diosgenin targets in NASH treatment
A flow chart illustrating the methodology employed in this research is presented in Fig. 1. To elucidate the potential targets of diosgenin in the treatment of NASH, we first conducted a search in the HERB database, PharmMapper database, and SwissTargetPrediction database to determine the action targets of diosgenin; subsequently, all the obtained targets were combined, and duplicate values were deleted. Finally, 329 diosgenin action targets were obtained. A PPI network was constructed to analyze the relationships between diosgenin and its targets (Fig. 2).
The search terms “non-alcoholic steatohepatitis” and “nonalcoholic steatohepatitis” were used to identify genes in the GeneCards database, OMIM database, TTD database, and DisGeNET database. After removing duplicates, 1240 NASH-related targets were identified (Table 1). To identify the key targets of NASH, we constructed a PPI network and performed network topology analysis. The findings revealed that the 5 targets with the highest degree values were EP300 (degree = 105), TP53 (degree = 100), RELA (degree = 91), STAT3 (degree = 89), and AKT1 (degree = 80) (Fig. 3).
Subsequently, mapping diosgenin targets (n = 329) to NASH targets (n = 1240) identified 114 overlapping targets (Table 2). Venn diagrams were generated for visualization purposes (Fig. 4A). The overlapping targets were imported into the STRING database to construct a PPI network. Additionally, network topology analysis was conducted using Cytoscape 7.3.2. As depicted in Fig. 4B, the PPI network contains a total of 114 nodes, and 9 key targets identified in the innermost circle based on the triple of the median degree value (≥ 57) are as follows: ALB (degree = 80), AKT1 (degree = 72), TP53 (degree = 66), VEGFA (degree = 64), MAPK3 (degree = 62), EGFR (degree = 59), STAT3 (degree = 59), CASP3 (degree = 58) and IGF1 (degree = 57). The above 9 targets are considered pivotal targets of diosgenin for NASH therapy.
GO and KEGG signaling pathway enrichment analysis
To further explore the potential mechanism of diosgenin in NASH treatment, an enrichment analysis of the 114 intersecting targets in the PPI network was performed employing the DAVID database. Using FDR < 0.05 and p-value < 0.05 as thresholds, 239 biological processes (BP), 32 cellular components (CC), and 66 molecular functions (MF) pathways were identified. The top 10 GO terms were ranked by Fold enrichment, and a bar graph was generated for visualization in Fig. 5A. The GO analysis revealed that the enriched BP pathways are primarily associated with metabolic regulation, oxidative stress and inflammation control (Table 3). The enriched CC pathways are mainly related to cell signaling, gene expression regulation and cellular secretion processes (Table 4). Meanwhile, the enriched MF pathways are predominantly involved in metabolic regulation, oxidative stress response, signal transduction, and gene expression regulation (Table 5). Among these, BP “positive regulation of fatty acid metabolic process (GO:0045923)” and “positive regulation of fatty acid oxidation (GO:0046321)” relate to Fatty acid metabolism, while BP “negative regulation of interferon-gamma-mediated signaling pathway (GO:0060336),” MF “superoxide dismutase activity (GO:0004784),” and “nitric-oxide synthase regulator activity (GO:0030235)” link to inflammation. Moreover, KEGG pathway enrichment analysis was performed to reveal the underlying signaling pathways involved. After removing the top 3 cancer-related pathways, the top 10 pathways were plotted in bubble diagrams according to the counts of hit targets and P values (Fig. 5B; Table 6). The primary pathway with the most enriched targets was the PI3K-Akt pathway (hsa04151). Based on this, we speculate that diosgenin could ameliorate fatty acid metabolism and inflammatory response in hepatocytes through the PI3K-Akt pathway to alleviate NASH progression.
Molecular docking results
To determine whether diosgenin can act on the nine core targets (ALB, AKT1, TP53, VEGFA, MAPK3, EGFR, STAT3, CASP3, IGF1), we conducted molecular docking studies. The results indicated that diosgenin exhibited binding affinity with all core targets. The other molecular docking results of diosgenin with target proteins are shown in Table 7 and the 3-dimensional map of the binding sites is shown in Fig. 6. The docking results demonstrate that diosgenin is capable of forming hydrogen bonds with these proteins, with bond lengths significantly shorter than the conventional 3.5 Å associated with standard hydrogen bonds. The binding energies are all below − 7 kcal/mol, which confirms a relatively strong binding ability and suggests potential interactions among them.
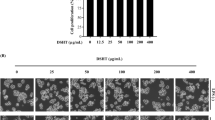
Assessment of HepG2 cell viability and morphology under FFA and diosgenin treatment
Since the signaling pathway with the most enriched targets in the KEGG enrichment analysis was the PI3K-Akt pathway, and all of the nine core targets (ALB45, AKT1, TP5346, VEGFA47, MAPK348, EGFR49, STAT350, CASP344, IGF151) were related to the PI3K-Akt pathway, we thus hypothesized that diosgenin affects NASH by alleviating triglyceride deposition and the inflammatory response through the PI3K-Akt pathway. To confirm this hypothesis, HepG2 cells were treated with free fatty acids (FFAs) at a 1:2 molar ratio of palmitic acid (PA) to oleic acid (OA), aiming to simulate NASH in vitro52. Initially, the effective concentrations of both FFA and diosgenin were determined using the CCK-8 assay. The FFA concentration gradient ranged from 0.1 to 0.5 mM. The findings demonstrated that the viability of HepG2 cells was significantly inhibited when the concentration of PA in the FFA solution exceeded 0.1 mM (Fig. 7A). The highest concentration surpassing the median lethal dose (LD50), namely, the FFA containing 0.2 mM PA, was chosen for the subsequent experiments. Subsequently, HepG2 cells were treated with different concentrations of diosgenin at 0, 5, 10, 25, 50, and 100 µM. Figure 7B shows that there was no significant effect on cell viability at diosgenin concentrations ranging from 0 to 25 µM. However, cell viability was dramatically inhibited when the concentration of diosgenin was greater than 25 µM. Therefore, 5, 10, and 25 µM diosgenin were used in the following experiments. The morphology of the HepG2 cells in the various groups is depicted in Fig. 7C. The cell number decreased, and the cell morphology changed after 24 h of FFA treatment. The cell condition improved after 5 µM diosgenin treatment and was further enhanced in the 10 µM treatment group. However, 25 µM diosgenin treatment did not ameliorate these changes.
Diosgenin alleviates triglyceride deposition and the inflammatory response in FFA-treated HepG2 cells
To verify the effects of diosgenin on lipid metabolism in hepatocytes, we measured the intracellular TG, TC and FC levels in diosgenin-treated HepG2 cells. The intracellular TG content was significantly greater in the FFA-treated group than in the control group. Additionally, the intracellular TG content tended to decrease after 5 µM diosgenin treatment and further decreased after 10 µM diosgenin treatment (Fig. 8B). However, the concentration of 25 µM diosgenin did not significantly affect the lowering of TG levels. Consistent with these findings, Oil Red O staining (Fig. 8A) revealed that the intracellular lipid content was significantly greater in HepG2 cells after 24 h of FFA treatment than in normal controls, and the lipid content was reduced by the addition of 5 µM diosgenin. The intracellular lipid droplet content was significantly reduced after 10 µM diosgenin treatment. Although FFA treatment for 24 h did not increase cholesterol levels in the NASH cell model, treatment with 10 µM diosgenin for 24 h resulted in notable decreases in total cholesterol (TC) and total fibrin (FC) levels in the cells (Fig. 8C).
To assess the ameliorative effect of diosgenin on NASH-related inflammation, we also measured the inflammatory factor IL-6 in the culture supernatant of a diosgenin-treated NASH cell model using an enzyme-linked immunosorbent assay. The IL-6 concentration in the culture supernatant of FFA-treated cells was elevated nearly 1.5-fold compared with that in the normal control group, and treatment with diosgenin significantly reduced the elevated IL-6 level (Fig. 8D).
Diosgenin ameliorates triglyceride deposition and inflammation through the PI3K-Akt pathway in FFA-treated HepG2 cells
The results of the network pharmacological analysis showed that diosgenin might attenuate fatty acid metabolism through the PI3K-Akt pathway in NASH therapy. To further verify whether diosgenin activates the PI3K-Akt pathway and thus plays a role in the treatment of NASH, we examined the protein expression levels of PI3K, p-AKT, and AKT by western blotting. These findings indicate that the phosphorylation level of AKT decreased by approximately 32.7% in HepG2 cells treated with FFA, while diosgenin significantly enhanced the phosphorylation of AKT. However, there was no significant change in the phosphorylation of PI3K between the groups (Fig. 9A, B). We observed that the protein expression of stearoyl-coenzyme A desaturase-1 (SCD1), which is closely related to fatty acid synthesis, was increased in the NASH cell line, and its expression was significantly decreased after treatment with 10 µM diosgenin (Fig. 8E, F). However, there was no significant difference in the expression of other key proteins involved in fatty acid metabolism, such as FASN, ACC, p-ACC, SREBP1c and CPT1α (Fig. 9C–F).
Discussion
In this study, we investigated the potential role that diosgenin plays in NASH remission. Network pharmacology was employed to predict the candidate therapeutic targets and signaling pathways of diosgenin in NASH, and then, experimental verification in HepG2 cells was conducted to further illustrate the pharmacological mechanism of diosgenin against NASH.
Based on the results of the PPI network topology analysis, 9 key genes related to the association of diosgenin with NASH were screened, the most important of which was AKT1. The protein kinase AKT is a serine threonine protein kinase that is activated in response to different stimuli through a phosphatidylinositol 3 kinase (PI3K)-dependent mechanism53 and plays a central role in promoting cell proliferation, migration and transcription and inhibiting apoptosis. Akt1 is a subtype of the Akt family54,55. Each Akt isoform plays a different role in metabolism and growth processes. However, Akt1 plays a key role in cell growth and survival56,57. In addition, several studies have shown that AKT plays an important role in the regulation of lipid metabolism. The results of KEGG pathway analysis showed that the signaling pathway with the most enriched targets was the PI3K-Akt pathway (hsa04151). The PI3K-Akt signaling pathway functions in organism growth and key cellular processes such as glucose homeostasis, lipid metabolism, protein synthesis, cell proliferation and survival by mediating growth factor signaling58. Several studies have shown that the PI3K-Akt pathway plays an important role in regulating lipid metabolism. Brg1 regulates lipid metabolism in hepatocellular carcinoma by mediating GLMP expression through the PIK3AP1/PI3K/AKT pathway59. LAMP3 regulates hepatic lipid metabolism through activation of the PI3K/Akt pathway60. Scutellaria baicalensis and Radix Scutellariae improve glycolipid metabolism in T2DM rats by modulating the metabolic profile and the MAPK/PI3K/Akt signaling pathway61. In addition, the PI3K-Akt pathway plays a crucial role in the development of inflammation. The PTX3/TIST1 feedback loop regulates lipopolysaccharide-induced inflammation via the PI3K/Akt signaling pathway62. Moreover, macrolides reduce pulmonary and systemic inflammation in COPD by modulating the PI3K/Akt-Nrf2 pathway63. Our experimental results suggest that diosgenin may ameliorate steatosis and attenuate the inflammatory response in HepG2 cells by activating the PI3K-Akt pathway.
Other core genes are discussed below. ALB is a carrier of fatty acids in the blood. It has been found that defective phosphorylation of TP53 at Ser312 leads to disruption of lipid metabolism, which causes fat accumulation and even the development of fatty liver64. Hepatocyte-derived VEGFA accelerates the progression of NAFLD to hepatocellular carcinoma through the activation of hepatic stellate cells65. MAPK3, or ERK1, plays key roles in many cell proliferation-related signaling pathways. EGFR is closely related to lipid rafts and plays an important role in the development of tumorigenesis66. Overexpression of STAT3 in the liver ameliorated hyperglycemia and hyperinsulinemia in insulin-resistant diabetic mice67. The protein encoded by the CASP3 gene is a cysteine-aspartate protease that plays a key role in the execution phase of apoptosis, and inhibition of CASP3 can reduce hepatocyte apoptosis and attenuate alcohol-induced liver injury68. It has been shown that IGF1 inhibits cholesterol accumulation in the liver of growth hormone-deficient mice through activation of ABCA169. The above proteins play important roles in the liver and are likely to play an equal role in the development of NASH; moreover, whether diosgenin exerts effects on NASH through these key targets needs to be further confirmed.
In recent years, diosgenin has received increasing attention for its efficacy in the treatment of various metabolic diseases and has been used to treat various cancers70, atherosclerosis71, skin diseases72, osteoporosis73, neurological diseases74, and metabolic diseases (obesity, diabetes, inflammation)75. It has been reported that diosgenin upregulates the expression of the caveolin-1 protein, which is closely related to cholesterol transport, and reduces intracellular cholesterol levels in human normal hepatocyte L02 cells76. In the present study, we investigated the triglyceride-lowering effect of diosgenin in a NASH cell model. Moreover, our experiments confirmed that diosgenin treatment significantly reduced intracellular cholesterol levels, which is consistent with the findings of the previous studies mentioned above.
Fatty acid synthesis is an important process in lipid metabolism. Stearoyl coenzyme A desaturase 1 (SCD1) is the rate-limiting enzyme in the biosynthesis of monounsaturated fatty acids77. Both systemic SCD1 knockout mice and liver-specific SCD1 knockout mice exhibit decreased hepatic triglyceride accumulation and resistance to high-fat diet (HFD)- or high-carbohydrate diet-induced steatosis78,79,80. Our experimental results suggest that diosgenin can reduce SCD1 protein expression in HepG2 cells and ameliorate hepatic lipids by reducing lipid synthesis. It is known that inflammatory cytokines play a key role in the pathogenesis and progression of NASH, leading to more severe fatty liver conditions81. It has been demonstrated that diosgenin can reduce proinflammatory and prosurvival signaling in cancer cells82. Our results showed that diosgenin significantly alleviated IL-6 secretion in a NASH cell model. These in vitro studies confirm diosgenin’s dual therapeutic effects: improving hepatocyte lipid metabolism and reducing inflammation in NASH.
Network pharmacology provides potential drug targets and pathways through computational predictions. However, it has certain limitations, as it excessively relies on network models that cannot fully capture the highly complex and dynamic nature of biological systems. The combination of network pharmacology and in vivo and in vitro experiments not only validates the accuracy of predictions but also provides deeper insights into the multi-component and multi-target characteristics of natural products, thereby enhancing the scientific value and translational potential of the research83,84.
In summary, this study investigated the effects and mechanisms of action of diosgenin on NASH at the systemic level through network pharmacological analysis plus cellular experimental validation. Studies suggest that diosgenin may reduce SCD1 expression by activating the PI3K-Akt pathway in the treatment of NASH. This study lays a good foundation for further in-depth study of the mechanism of diosgenin-induced NASH and provides an important scientific basis for broader clinical application. However, further studies of the mechanism of diosgenin in the treatment of NASH are needed, and an animal model is needed for further research and exploration.
Conclusion
In this study, we employed a network pharmacology approach to investigate the effects and underlying mechanisms of diosgenin on NASH. Through screening of potential targets, construction of protein-protein interaction networks, and bioenrichment analysis, we found that diosgenin may alleviate NASH progression by enhancing fatty acid metabolism in hepatocytes via the PI3K-Akt pathway. Molecular docking results revealed that nine core targets not only exhibited strong binding affinity with diosgenin but also were reported to be associated with the PI3K-Akt pathway. Cellular experiments further confirmed our predictions, demonstrating that diosgenin reduced triglyceride accumulation and inflammatory responses in FFA-treated HepG2 cells through the PI3K-Akt pathway. This study lays a solid foundation for further exploration of the intrinsic mechanisms by which diosgenin ameliorates NASH and provides critical scientific support for its broader clinical application. As a natural compound, diosgenin holds promise as a novel therapeutic approach for NASH.
Target screening of the effect of diosgenin on NASH. (A) Venn diagram of Diosgenin and NASH targets; (B) PPI network of Diosgenin and NASH intersection targets. The targets are sorted incrementally by degree-value, and the node colors and sizes are adjusted according to the degree values. Node colors from blue to orange and from small to large indicate progressively higher degree values.
GO and KEGG enrichment analyses of the overlapping targets of diosgenin in patients with NASH. (A) GO analysis revealed three aspects: biological process (BP), cellular component (CC), and molecular function (MF). The enriched targets were sorted in descending order according to Fold enrichment. (B) KEGG analysis was performed in descending order according to the number of potential targets enriched in the pathway.
The 3-dimensional map of the binding sites between diosgenin and target proteins. (A) ALB. (B) AKT1, (C) TP53, (D) VEGFA, (E) MAPK3, (F) EGFR, (G) STAT3, (H) CASP3, (I) IGF1. Diosgenin is shown in green. Target proteins are displayed as white. The places where diosgenin and the target proteins are connected represent specific docking sites between diosgenin and target proteins.
Free fatty acid and diosgenin concentration screening. (A) Effects of different concentrations of free fatty acids (FFA, PA-to-OA molar ratio of 1:2) on cell proliferation and toxicity; (B) cell survival after 24 h of incubation with different concentrations of diosgenin (µM); (C) cell morphology of HepG2 cells in different intervention states (× 50).
Diosgenin reduces cellular triglyceride deposition and attenuates FFA-induced inflammation. (A) Cellular Oil Red O staining (× 40) of HepG2 cells treated with FFA and different concentrations of diosgenin. (B) Determination of cellular triglyceride (TG) levels in HepG2 cells treated with FFA and different concentrations of diosgenin; (C) Determination of cellular total cholesterol (TC) and free cholesterol (FC) levels in HepG2 cells treated with FFA and different concentrations of diosgenin; (D) Determination of IL-6 levels in cell culture supernatants in HepG2 cells treated with FFA and different concentrations of diosgenin; (D) Determination of IL-6 in cell culture supernatant after treatment of HepG2 cells with FFA and different concentrations of diosgenin. The data are expressed as the mean ± standard deviation; * indicates P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001, and **** indicates P < 0.0001.
Activation of the PI3K-Akt pathway by diosgenin activates lipid metabolism in hepatocytes. (A) Western blotting was used to detect PI3K, p-AKT, and AKT protein expression. (C) Western blotting was used to detect the protein expression of CPT1α, an indicator of fatty acid oxidation. (E) protein expression of other fatty acid metabolism-related indicators determined by Western blotting. (B, D, F) the data shown in the bar graphs indicate the mean ± standard deviation of three independent experiments. * indicates P < 0.05, ** indicates P < 0.01.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Kanwal, F. et al. Preparing for the NASH epidemic: A call to action. Gastroenterology 161(3), 1030–42e8 (2021).
Chalasani, N. et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American association for the study of liver diseases. Hepatology 67(1), 328–357 (2018).
Sheka A C, Adeyi, O. et al. Nonalcoholic Steatohepatitis Jama 323(12) (2020).
Bugianesi, E. Nafld/Nash. J. Hepatol. 77(2), 549–550 (2022).
Ditah, I. C. et al. Changes in the prevalence of hepatitis C virus infection, nonalcoholic steatohepatitis, and alcoholic liver disease among patients with cirrhosis or liver failure on the waitlist for liver transplantation. Gastroenterology 152(5), 1090–9e1 (2017).
Lambert, J. E. et al. Increased de Novo lipogenesis is a distinct characteristic of individuals with nonalcoholic fatty liver disease. Gastroenterology 146(3), 726–735 (2014).
Kawano, Y. & Cohen, D. E. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J. Gastroenterol. 48(4), 434–441 (2013).
Koyama, Y. & Brenner, D. A. Liver inflammation and fibrosis. J. Clin. Invest. 127(1), 55–64 (2017).
Muthiah, M. D. & Sanyal A. J. Current management of non-alcoholic steatohepatitis. Liver Int. 40(Suppl 1), 89–95 (2020).
Harrison S A, Allen A M, Dubourg, J. et al. Challenges and opportunities in NASH drug development. Nat. Med. 29(3), 562–573 (2023).
Li, C. et al. Vitexin ameliorates chronic stress plub high fat diet-induced nonalcoholic fatty liver disease by inhibiting inflammation. Eur. J. Pharmacol. 882, 173264 (2020).
Wu, C. & Jing, M. Alisol A 24-acetate ameliorates nonalcoholic steatohepatitis by inhibiting oxidative stress and stimulating autophagy through the AMPK/mTOR pathway. Chem. Biol. Interact. 291, 111–119 (2018).
Zhao, Z., Deng, Z. T., Huang, S. et al. Alisol B alleviates hepatocyte lipid accumulation and lipotoxicity via regulating RARalpha-PPARgamma-CD36 cascade and attenuates non-alcoholic steatohepatitis in mice. Nutrients 14(12). (2022).
RAJU, J. Cancer chemopreventive and therapeutic effects of diosgenin, a food saponin. Nutr. Cancer 61(1), 27–35 (2009).
Huang, C. H. et al. Oral administration with Diosgenin enhances the induction of intestinal T helper 1-like regulatory T cells in a murine model of food allergy. Int. Immunopharmacol. 42, 59–66 (2017).
Kim, J. K. & Park, S. U. An update on the biological and pharmacological activities of diosgenin. Excli J. 17, 24–28 (2018).
Gong, J. et al. Effect of Fenugreek on hyperglycaemia and hyperlipidemia in diabetes and prediabetes: A meta-analysis. J. Ethnopharmacol. 194, 260–268 (2016).
Wu, S. et al. Dioscin improves postmenopausal osteoporosis through inducing bone formation and inhibiting apoptosis in ovariectomized rats. Biosci. Trends 13(5), 394–401 (2019).
Li, R. et al. Diosgenin regulates cholesterol metabolism in hypercholesterolemic rats by inhibiting NPC1L1 and enhancing ABCG5 and ABCG8. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids. 1864(8), 1124–1133 (2019).
Zhang, S. Z. et al. Therapeutic potential and research progress of Diosgenin for lipid metabolism diseases. Drug Dev. Res. 83(8), 1725–1738 (2022).
Sun, F. & Yang, X. The effects of Diosgenin on hypolipidemia and its underlying mechanism: A review. Diabetes Metab. Syndr. Obes. 14, 4015–4030 (2021).
Cheng, S. et al. Diosgenin prevents high-fat diet-induced rat non-alcoholic fatty liver disease through the AMPK and LXR signaling pathways. Int. J. Mol. Med. 41(2), 1089–1095 (2018).
Yan, M. et al. Diosgenin alleviates nonalcoholic steatohepatitis through affecting liver-gut circulation. Pharmacol. Res., 187 (2023).
Hirai, S. et al. Diosgenin attenuates inflammatory changes in the interaction between adipocytes and macrophages. Mol. Nutr. Food Res. 54(6), 797–804 (2010).
Khateeb, S. & Albalawi, A. Diosgenin modulates oxidative stress and inflammation in high-fat diet-induced obesity in mice. Diabetes Metab. Syndr. Obes. 15, 1589–1596 (2022).
Nogales, C. et al. Network pharmacology: Curing causal mechanisms instead of treating symptoms. Trends Pharmacol. Sci. 43(2), 136–150 (2022).
Lai, X. et al. Editorial: Network pharmacology and traditional medicine. Front. Pharmacol. 11, 1194 (2020).
Fang, S. et al. HERB: A high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Res. 49(D1), D1197–d206 (2021).
Kim, S. et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 49(D1), D1388–d95 (2021).
Wang, X. et al. Enhancing the enrichment of pharmacophore-based target prediction for the polypharmacological profiles of drugs. J. Chem. Inf. Model. 56(6), 1175–1183 (2016).
Wang, X. et al. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 45(W1), W356–w60 (2017).
Daina, A. & Michielin, O. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 47(W1), W357–w64 (2019).
UniProt. A worldwide hub of protein knowledge. Nucleic Acids Res. 47(D1), D506–d15 (2019).
Stelzer, G. et al. The GeneCards Suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 54 1.30.1–1.3 (2016).
PIñero, J. et al. The disgenet knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 48(D1), D845–d55 (2020).
Boyadjiev, S. A. & Jabs E. W. Online Mendelian inheritance in man (OMIM) as a knowledgebase for human developmental disorders. Clin. Genet. 57(4), 253–266 (2000).
Chen, X. et al. Therapeutic target database. Nucleic Acids Res. 30(1), 412–415 (2002).
LUO, W. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29(14), 1830–1831 (2013).
Huang Da, W., Sherman, B. T., Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4(1), 44–57 (2009).
Shi, L. et al. Gene expression profiling and functional analysis reveals that p53 pathway-related gene expression is highly activated in cancer cells treated by cold atmospheric plasma-activated medium. PeerJ 5, e3751 (2017).
Szklarczyk, D. et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45(D1), D362–d8 (2017).
Ru, J. et al. TCMSP: A database of systems Pharmacology for drug discovery from herbal medicines. J. Cheminform. 6, 13 (2014).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11), 2498–2504 (2003).
Wang, C. et al. PI3K-AKT-mediated phosphorylation of Thr260 in CgCaspase-3/6/7 regulates heat-induced activation in oysters. Commun. Biol. 7(1), 1459 (2024).
Bao, X. Exploring the protective mechanism of Gankang tablets on liver function in rats with carbon tetrachloride-induced liver fibrosis based on PI3K/Akt signaling pathway. Mod. Dig. Interv. Diagn.Treat. 25(02), 188–192 (2020).
Alvarado-ortiz, E. et al. Mutant p53 gain-of-function stimulates canonical Wnt signaling via PI3K/AKT pathway in colon cancer. J. Cell. Commun. Signal. 17(4), 1389–1403 (2023).
Dai, W. et al. Human umbilical cord-derived mesenchymal stem cells (hUC-MSCs) alleviate excessive autophagy of ovarian granular cells through VEGFA/PI3K/AKT/mTOR pathway in premature ovarian failure rat model. J. Ovarian Res. 16(1), 198 (2023).
Prochazka, R. et al. The role of MAPK3/1 and AKT in the acquisition of high meiotic and developmental competence of Porcine oocytes cultured in vitro in FLI medium. Int. J. Mol. Sci. 22(20) (2021).
Novoplansky, O., Shnerb, A. B., Marripati, D. et al. Activation of the EGFR/PI3K/AKT pathway limits the efficacy of Trametinib treatment in head and neck cancer. Mol. Oncol. 17(12), 2618–2636 (2023).
Sun, Z. & Jiang, Q. AKT blocks SIK1-mediated repression of STAT3 to promote breast tumorigenesis. Cancer Res. 83(8), 1264–1279 (2023).
Zhao, J. et al. CircCCDC91 regulates chicken skeletal muscle development by sponging miR-15 family via activating IGF1-PI3K/AKT signaling pathway. Poult. Sci. 101(5), 101803 (2022).
Shi, H. et al. Baicalin attenuates hepatic injury in non-alcoholic steatohepatitis cell model by suppressing inflammasome-dependent GSDMD-mediated cell pyroptosis. Int. Immunopharmacol. 81, 106195 (2020).
Heron-Milhavet, L. & Khouya, N. Akt1 and Akt2: Differentiating the Aktion. Histol. Histopathol. 26(5), 651–662 (2011).
Vivanco I. & Sawyers, C. L. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer 2(7), 489–501 (2002).
Yoeli-Lerner, M. et al. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol. Cell. 20(4), 539–550 (2005).
Chen, W. S. et al. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 15(17), 2203–2208 (2001).
Cho, H. et al. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J. Biol. Chem. 276(42), 38349–38352 (2001).
Abeyrathna, P. & su, Y. The critical role of Akt in cardiovascular function. Vascul Pharmacol. 74, 38–48 (2015).
Liu, G. et al. BRG1 regulates lipid metabolism in hepatocellular carcinoma through the PIK3AP1/PI3K/AKT pathway by mediating GLMP expression. Dig. Liver Dis. 54(5), 692–700 (2022).
Liao, X. et al. LAMP3 regulates hepatic lipid metabolism through activating PI3K/Akt pathway. Mol. Cell. Endocrinol. 470, 160–167 (2018).
Cui, X. et al. Scutellariae radix and coptidis rhizoma improve glucose and lipid metabolism in T2DM rats via regulation of the metabolic profiling and MAPK/PI3K/Akt signaling pathway. Int. J. Mol. Sci., 19(11). (2018).
Li, A. et al. PTX3/TWIST1 feedback loop modulates Lipopolysaccharide-Induced inflammation via PI3K/Akt signaling pathway. J. Interferon Cytokine Res. 42(4), 161–169 (2022).
Sun, X., Chen, L. & He, Z. PI3K/Akt-Nrf2 and Anti-Inflammation effect of macrolides in chronic obstructive pulmonary disease. Curr. Drug Metab. 20(4), 301–304 (2019).
He, M. et al. Defect in Ser312 phosphorylation of Tp53 dysregulates lipid metabolism for fatty accumulation and fatty liver susceptibility: Revealed by lipidomics. J. Chromatogr. B Analyt Technol. Biomed. Life Sci. 1211, 123491 (2022).
Shen, H. et al. Hepatocyte-derived VEGFA accelerates the progression of non-alcoholic fatty liver disease to hepatocellular carcinoma via activating hepatic stellate cells. Acta Pharmacol. Sin. 43(11), 2917–2928 (2022).
Guéguinou, M. et al. Lipid rafts, KCa/ClCa/Ca2 + channel complexes and EGFR signaling: novel targets to reduce tumor development by lipids?. Biochim. Biophys. Acta 1848(10 Pt B), 2603–2620 (2015).
Kinoshita, S. et al. Role of hepatic STAT3 in the regulation of lipid metabolism. Kobe J. Med. Sci. 54(4), E200–E208 (2008).
Ma, X. Y. et al. Ursolic acid reduces hepatocellular apoptosis and alleviates alcohol-induced liver injury via irreversible Inhibition of CASP3 in vivo. Acta Pharmacol. Sin. 42(7), 1101–1110 (2021).
Fukunaga, K. et al. IGF1 suppresses cholesterol accumulation in the liver of growth hormone-deficient mice via the activation of ABCA1. Am. J. Physiol. Endocrinol. Metab. 315(6), E1232–e41 (2018).
Sethi, G. et al. Pro-apoptotic and anti-cancer properties of diosgenin: A comprehensive and critical review. Nutrients 10(5) (2018).
Lv, Y. C., Yang, J., Yao, F. et al. Diosgenin inhibits atherosclerosis via suppressing the MiR-19b-induced downregulation of ATP-binding cassette transporter A1. Atherosclerosis 240(1), 80–89 (2015).
Kim, J. E. et al. Diosgenin effectively suppresses skin inflammation induced by phthalic anhydride in IL-4/Luc/CNS-1 transgenic mice. Biosci. Biotechnol. Biochem. 80(5), 891–901 (2016).
Chiang, S. S., Chang, S. P. & Pan, T. M. Osteoprotective effect of Monascus-fermented Dioscorea in ovariectomized rat model of postmenopausal osteoporosis. J. Agric. Food Chem. 59(17), 9150–9157 (2011).
Chen, Y. et al. Advances in the pharmacological activities and mechanisms of diosgenin. Chin. J. Nat. Med. 13(8), 578–587 (2015).
Basch, E. et al. Therapeutic applications of fenugreek. Altern. Med. Rev. 8(1), 20–27 (2003).
Wang, T. et al. Antihyperlipidemic effect of protodioscin, an active ingredient isolated from the rhizomes of Dioscorea Nipponica. Planta Med. 76(15), 1642–1646 (2010).
Am, A. L., Syed, D. N. & Ntambi, J. M. Insights into stearoyl-CoA desaturase-1 regulation of systemic metabolism. Trends Endocrinol. Metab. 28(12), 831–842 (2017).
Ntambi, J. M. et al. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc. Natl. Acad. Sci. U S A 99(17), 11482–11486 (2002).
Sampath, H. & Ntambi, J. M. The role of stearoyl-CoA desaturase in obesity, insulin resistance, and inflammation. Ann. N Y Acad. Sci. 1243, 47–53 (2011).
Miyazaki, M. et al. Hepatic stearoyl-CoA desaturase-1 deficiency protects mice from carbohydrate-induced adiposity and hepatic steatosis. Cell. Metab. 6(6), 484–496 (2007).
Lee, J. J. et al. Palmitoleic acid is elevated in fatty liver disease and reflects hepatic lipogenesis. Am. J. Clin. Nutr. 101(1), 34–43 (2015).
Karami-Mohajeri, S. et al. Diosgenin: Mechanistic insights on its anti-inflammatory effects. Antiinflamm. Antiallergy Agents Med. Chem. 21(1), 2–9 (2022).
Li, S. et al. Suppressive effects of a Chinese herbal medicine qing-luo-yin extract on the angiogenesis of collagen-induced arthritis in rats. Am. J. Chin. Med. 31(5), 713–720 (2003).
Wang, Z. Y. et al. Mechanism of action of daqinjiao decoction in treating cerebral small vessel disease explored using network pharmacology and molecular docking technology. Phytomedicine 108, 154538 (2023).
Funding
This work was supported by the National Natural Science Foundation (82070818 and 82370868), Taishan Scholar Project of Shandong Province (tsqn202211330), the Natural Science Foundation of Shandong Province (ZR2024MH086, ZR2020MH037), together with Postdoctoral Innovation Project of Shandong Province (SDCX-ZG-202203058).
Author information
Authors and Affiliations
Contributions
P.Y.G.: Writing—original draft, Validation, Data curation, Visualization. J.C.: Writing—original draft, Methodology, Investigation, Data curation. J.X.X.: Methodology, Data curation, Validation. H.Q.C.: Investigation, Validation. R.Z.: Methodology, Formal analysis. D.C.: Formal analysis, Investigation. Y.H.Z.: Writing—review & editing, Methodology, Resources, Supervision. S.S.S.: Writing—review & editing, Supervision, Funding acquisition, Resources, Conceptualization. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gu, P., Chen, J., Xin, J. et al. Network pharmacology-based investigation of the pharmacological mechanisms of diosgenin in nonalcoholic steatohepatitis. Sci Rep 15, 10351 (2025). https://doi.org/10.1038/s41598-025-95154-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-95154-z