Abstract
Metabolic syndrome (MetS) and Type-2 diabetes mellitus (T2DM), often termed “glucolipotoxicity” or insulin resistance syndrome, are complex metabolic disorders typically managed by lifestyle interventions and oral hypoglycemic agents. While conventional drugs, including metformin, sulfonylureas, and sodium-glucose cotransporter-2 (SGLT2) inhibitors, show efficacy, they also present risks such as hypoglycemia and weight gain. Among these, metformin remains the preferred first-line treatment due to its safety, low cost, and minimal side effects. However, the multifactorial nature of MetS/T2DM, coupled with increased cardiovascular risk, demands novel therapeutic strategies targeting broader disease-specific mediators. This study employs network pharmacology to identify potential gene targets linked to MetS/T2DM. We constructed a protein-protein interaction (PPI) network of 97 genes involved in the pathophysiology of these disorders, identifying 89 interlinked genes. The top 10 crucial genes including insulin receptor substrate 1 (IRS1), and interleukin-6 (IL6) are highlighted as key contributors to disease progression. Gene Ontology (GO) analysis revealed their involvement in essential biological processes like peptide synthesis, lipid regulation, and glucose homeostasis. Interestingly, nine of these genes are influenced by metformin, suggesting its broader mechanism in modulating metabolic pathways. Additionally, scrutiny of transcription factors for the considered dataset shows that forkhead box protein O1 (FOXO1), sirtuin 1 (SIRT1), and peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α) play crucial roles in insulin signalling, mitochondrial function, and glucose metabolism, further elucidating the molecular complexity of T2DM. Through a network biology framework, this study attempts to highlight the potential for prospective multi-targeting therapeutic strategies that may exhibit improved efficacy and safety, for the management of MetS/T2DM.
Similar content being viewed by others
Introduction
The term “metabolic diseases” refers to a class of illnesses marked by aberrant metabolism or the chemical processes that take place inside cells to transform nutrients into energy and other necessary chemicals. Some of the prevalent metabolic diseases include obesity, type 2 diabetes mellitus (T2DM), non-alcoholic fatty liver disease (NAFLD), and metabolic syndrome (MetS). A combination of genetic, lifestyle, and endocrinological factors play an influential role in these metabolic diseases1,2,3. Indeed, sedentary habits and overnutrition promote abnormal adiposity culminating in insulin resistance (IR) that is closely associated with MetS2,4. Particularly, MetS, often known as syndrome X, is not a single entity, but a complex cluster of cardiovascular disease risk factors. This non-communicable disease has emerged as the leading global health hazard5,6,7. Though MetS diagnosing criteria have evolved, the International diabetes federation (IDF) and American Heart Association/National Heart, lung, and Blood Institute (AHA/ NHLBI) established consensus criteria, which remain applicable even today8,9. Indeed, MetS patients are identified if they exhibit at least three of the following interconnected cardiometabolic symptoms: raised blood pressure, increased body mass index (BMI) hypertriglyceridemia, decreased high-density lipoprotein (HDL) cholesterol, and glucose intolerance4,10,11,12. The World Health Organization (WHO) proposed the definition of MetS for the first time in 1999, highlighting hyperglycemia and insulin resistance as the primary diagnostic criteria13. The global prevalence of MetS ranges from 12.5% (95%CI: 10.2–15.0) to 31.4% (29.8–33.0), and its incidence is quite like the incidence of its associated risk factors, T2DM, and obesity. Particularly, T2DM is considered an integral component of MetS2,6,14. According to numerous epidemiological studies, MetS is linked to a five-fold greater risk of T2DM, a two- to four-fold risk of stroke, and a three- to four-fold risk of myocardial infarction8,11,15. In addition, visceral obesity is identified as one of the major and inherent risk factors for MetS15.
An abnormal interplay between environmental (lifestyle changes) and genetic factors alters cellular energy homeostasis and culminates in inherent metabolic derangements. This concomitantly triggers intracellular stress and systemic inflammation that contribute to alarming consequences including cardiovascular disease, progression of NAFLD to hepatocellular carcinoma, β-cell dysfunction, IR, IR-mediated nephropathy, neuropathy, retinopathy, certain cancers like breast, colorectal, and prostate4,16,17,18. Indeed, chronic low-grade inflammation is often driven by obesity, which eventually leads to IR and metabolic dysfunction19. This complex interaction between immunological processes and metabolic defects rightly deserves the term “immunometabolism”20,21. Generally speaking, the adipose tissue acts as a reservoir of energy by supplying fatty acids during starvation. However, on the other hand, an excess supply of nutrients triggers inflammatory pathways via significant infiltration of activated M1 macrophages in tissues especially including adipose, skeletal muscle, liver, and pancreas, that in turn secrete a broad array of pro-inflammatory cytokines and chemokines leading to inflammation and IR20,22,23,24. Over-nutrition also promotes reactive oxygen species (ROS) generation, hypoxia, endoplasmic reticulum (ER) stress, and unfolded protein response in adipose tissue that stimulates inflammation21,25,26,27,28.
However, the precise factors that are responsible for over-nutrition or obesity-associated inflammation are quite incomprehensible29. Interestingly, adipose tissue (AT) also secretes a specialized class of cytokines, also known as adipocytokines or adipokines, including adiponectin, apelin omentin-1, interleukin-10 (IL-10), Sfrp5, resistin, angiotensinogen, leptin, omentin, visfatin, tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), and chemerin. A healthy AT secretes anti-inflammatory adipokines including adiponectin, omentin, secreted frizzled-related protein 5 (Sfrp5), IL-10, and members of C1q/TNF-related protein family (CTRP) that are known to orchestrate lipid metabolism, insulin sensitivity, and vascular homeostasis., However, pro-inflammatory adipokines such as leptin, resistin, TNF-α, and IL-6 are produced by hypertrophic adipocytes, particularly, in obesity- induced endoplasmic reticulum (ER) stress or oxidative stress7,30,31.
The poorly vascularized, hypoxic, and hypertrophic adipocytes mediate cytokine-induced inflammatory signalling exclusively through c-Jun, N-terminal kinases/mitogen activated-protein kinases (JNKs/MAPKs), Janus kinase-signal transducer of activation (JAK-STAT), an inhibitor of nuclear factor–κB kinase/ nuclear factor kappa light chain enhancer of activated-B cells (IKK/NF-κB), and protein kinases C (PKCs) pathways in turn contributing to endothelial dysfunction, chronic local and systemic IR, T2DM, and atherogenic dyslipidemia20,31,32,33,34. JNK and IKK/NF-κB are known to be the two principal signalling pathways that are linked to the development of IR, and they markedly affect crucial metabolic organs including the liver, skeletal muscle, adipose tissue, and pancreas21,35,36,37. Essentially, IR is a multifaceted pathological state characterized by a reduced ability to suppress glucose production and promote peripheral glucose utilization. This kick-starts the overwhelming production of insulin by the pancreatic β-cells as an act of compensation, eventually causing β- cell dysfunction. This condition primarily culminates in T2DM in addition to dyslipidemia and cerebrocardiovascular disease. The onset of T2DM is linked to a progressive decline in insulin’s action, impairing tissue glucose disposal. The resulting hyperglycemia, along with pro-inflammatory cytokines, and free fatty acids (FFAs), creates an inflammatory milieu, triggering a cascade of destructive events, including β-cell hypertrophy, apoptosis, and fibrosis, ultimately leading to β-cell death18,37,38,39.
T2DM, in a simplified gluco-centric context, is characterized by insulin resistance-induced hyperglycemia that needs critical glycemic control as a therapeutic strategy. However, the aspect of glycemic control is just a shallow view, as approximately 90% of T2DM patients are obese and exhibit a combination of micro- and macro-vascular symptoms, known to account for 50% of deaths, particularly affecting the ocular, renal, cardiovascular, and nervous systems. In addition, T2DM also increases the incidence and risk for a set of non-glycemic conditions, including Alzheimer’s disease, dementia, polycystic ovarian syndrome (PCOS), and various cancers. Hence, realizing this and putting it into a broader context of MetS/glucolipotoxicity seems rational. Moreover, the standard-of-care (SOC) treatments, that have been primarily intended to manage hyperglycemia, now attempt to mainly target cardiovascular and other non-glycemic diseases apart from glycemic control. This clearly indicates the need for a different comprehensive therapeutic approach for targeting MetS/T2DM41. Lifestyle modifications, accompanied by regular physical exercise, are considered the first-line, non-pharmacological approach for effectively managing MetS/T2DM and its co-morbidities. However, most of the time, lifestyle changes are not enough to improve the multiple risk factors in individuals, necessitating a multi-drug or polypharmacy strategy, involving the usage of a variety of drug classes, including lipid-lowering, anti-hypertensive/ heart-failure, anti-diabetic, and anti-obesity medications4,40. Nevertheless,, a multi-drug regimen is not always an ideal option due to potential side effects, drug-drug interactions, and poor patient compliance4,41,42. Since inflammation plays a pivotal role in the progression of MetS and its complications, targeting it would certainly be a judicious therapeutic approach. Furthermore, several anti-diabetic medications have demonstrated anti-inflammatory properties and encouraging findings from preclinical studies suggest the potential effectiveness of anti-inflammatory drugs in treating IR43.
For more than 70 years, metformin (1,1-dimethylbiguanide) has been routinely prescribed oral hypoglycemic medication. Despite the emergence of a variety of novel anti-diabetic drug classes, metformin is still the preferred oral anti-hyperglycemic drug. Also, several clinical guidelines advocate it as the first-line treatment for T2DM, particularly in those who have recently been diagnosed. Indeed, interest in metformin is continuously rising even after being used for such a long time and ample research. Apart from gastrointestinal adverse effects, it is known to increase the risk of lactic acidosis, due to which it is contraindicated in patients with acute kidney failure. However, metformin-associated lactic acidosis (MALA) may go underreported and seems to be uncommon with high mortality44,45,46,47. Reports highlight that metformin combats the gluconeogenic pathway by either directly or indirectly activating AMPK in the liver, leading to the suppression of hepatic glucose production. Additionally, metformin exerts its hypoglycemic and metabolic effects via other important routes, which include promoting translocation of skeletal muscle glucose transporter GLUT4, enhancing autophagy in β-cells under hyperglycemia conditions, inhibiting mitochondrial complex I, and altering the gut microbiome. Moreover, it has shown effectiveness against multiple cancers, cardiovascular diseases, liver disorders, neurodegenerative conditions, and renal diseases. Owing to metformin’s multifaceted actions, research is still being pursued to identify many novel aspects, particularly regarding metabolic regulation46,48,49.
Conclusively, the potential therapeutic intervention for MetS/T2DM involves either the administration of multiple drugs in combination or the development of new pharmaceuticals with a multitude of activities. There is an escalating interest and dire need for the development of novel drug classes that could effectively target multiple components in each disease pathway and offer a thorough and holistic therapy4. The present study attempts to decode the intricate network of selected genes concerning the pathogenesis of MetS/T2DM and understand whether metformin could influence some or many of these genes that are known to regulate a broad range of metabolic pathways, through a rational and systematic network biology-based approach. We hypothesize that this approach could reliably identify potential disease pathways and intrinsically associated protein regulators, which could be collectively targeted by prospective candidate molecules as part of a multi-targeting approach to reduce the incidence and effectively manage MetS/T2DM-related morbidities.
Methodology
Building the sequence dataset
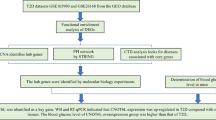
To facilitate downstream analysis and increase prediction accuracy, extensive data mining and a thorough assessment of the literature are conducted using some of the publicly available databases, including PubMed, String50, NCBI gene51, the human protein atlas52, and Reactome53. For building a non-redundant and accurate resource for subsequent analyses, duplicate entries and isoform proteins are carefully screened to derive the final sequence dataset.
Network Pharmacology studies
For the selected genes, the protein-protein interaction network is constructed primarily to identify potential disease markers related to MetS/T2DM. Utilizing the search tool for the retrieval of interacting genes/protein (STRING), the physical and functional interactions of the protein-protein interactions (PPI) are constructed with a high confidence threshold of 0.952. Excluding the non-interacting proteins (nodes), the remaining PPI data is input into Cytoscape (version 3.10.1) for network topology analysis. Subsequently, the 10 top-ranked target genes are prioritized using the Cytohubba extension in Cytoscape54. To further elucidate the biological significance, the gene enrichment analysis of the constructed PPI network is performed using ShinyGO 0.80 (http://bioinformatics.sdstate.edu/go/). The genomic information of the network is subsequently analyzed for biological processes, molecular functions, and cellular components using gene set enrichment analysis with a significance threshold of p-value < 0.05. This p-value is set to minimize the likelihood of gene enrichment occurring by chance, based on false discovery rate (FDR)55. Additionally, KEGG pathways are utilized to explore the underlying metabolic pathways associated with the target genes relevant to MetS /T2DM. For the identification of genes influenced by metformin, the Genecards (https://www.genecards.org) server is used to extract reliable genes. Subsequently, common genes, linked to both MetS/T2DM and metformin, are identified using Venny 2.0.
Gene enrichment analysis of selected genes related to MetS and T2DM
The gene enrichment analysis is performed using DAVID (https://david.ncifcrf.gov/tools.jsp) by preparing a list of genes formatted with recognized gene identifiers such as gene symbols or ENSEMBL IDs. The list is fed to the DAVID server, and various annotation categories, including the GO terms such as biological processes, molecular function, and cellular components, and KEGG pathways are analyzed. Parameters such as P-value and FDR cutoff and pathway size limits are set to define the enrichment analysis criteria, and the results are filtered based on statistical significance and fold enrichment, focusing on the key pathways or terms. The visualization tool Cytoscape 3.10.2 version is utilized to construct the optimal PPI network between the analysed proteins.
Gene expression analysis
To understand the over- or under-expression of gene sets, their heatmap is computed using the g: Profiler tool (https://biit.cs.ut.ee/gprofiler/gost), a web-based platform for gene list analysis56. The Benjamini-Hochberg procedure is applied to adjust p-values, with a significance threshold of 0.05, to filter out statistically insignificant data. Differentially expressed genes (DEGs) are then analyzed within specific pathways to determine their roles in key biological processes and regulatory mechanisms. This approach provides insights into pathway interactions and the functional consequences of gene expression changes. The major hub genes, functionally associated with MetS/T2DM and metformin, are screened for their interaction with kinases and transcription factors (TFs) using the X2K web server57, as their binding would implicate their substantial role in regulating MetS/T2DM and metformin’s pharmacological or therapeutic activity. Based on Fischer’s t-test, the twenty most significant kinases based on the enrichment scores and TFs are analyzed, and their functional roles are interpreted.
Results and discussion
Building the sequence dataset
Critically mining the sequence databases, a dataset of 97 proteins is found to play a subtle role in the onset or culmination of MetS/T2DM. By excluding the two redundant hits, a derived dataset of 97 potential entries is constructed, as detailed in Supplementary Table S1.
Network Pharmacology studies
The common genes of MetS/T2DM and Metformin
The common genes of MetS/T2DM and metformin are depicted in Fig. 1. Out of the 42 and 1414 genes, orderly associated with T2DM and Metformin, respectively, the Venn diagram reveals an overlap of 54 genes, indicating that these genes are commonly linked to both the pathogenesis of MetS/T2DM and the therapeutic mechanism of Metformin.
Protein-Protein interaction (PPI) analysis
The PPI interactions among the selected 97 genes and their association with metformin is constructed using STRING, as illustrated in Fig. 2. The STRING-based PPI interactions at high confidence (0.9) reveal the PPI network consisting of 89 nodes (proteins) and 123 edges (interactions), with an average local clustering coefficient of 0.502. The expected number of edges is 24; however, we observed a significantly higher number of 123 interactions, suggesting that the STRING network contains more interactions than anticipated. The PPI enrichment p-value is observed to be less than 0.05, indicating that the proteins are at least partially biologically connected, as a group. Further, the top-10 genes associated with MetS/T2DM are found to be insulin receptor substrate 1 (IRS1), interleukin 6 (IL-6), forkhead box O1 (FOXO1), peroxisome proliferator-activated receptor γ coactivator-1 α (PPARGC1A/ PGC-1α), mammalian target of rapamycin (mTOR), Leptin (LEP), peroxisome proliferator-activated receptor-γ(PPARG), adiponectin, C1Q and collagen domain containing (ADIPOQ), protein kinase AMP-activated catalytic subunit alpha1 (PRKAA1), and silent mating type information regulation 2 homolog 1 (SIRT1). Moreover, metformin is predicted to associate with or possibly influence various key genes, including, PPARGC1A/PGC-1α, mTOR, PPARG, ADIPOQ, LEP, IL-6, IRS1, FOXO1, and PIK3CA, which affect the major biological pathways like insulin sensitivity, energy balance, inflammation, glucose metabolism, and mitochondrial biogenesis, related to MetS/T2DM, indicating their strong connection.
The PPI networks of the selected genes. Figures (A) and (B) orderly represent the PPI networks of the selected genes and the top 10 genes that are associated with MetS/T2DM. Figures C and D represent the PPI networks of the selected genes and the top 10 genes that are associated with MetS/T2DM and metformin, respectively.
Kyoto encyclopedia of genes and genomes (KEGG) pathway
The KEGG pathway analysis is illustrated in Fig. 3. As per the information retrieved from the KEGG database58,59, the pathway for the selected genes shows that several pathways, including the insulin signaling pathway, adipocytokine pathway, glucose transporters, and apoptosis, are associated with the pathogenesis of MetS/T2DM. Moreover, it also involves the phosphoinositide 3-kinases, which are involved in a variety of cellular functions including cell growth, differentiation, motility, and especially insulin-mediated glucose uptake.
Gene enrichment analysis of selected genes related to MetS and T2DM
Gene Ontology is a framework for the standardized representation of gene and gene product attributes across species and databases. It consists of three main categories: biological process refers to the biological objectives to which the gene or gene product contributes. This includes pathways or larger processes made up of the ordered assemblies of molecular functions; molecular function describes the elemental activities of a gene product at the molecular level, such as binding or catalysis, and cellular component indicates where in the cell the gene product is active, such as organelles or membrane regions.
The study utilizes the gene ontology server for the required genes, and their associated biological processes are analyzed. The analysis revealed the biological processes along with the functionally involved genes and provided an estimated p-value, the lower being the better. Quite interestingly, it is observed that a set of 10 genes: IL-6, IRS1, LEP, ADIPOQ, UCP1, SIRT1, PPARG, PPARGC1A, FOXO1, and FGF21 majorly govern 13 different biological pathways including homeostatic process, cellular response to chemical stimulus, positive regulation of the metabolic process, response to chemical, regulation of primary metabolic process, regulation of macromolecule biosynthetic process, regulation of cellular biosynthetic process, regulation of macromolecule metabolic process, regulation of cellular metabolic process, and cellular response to stimulus (Supplementary Table S2). Hence, the extracted data clearly illustrates the influence of all the genes on a variety of biological processes, crucial for the homeostatic state of the human body. The extremely low p-value indicates a strong association of gene set with glucose homeostasis, a critical process in maintaining stable blood glucose levels. Genes like IRS1 and PPARG have already been shown to be involved in insulin signalling and glucose and lipid metabolism.
Further, visualizing the KEGG information of 97-gene and 9-gene datasets using Cytoscape allowed us to extract the cellular component, molecular function, and biological process information, as represented in Figs. 4 and 5, respectively. However, it hereby reveals that even the nine most crucial genes of MetS/T2DM and metformin do not encompass all entities within each specific group, indicating that metformin may not modulate or influence most of the target proteins, underlying the pathogenesis of MetS/T2DM.
The top 10 GO biological, cellular, and molecular processes for the selected genes with respect to the disease and the drug (metformin) are depicted in Fig. 6. The selected genes are associated with various biological processes such as hormone and lipid regulation, peptide hormone stimulation, regulation of glucose, and lipid metabolism (Figs. 6A & B). Cellular components are primarily associated with the phosphatidylinositol-3-kinase complexes (Figs. 6C & D). Further, molecular functions are particularly related to the signalling events, kinase activity, and nucleotide binding (Figs. 6E & F). It is apparent from the figure that both disease and metformin share significant commonalities in terms of biological processes, cellular components, and molecular functions, respectively.
Gene enrichment analysis of the selected genes. Figures (A), (C) and E represent the biological process, molecular functions, and cellular components associated with MetS/T2DM, respectively. In contrast, Figures (B), (D), and F depict the corresponding biological processes, molecular functions, and cellular components related to the effects of metformin. The abbreviation “DM” indicates type 2 diabetes mellitus.
Gene expression analysis
For the 97- and 9-gene datasets, the g: Profiler resultant heatmap cluster analysis reveals the involvement of these genes in MetS/T2DM. In the heatmap, rows depict genes, proteins, or biological activities, along with additional details such as identifiers and pathways, while columns represent different experimental conditions, datasets, and biological annotations, including gene ontology terms, pathways, and diseases.
For the 97-gene dataset, the heatmap (Fig. 7) clusters 22 specific molecular functions: NAD-dependent protein deacetylase activity, acetyl nucleotide binding, identical protein binding, protein kinase activity, receptor ligand activity, 1-phosphatidylinositol-4-5-bisphosphate 3-kinase activity, 1-phosphatidylinositol-4-bisphosphate 3-kinase activity, DNA-binding transcription-factor, fatty-acid transmembrane transporter activity, very long-chain fatty acid CoA-ligase activity, 1-phosphatidylinositol-3-kinase activity, D-glucose transmembrane transporter activity, NAD-dependent protein demyristoylase activity, monocarboxylic acid binding, long-chain fatty-acid CoA-ligase activity, long-chain fatty-acid binding, long-chain fatty-acid transporter activity, acetyl-CoA carboxylase activity, pentosyltransferase activity, insulin-receptor activity, adipokinetic hormone receptor activity, angiotensin type-II receptor activity in light blue, ranging from the statistical p-value 1.24 × 10− 8 to 4.993 × 10− 2, clinically proven to have a strong linkage to MetS/T2DM. The involvement of fatty acid metabolism and DNA-binding transcription factor-based gene-regulatory and signalling activities, highlights the widespread metabolic disruption caused by T2DM, as evidenced by the association with metabolic disorders related to insulin and angiotensin-II, demonstrating the clinical relevance.
However, the heatmap of the nine hub genes, associated with MetS/T2DM and metformin, depicts the expression or activity levels of these genes across various cellular compartments and human tissues (Fig. 8). Here, while the X-axis shows different compartments, such as the cytosol, endosome, mitochondrion, and nucleus, and various tissues, comprising blood, liver, brain, muscle, and skin, the Y-axis features the nine genes: IL-6, IRS1, LEP, mTOR (Mechanistic Target of Rapamycin), FOXO1, PPARG, PPARGC1A, SIRT1 and ADIPOQ. The color gradient, ranging from yellow to dark purple, indicates varying levels of gene expression or activity, with yellow representing higher levels and dark purple indicating lower levels. IL6 shows higher activity in the extracellular space and certain tissues like blood, indicating its involvement in immune responses and inflammation. LEP is found to be more active in the liver and the extracellular compartment, reflecting its role in metabolism and energy regulation. In contrast, genes like SIRT1 and ADIPOQ exhibit moderate to low expression across many compartments and tissues, with some areas showing distinct levels, suggesting a more specific or context-dependent role in cellular processes. These initial findings have sparked our interest in identifying these key genes and their associated proteins as significant prognostic markers to diagnose and target MetS/T2DM.
Robustly analyzing the key 97 genes, potentially associated with MetS and T2DM, this study presents a comprehensive pathway enrichment analysis. It is observed that molecular functions and the associated biological pathways of these genes majorly underscore the complex interactions that provide valuable insights into the genetic underpinnings, contributing to the onset/progression of MetS and T2DM. The study thus offers crucial pathways and potential targets for a credible therapeutic intervention, necessitating experimental validation, and highlights the importance of complex genetic contributions behind the disease pathogenesis.
Analysis of all the nine hub genes, associated with the functional roles of MetS/T2DM and metformin, based on p-value confirms their cruciality in the preliminary diagnostics and prediction of MetS/T2DM, necessitating experimental validation. As shown in Fig. 9, the survival plot of hub genes, estimated by the KMPlot server, underscores the results of transcription factor enrichment analysis. Specifically, Fig. 9A reveals that a set of 20 top-ranked transcription factors are intricately associated with MetS/T2DM, as well as the therapeutic effects of metformin. The PPI interaction network of all these key proteins is shown in Fig. 9B. Here, peroxisome proliferator-activated receptor- γ (PPARG) and peroxisome proliferator-activated receptor-Δ (PPARD) are majorly involved in lipid metabolism and insulin sensitivity, with PPARG being the key target for anti-diabetic drugs like thiazolidinediones60. As the hallmark of MetS/T2DM, androgen receptor (AR) is functionally associated with insulin resistance, as its disruption leads to dysregulation of glucose uptake and storage in liver and muscle tissues, resulting in elevated glucose levels61. Moreover, four transcription factors viz. GATA binding protein 1 (GATA1), nuclear transcription factor y subunit-α (NFYA), specificity protein 1 (SP1), and SRY-box transcription factor2 (SOX2) regulate genes associated with pancreatic β-cell function, and glucose homeostasis, which get impaired in T2DM62. Insulin signalling and glucose metabolism are also influenced by Pre-B-cell leukemia homeobox 3 (PBX3), and mothers against decapentaplegic homolog 4 (SMAD4)63. As key factors in MetS/T2DM pathogenesis, interferon regulatory factor-3 (IRF3) and ‘fos’ proto-oncogene (FOS) are associated with the inflammatory response64. To worsen the condition, BCL2-associated transcription factor-1 (BCLAF1), and upstream stimulatory factor-2 (USF2) further dysregulate the apoptotic pathways and cellular stress responses65. Epidermal growth factor receptor-1 (EGFR1) has been linked to insulin resistance, and its inhibition may enhance insulin sensitivity, the mechanism potentially targeted by metformin66. Another protein (zinc finger miz-type containing-1) is also shown to modulate the AR activity, indirectly affecting glucose metabolism and insulin sensitivity67. Altogether, these genes highlight the complex molecular interactions underlying T2DM and suggest the major pathways through which metformin exerts its therapeutic effects. PPI analysis reveals that PPARG, SMAD4, FOS, PPARD, UBTF, CTCF, EGR1, BHLHE40, and PBX3 play central roles in regulating the intricate interplay of regulatory molecules involved in metabolic syndrome (MetS/T2DM).
(A) The predicted transcription factors for the common nine hub genes between MetS/T2DM and metformin. The horizontal sliding bar highlights the hypergeometric p-value and the scores for the top 20 transcription factors. (B) The network of transcription factors (pink) and their major interaction partners (grey), are represented as a ball-and-stick diagram, with node size proportional to their degree.
Discussion and Conclusion
MetS/T2DM is a complex metabolic abnormality for which lifestyle modifications with concurrent administration of oral hypoglycaemic agents constitute the conventional treatment strategy to date. The drug classes act by a variety of mechanisms such as inhibition of hepatic gluconeogenesis (metformin), improving insulin secretion (sulfonylureas) and insulin sensitivity (glitazones), discouraging glucose absorption (alpha-glucosidase inhibitors), and promoting glucose excretion (SGLT2 inhibitors). Despite their potency and efficacy, most of them are associated with adverse complications, including hypoglycemia, pancreatitis, weight gain, and skin disorders. However, among these, metformin is used as a front-line drug either in mono or combinatorial therapy for treating T2DM. Moreover, because of its low cost, better efficacy, low risk of hypoglycemia and above all long-term safety with minimal side effects, it has become a preferable drug for T2DM patients18,47,68,69,70. However, its risk-to-benefit ratio should be thoroughly ascertained in comparison with the recently emerged glucose-lowering agents that are known to exhibit protective effects on diabetes complications71. Furthermore, MetS/T2DM is characterized by a complex cluster of symptoms with an increased risk for cardiovascular disorders. Hence, finding agents that could either modulate certain primary mediators/pathways, in turn influencing the downstream secondary mediators/pathways or concurrently target a broad range of disease-specific mediators/pathways with better efficacy and safety, would be a feasible futuristic therapeutic approach.
Network pharmacology approach is commonly employed to explore different candidate proteins or genes that could serve as targets or mediators for the therapeutic actions of drugs or novel chemical entities against specific diseases72. In the present study, we have developed a PPI network of the selected genes that are known to be critically involved in the pathophysiology of MetS/T2DM. Among 97 putative genes identified through extensive data mining, 89 genes are found to be interlinked within the PPI network, forming 123 connections. Further, this study has predicted 10 genes including ─ IRS1, IL6, FOXO1, PPARGC1A, MTOR, LEP, PPARG, ADIPOQ, PRKAA1, and SIRT1─ as the most crucial and key contributors to disease pathogenesis. Interestingly, Notably, these predictions are strongly supported by the existing literature evidence. Further, GO analysis of our study indicates that several biological, molecular, and cellular events are closely associated with the selected genes of MetS and T2DM.
Apart from this, interestingly, out of the above ten crucial MetS/T2DM-related genes, nine are predicted to be associated with metformin as well, except PIK3CA (as evident from the PPI network in Fig. 2). This overlap suggests that these genes may either have a significant direct or indirect role, especially from the standpoint of metformin’s therapeutic mechanism, which is also clearly evident from the literature. IL-6, LEP, and ADIPOQ are well-known adipocytokines or adipokines, with ADIPOQ being a prevalent anti-inflammatory adipokine secreted by adipocytes. It exhibits a negative correlation with insulin resistance and obesity and is also known to exhibit cardioprotective effects. In contrast, IL-6 and leptin (LEP), while regulating immune function and food intake under physiological conditions, are pre-dominantly pro-inflammatory, contributing to adipocyte hypertrophy, obesity, insulin resistance, and atherogenesis31,73. Sirtuins constitute a family of NAD + deacetylases with differential subcellular localization, and regulate a variety of crucial metabolic functions such as mitochondrial homeostasis, oxidative stress, autophagy, inflammation, and apoptosis. Among various Sirtuins, playing a substantial role in aging, obesity, T2DM, cancers, and cardiovascular diseases, SIRT1 and SIRT6 have gained considerable attention for their protective effects on insulin resistance and oxidative stress. Especially, through deacetylation, SIRT 1 regulates various non-histone proteins comprising PPARγ, PGC1α, liver X receptor (LXR), sterol regulatory element binding protein (SREBP), farnesoid X receptor (FXR), also known to protect pancreatic beta cells, promote myogenesis, adipogenesis, glucose and lipid utilization74,75.
Activation of adapter proteins, primarily, insulin receptor substrate (IRS) proteins, PI3-kinase (PI3-K), and Akt, Src-homology2 (SH2), and protein-tyrosine phosphatase1B (PTB1B) is the prerequisite for downstream insulin signalling and eventually glucose homeostasis. However, serine/threonine phosphorylation of IRS proteins by IRS kinases is found to be implicated in insulin resistance which further propels T2DM and various other comorbidities. Reports emphasize IRS 1 as a major target for IRS kinases76,77. The forkhead box O1 (FOXO1) is one of the important transcription factors that, upon phosphorylation by Akt, turns off transcription factor activity and and activates gluconeogenesis eventually leading to insulin resistance77,78. Similarly, mTORC1 is also one of the substrates of Akt and regulates cell cycle, mitochondrial biogenesis, adipogenesis, glycolysis, and autophagy. It is related to the mammalian target of rapamycin (mTOR), which is a serine/ threonine kinase. Through negative regulations of autophagy, mTORC1 favors protein misfolding, organelle impairment, and aging-associated disorders, including T2DM. In T2DM, hyperactivation of mTORC1 is known to contribute to pancreatic beta cell death68,79,80. AMP-activated protein kinase (AMPK), or PRKA, is a classical cellular energy sensor that is activated when the intracellular ATP levels are modest to low. It constitutes a catalytic α and two regulatory β and γ subunits. These subunits exist in different tissue specific isoforms, including PRKAA1 and PRKAA2 (α1 and α2 subunits); PRKAB1 and PRKAB2 (β1 and β2subunits); PRKAG1, PRKAG2, and PRKAG3 (γ1, γ2 and γ3 subunits). It chiefly regulates glucose, lipid, protein metabolism, mitochondrial homeostasis, and autophagy. AMPK facilitates muscle glucose uptake via positive modulation of glucose transporter 4 (GLUT4). Hence, the AMPK/ GLUT4 axis could be feasibly targeted for the therapeutic intervention of T2DM81,82. Peroxisome proliferator-activated receptors (PPARs) are affiliated with the nuclear receptor superfamily of transcription factors and exist as three subtypes (PPARα, PPARβ/Δ, and PPARγ) in humans. These are known to regulate the gene expression of a variety of mediators involved in metabolic homeostasis. Among the three types, PPARγ is the most frequently researched one which is expressed in both brown and white adipose tissues, in turn contributing to glucose and lipid metabolism, particularly regulating adipogenesis. PPARγ is one of the (crucial mediators of adipocyte differentiation and its activation by synthetic agonists, especially, thiazolidine-2,4-diones (TZDs) such as troglitazone and pioglitazone improves whole-body insulin sensitivity, glucose, and lipid metabolism58,83,84. In addition, peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α) is one of the members of peroxisome proliferator-activated receptor γ coactivator- 1 (PGC-1) involved in assisting the transcriptional regulation of genes related to metabolic homeostasis. It is considered a primary regulator of mitochondrial physiology and reported to be influenced by many different metabolic mediators such as mTORC, AMPK, SIRT1, cyclic AMP response element binding protein (CREB), Akt, and glycogen synthase kinase-3β (GSK-3β). On the other hand, there is a high probability of its association with disorders including obesity, T2DM, cardiomyopathy, and different cancers. Because of its regulatory role in mitochondrial biogenesis and function, it promotes fatty acid oxidation, thereby minimizing its intracellular accumulation. It also regulates glucose metabolism under the influence of hormones and nutrients. Hence, a defective status of PCG1α could act in favour of conditions like obesity and insulin resistance, thus, serving as a desirable pharmacological target85,86,87.
Based on the above findings, we believe that our study has fairly succeeded in identifying some valid hub genes that are predicted to contribute either to the progression or amelioration of the symptoms of MetS/T2DM In addition, intricate connections of the 10 hub genes with several other genes, indicate a feasible role of these broad networks at some point in the disease pathogenesis. Finally, we presume that a thorough insight into these networks supported by a systematic validation in suitable experimental models could certainly help decipher more reliable avenues for the diagnosis, and in particular, management of MetS/T2DM, thereby providing a new paradigm for rational and effective multi-targeting therapy.
Data availability
The supporting data for this study are available when reasonably requested from the corresponding author.
References
Heindel, J. J. et al. Vandenberg, L. N. Metabolism disrupting chemicals and metabolic disorders. Reprod. Toxicol. 68, 3–33 (2017).
Noubiap, J. J. et al. Geographic distribution of metabolic syndrome and its components in the general adult population: A meta-analysis of global data from 28 million individuals. Diabetes Res. Clin. Pract. 188, 109924 (2022).
Clemente-Suárez, V. J. et al. New insights and potential therapeutic interventions in metabolic diseases. Int. J. Mol. Sci. 24, 10672 (2023).
Lillich, F. F., Imig, J. D. & Proschak, E. Multi-target approaches in metabolic syndrome. Front. Pharmacol. 11, 554961 (2021).
M. Reaven, G. Banting lecture 1988: role of insulin resistance in human disease. Diabetes 37, 1595–1607 (1988).
Saklayen, M. G. The global epidemic of the metabolic syndrome. Curr. Hypertens. Rep. 20, 1–8 (2018).
Fahed, G. et al. I. Metabolic syndrome: updates on pathophysiology and management in 2021. Int. J. Mol. Sci. 23, 786 (2022).
Alberti, K. G. et al. Harmonizing the metabolic syndrome: a joint interim statement of the international diabetes federation task force on epidemiology and prevention; National heart, lung, and blood institute; American heart association; world heart federation; international atherosclerosis society; and international association for the study of obesity. Circulation 120, 1640–1645 (2009).
Zujko, M. E., Rożniata, M. & Zujko, K. Individual diet modification reduces the metabolic syndrome in patients before Pharmacological treatment. Nutrients 13, 2102 (2021).
Grundy, S. M., Brewer, H. B., Cleeman, J. I., Smith, S. C. & Lenfant, C. Definition of metabolic syndrome: report of the National heart, lung, and blood institute/american heart association conference on scientific issues related to definition. Circulation 109, 433–438 (2004).
Alberti, K. G. M., Zimmet, P. & Shaw, J. The metabolic syndrome—a new worldwide definition. Lancet 366, 1059–1062 (2005).
Grundy, S. M. Drug therapy of the metabolic syndrome: minimizing the emerging crisis in polypharmacy. Nat. Rev. Drug Discov. 5, 295–309 (2006).
Rus, M. et al. Prevalence and risk factors of metabolic syndrome: A prospective study on cardiovascular health. Medicina 59, 1711 (2023).
Tenenbaum, A., Fisman, E. Z. & Motro, M. Metabolic syndrome and type 2 diabetes mellitus: focus on peroxisome proliferator activated receptors (PPAR). Cardiovasc. Diabetol. 2, 1–7 (2003).
Jha, B. K. et al. Progress in Understanding metabolic syndrome and knowledge of its complex pathophysiology. Diabetology 4, 134–159 (2023).
Dandona, P., Aljada, A., Chaudhuri, A., Mohanty, P. & Garg, R. Metabolic syndrome: a comprehensive perspective based on interactions between obesity, diabetes, and inflammation. Circulation 111, 1448–1454 (2005).
Esser, N., Legrand-Poels, S., Piette, J., Scheen, A. J. & Paquot, N. Inflammation as a link between obesity, metabolic syndrome and type 2 diabetes. Diabetes Res. Clin. Pract. 105, 141–150 (2014).
Zhao, X. et al. The crucial role and mechanism of insulin resistance in metabolic disease. Front. Endocrinol. 14, 1149239 (2023).
Gregor, M. F. & Hotamisligil, G. S. Inflammatory mechanisms in obesity. Annu. Rev. Immunol. 29, 415–445 (2011).
Li, H. et al. Macrophages, chronic inflammation, and insulin resistance. Cells 11, 3001 (2022).
Rohm, T. V., Meier, D. T., Olefsky, J. M. & Donath, M. Y. Inflammation in obesity, diabetes, and related disorders. Immunity 55, 31–55 (2022).
Charo, I. F. Macrophage polarization and insulin resistance: PPARγ in control. Cell. Metab. 6, 96–98 (2007).
Lumeng, C. N., Bodzin, J. L. & Saltiel, A. R. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 117, 175–183 (2007).
Subramanian, V., Ferrante, A. W. & Obesity Inflammation, and Macrophages. In Emerging Societies-Coexistence of Childhood Malnutrition and Obesity 63, 259–268Karger, Switzerland, (2009).
Houstis, N., Rosen, E. D. & Lander, E. S. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature 440, 944–948 (2006).
Hotamisligil, G. S. & Erbay, E. Nutrient sensing and inflammation in metabolic diseases. Nat. Rev. Immunol. 8, 923–934 (2008).
Halberg, N. et al. Hypoxia-inducible factor 1α induces fibrosis and insulin resistance in white adipose tissue. Mol. Cell. Biol. 29, 4467–4483 (2009).
Lee, Y. S. et al. Increased adipocyte O2 consumption triggers HIF-1α, causing inflammation and insulin resistance in obesity. Cell 157, 1339–1352 (2014).
Saltiel, A. R. & Olefsky, J. M. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Invest. 127, 1–4 (2017).
Trayhurn, P. & Wood, I. S. Adipokines: inflammation and the pleiotropic role of white adipose tissue. Br. J. Nutr. 92, 347–355 (2004).
Roy, P. K., Islam, J. & Lalhlenmawia, H. Prospects of potential adipokines as therapeutic agents in obesity-linked atherogenic dyslipidemia and insulin resistance. Egypt. Heart J. 75, 24 (2023).
Trayhurn, P. Hypoxia and adipose tissue function and dysfunction in obesity. Physiol. Rev. 93, 1–21 (2013).
Wu, H. & Ballantyne, C. M. Metabolic inflammation and insulin resistance in obesity. Circ. Res. 126, 1549–1564 (2020).
Fatima, K., Hussain, Z. & Hamid, R. Oxidative stress and diabetic complication: A systematic review. Chem. Biol. Interface. 11, 182 (2021).
Shoelson, S. E., Herrero, L. & Naaz, A. Obesity, inflammation, and insulin resistance. Gastroenterology 132, 2169–2180 (2007).
Olefsky, J. M. & Glass, C. K. Macrophages, inflammation, and insulin resistance. Annu. Rev. Physiol. 72, 219–246 (2010).
Hameed, I. et al. Type 2 diabetes mellitus: from a metabolic disorder to an inflammatory condition. World J. Diabetes. 6, 598 (2015).
Dilworth, L., Facey, A. & Omoruyi, F. Diabetes mellitus and its metabolic complications: the role of adipose tissues. Int. J. Mol. Sci. 22, 7644 (2021).
Hayden, M. R. Overview and new insights into the metabolic syndrome: risk factors and emerging variables in the development of type 2 diabetes and cerebrocardiovascular disease. Medicina 59, 561 (2023).
Bianchi, C., Penno, G., Romero, F., Del Prato, S. & Miccoli, R. Treating the metabolic syndrome. Expert Rev. Cardiovasc. Ther. 5, 491–506 (2007).
Noale, M. et al. Reply to letter to the editor polypharmacy in elderly people with diabetes admitted to hospital. Acta Diabetol. 53, 859–860 (2016).
Alwhaibi, M. et al. Polypharmacy among patients with diabetes: A cross-sectional retrospective study in a tertiary hospital in Saudi Arabia. BMJ Open. 8, e020852 (2018).
Kothari, V., Galdo, J. A. & Mathews, S. T. Hypoglycemic agents and potential anti-inflammatory activity. J. Inflamm. Res. 9, 27–38 (2016).
Drzewoski, J. & Hanefeld, M. The current and potential therapeutic use of metformin—the good old drug. Pharmaceuticals 14, 122 (2021).
Ott, M. & Werneke, U. Metformin-associated lactic acidosis May be treatable with thiamine. Med. Hypotheses. 189, 111416 (2024).
Szymczak-Pajor, I., Wenclewska, S. & Śliwińska, A. Metabolic action of Metformin. Pharmaceuticals 15, 810 (2022).
Foretz, M., Guigas, B., Viollet, B. & Metformin Update on mechanisms of action and repurposing potential. Nat. Rev. Endocrinol. 19, 460–476 (2023).
Lv, Z. & Guo, Y. Metformin and its benefits for various diseases. Front. Endocrinol. 11, 191 (2020).
Kaneto, H., Kimura, T., Obata, A., Shimoda, M. & Kaku, K. Multifaceted mechanisms of action of Metformin which have been unraveled one after another in the long history. Int. J. Mol. Sci. 22, 2596 (2021).
Szklarczyk, D. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2021).
Brown, G. R. et al. A gene-centered information resource at NCBI. Nucleic Acids Res. 43, D36–42 (2015).
Pontén, F., Jirström, K. & Uhlen, M. The human protein Atlas—a tool for pathology. J. Pathol. 216, 387–393 (2008).
Milacic, M. et al. D’Eustachio, P. The reactome pathway knowledgebase 2024. Nucleic Acids Res. 52, D672–678 (2024).
Kohl, M., Wiese, S., Warscheid, B. & Cytoscape Software for visualization and analysis of biological networks. Methods Mol. Biol. 696, 291–303 (2011).
Shreya, S. et al. Tracing the anti-cancer mechanism of Pleurotus osteratus by the integrative approach of network Pharmacology and experimental studies. Appl. Biochem. Biotechnol. 195, 152–171 (2023).
Reimand, J., Kull, M., Peterson, H., Hansen, J. & Vilo, J. g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35, W193–200 (2007).
Clarke, D. J. B. et al. Ma’ayan, A. eXpression2Kinases (X2K) web: linking expression signatures to upstream cell signaling networks. Nucleic Acids Res. 46, W171–W179 (2018).
Kanehisa, M. & Goto, S. K. E. G. G. Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–D677 (2025).
Ahmadian, M., Suh, J. M. & Hsiao, J. J. PPARγ signaling and metabolism: the good, the bad and the future. Nat. Med. 19, 557–566 (2013).
Ghaibour, K. et al. Androgen receptor coordinates muscle metabolic and contractile functions. J. Cachexia Sarcopenia Muscle. 14, 1707–1720 (2023).
Kaimala, S., Kumar, C. A., Allouh, M. Z., Ansari, S. A. & Emerald, B. S. Epigenetic modifications in pancreas development, diabetes, and therapeutics. Med. Res. Rev. 42, 1343–1371 (2022).
Papagiannakopoulos, T. et al. Circadian rhythm disruption promotes lung tumorigenesis. Cell. Metab. 24, 324–331 (2016).
Morone, B. & Grimaldi, G. PARP enzymes and mono-ADP-ribosylation: advancing the connection from interferon-signalling to cancer biology. Exp. Rev. Mol. Med. 26, e17 (2024).
Zhao, H. et al. The regulation of circrna and LncRNA protein binding in cardiovascular diseases: emerging therapeutic targets. Biomed. Pharmacother. 165, 115067 (2023).
Dalle, S. & Abderrahmani, A. Receptors and signaling pathways controlling beta-cell function and survival as targets for anti-diabetic therapeutic strategies. Cells 13, 1244 (2024).
Szczerbinska, I. et al. Large-scale functional genomics screen to identify modulators of human β-cell insulin secretion. Biomedicines 10, 103 (2022).
Bar-Tana, J. mTORC1 syndrome (TorS): unified paradigm for diabetes/metabolic syndrome. Trends Endocrinol. Metab. 34, 135–145 (2023).
Ngoei, K. R. et al. Structural determinants for small-molecule activation of skeletal muscle AMPK Α2β2γ1 by the glucose importagog SC4. Cell. Chem. Biol. 25, 728–737e9 (2018).
Kowalska, J. & Wrześniok, D. Skin-related adverse reactions induced by oral antidiabetic drugs—A review of literature and case reports. Pharmaceuticals 17, 847 (2024).
Baker, C. et al. Should Metformin remain the first-line therapy for treatment of type 2 diabetes? Ther. Adv. Endocrinol. Metab. 13, 2042018820980225 (2021).
Hopkins, A. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 4, 682–690 (2008).
Kim, J. E. et al. The roles and associated mechanisms of adipokines in development of metabolic syndrome. Molecules 27, 334 (2022).
Zhou, S., Tang, X. & Chen, H. Z. Sirtuins and insulin resistance. Front. Endocrinol. 9, 748 (2018).
Kanwal, A. & Dsouza, L. A. Sirtuins and diabetes: optimizing the sweetness in the blood. Transl Med. Commun. 4, 3 (2019).
Boura-Halfon, S. & Zick, Y. Phosphorylation of IRS proteins, insulin action, and insulin resistance. Am. J. Physiol. Endocrinol. Metab. 296, E581–E591 (2009).
Li, M. et al. Trends in insulin resistance: insights into mechanisms and therapeutic strategy. Signal. Transduct. Target. Ther. 7, 216 (2022).
Puigserver, P. et al. Insulin-regulated hepatic gluconeogenesis through FOXO1–PGC-1α interaction. Nature 423, 550–555 (2003).
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017).
Guillén, C. & Benito, M. mTORC1 overactivation as a key aging factor in the progression to type 2 diabetes mellitus. Front. Endocrinol. 9, 621 (2018).
Herzig, S. & Shaw, R. J. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell. Biol. 19, 121–135 (2018).
Entezari, M. et al. N. AMPK signaling in diabetes mellitus, insulin resistance and diabetic complications: A pre-clinical and clinical investigation. Biomed. Pharmacother. 146, 112563 (2022).
Leonardini, A., Laviola, L., Perrini, S., Natalicchio, A. & Giorgino, F. Cross-talk between PPARγ and insulin signaling and modulation of insulin sensitivity. PPAR Res. 818945 (2009). (2009).
Sun, C., Mao, S., Chen, S., Zhang, W. & Liu, C. PPARs-orchestrated metabolic homeostasis in the adipose tissue. Int. J. Mol. Sci. 22, 8974 (2021).
Liang, H. & Ward, W. F. PGC-1α: a key regulator of energy metabolism. Adv. Physiol. Educ. 30, 145–151 (2006).
Abu, S. O., Arroum, T., Morris, S. & Busch, K. B. PGC-1α is a master regulator of mitochondrial lifecycle and ROS stress response. Antioxidants 12, 1075 (2023).
Qian, L. et al. Peroxisome proliferator-activated receptor gamma coactivator-1 (PGC-1) family in physiological and pathophysiological process and diseases. Signal. Transduct. Target. Ther. 9, 50 (2023).
Acknowledgements
The authors acknowledge their institute, and colleagues for the support required to complete this paper.
Author information
Authors and Affiliations
Contributions
AR, SRV, and KN conceived the idea and started building the initial datasets. AR, SK, RP, and KR performed the experimental steps, and with the contribution of SRV, wrote the first draft. AR, SRV, and MS reviewed the paper for grammatical errors. All authors have reviewed and proofread the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Runthala, A., Vulichi, S.R., Kosanam, S. et al. Exploring possible hub genes of metabolic syndrome and type 2 diabetes mellitus: a systematic network biology study. Sci Rep 15, 23108 (2025). https://doi.org/10.1038/s41598-025-95439-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-95439-3