Abstract
Breast cancer remains a formidable global health challenge, emphasizing the critical importance of accurate and early diagnosis for improved patient outcomes. In recent years, machine learning, particularly deep learning, has shown substantial promise in assisting medical practitioners with breast cancer classification tasks. However, achieving consistently high accuracy and robustness in the classification process remains a significant challenge due to the inherent complexity and heterogeneity of breast cancer data. This study introduces an innovative approach to optimize breast cancer classification using the CS-EENN Model by harnessing the combined power of Cat Swarm Optimization (CSO) and an Enhanced Ensemble Neural Network approach. The ensemble approach capitalizes on the strengths of EfficientNetB0, ResNet50, and DenseNet121 architectures, known for their superior performance in computer vision tasks, to achieve a multifaceted understanding of breast cancer data. CSO employed to optimize the architecture and hyperparameters of these neural networks, enhancing their performance by facilitating convergence and preventing overfitting. Experimental evaluations conducted on the publicly available ‘Breast Histopathology Images’ dataset from Kaggle demonstrate the effectiveness of the proposed approach. The CS-EENN model achieved an impressive accuracy of 98.19%, significantly outperforming conventional methods. These advancements expected to have a direct and favourable impact on the accuracy of breast cancer detection and subsequent treatment decisions.
Similar content being viewed by others
Introduction
Breast cancer is a global epidemic that poses a substantial danger to the lives of women everywhere. With an expected 2.3 million additional cases identified in 2020 alone, it is a worldwide public health problem1,2. The impact of breast cancer extends beyond statistics; it touches the lives of countless individuals, families, and communities, highlighting the critical need for early detection, accurate diagnosis, and effective treatment. The journey to combat breast cancer begins with a fundamental understanding of its complexities. Breast cancer is a heterogeneous disease, meaning it can manifest in various forms, exhibiting distinct genetic, molecular, and pathological characteristics. These variations in breast cancer’s subtypes necessitate precise classification and stratification to tailor treatment plans to individual patients, ensuring the best possible outcomes.
Traditionally, the classification of breast cancer has relied heavily on the expertise of pathologists, who examine tissue samples under a microscope to determine the cancer’s subtype and grade3,4. While this approach has served as the gold standard for decades, it comes with inherent limitations, including subjectivity, inter-observer variability, and the time-consuming nature of manual analysis. In response to these challenges, there has been a drive to explore ground-breaking solutions, with a particular focus on the realms of artificial intelligence (AI) and machine learning. The incorporation of AI and machine learning methodologies into the classification of breast cancer marks a transformative shift in this domain. These technologies have the potential to revolutionize the way we diagnose and treat breast cancer, offering new avenues for early detection, precision medicine, and improved patient outcomes5,6,7. The power of AI lies in its ability to analyse vast amounts of data, uncover intricate patterns, and make predictions based on objective criteria, transcending the limitations of manual analysis.
The journey of breast cancer classification through AI begins with the acquisition of medical imaging data, primarily in the form of mammograms, ultrasounds, and histopathological slides. These images serve as the foundation for AI-driven analysis, providing crucial information about the size, location, and characteristics of breast tumours8,9. Artificial intelligence models, especially deep learning architectures such as AlexNet, have exhibited remarkable proficiency in analysing these images10. They frequently surpass human experts in tasks like lesion identification and feature extraction. Beyond medical imaging, AI also delves into the realm of genomics, analysing the genetic makeup of breast cancer cells. Figure 1 shows the breast cancer. This enables the identification of specific mutations and biomarkers associated with different breast cancer subtypes, guiding treatment decisions and predicting patient outcomes. Additionally, AI-driven analysis of clinical data, such as patient histories and treatment responses, contributes to a holistic understanding of breast cancer, further enhancing the accuracy of classification.
One of the key advantages of AI-driven breast cancer classification is its potential for automation and standardization. Unlike human pathologists who may exhibit variability in their interpretations, AI models provide consistent and reproducible results. This consistency is vital in reducing diagnostic errors, ensuring timely interventions, and ultimately saving lives. Moreover, the automation of routine tasks frees up healthcare professionals to focus on more complex and patient-centric aspects of care. In recent years, the field of AI-based breast cancer classification has witnessed remarkable advancements, driven by the availability of large-scale datasets, increased computational power, and breakthroughs in deep learning11,12. These advancements have paved the way for the development of sophisticated AI models capable of not only classifying breast cancer subtypes but also predicting disease progression, treatment responses, and survival outcomes. AI’s potential in breast cancer extends beyond diagnosis to personalized treatment recommendations, risk assessment, and early intervention strategies. Computer-aided detection (CAD) technologies have become the subject of much study for their potential in spotting and categorising lung cancer. These CAD algorithms outperform human radiologists at spotting lung nodules as well as cancer in medical imagery. Image pre-processing, region of interest (ROI) mining, feature selection, as well as categorisation are the typical four processes of CAD-based lung cancer identification systems. Feature selection as well as categorisation, which rely on image processing to collect trustworthy characteristics, play crucial roles in improving the accuracy and sensitivity of CAD systems. Still, it’s not easy to tell which nodules are benign and which are cancerous13,14.
To help radiologists make more accurate diagnoses, researchers have resorted to deep learning approaches to solve this issue. The speed and precision of medical diagnosis, especially for prevalent diseases like lung and breast, have been shown to be greatly improved by the use of deep learning-based CAD systems, according to previous research. When opposed to typical CAD systems, deep learning-based systems’ unique network architectures allow them to effortlessly extract high-level characteristics from source images15,16. The sensitivity, false-positive rate, and processing time of CAD systems trained using deep learning are only a few of its weaknesses. As a result, there is a pressing need to create a quick, low-cost, and highly sensitive CAD system based on deep learning for detecting lung cancer. Nevertheless, there are obstacles on the road to using AI’s full potential in breast cancer categorisation. Ensuring the ethical and responsible use of AI, addressing data privacy concerns, and validating AI models in diverse patient populations are essential considerations17,18. In addition, AI should be seen as a supplement to, instead of a substitute for, human healthcare workers. Better, more patient-centred treatment is possible when AI is combined with human knowledge. This comprehensive introduction to breast cancer classification through AI sets the stage for a deep exploration of the subject. It underscores the urgency of finding innovative solutions to combat breast cancer’s multifaceted challenges and highlights the transformative impact that AI can have in achieving this goal. As we delve further into the realm of AI-driven breast cancer classification, we will uncover the intricacies of the technology, its applications in medical imaging and genomics, its role in precision medicine, and the ethical considerations that guide its integration into healthcare. The end aim is quite clear: using AI to better diagnose breast cancer earlier, optimise treatment techniques, and lessen the impact the disease has on individuals and communities.
Contributions of the work
-
Integration of Diverse designs: By combining the designs of EfficientNetB0, ResNet50, and DenseNet121, breast cancer data may be understood comprehensively improving classification accuracy by collecting a variety of variables.
-
By combining predictions from several models, the ensemble method lowers the possibility of false positives and negatives, increasing the precision of diagnosis.
-
CSO improves model performance by promoting convergence and avoiding overfitting, resulting in more dependable classifications. It does this by optimizing the architecture and hyperparameters.
-
The suggested CS-EENN model provides a flexible solution that can be adjusted to varied datasets and situations, making it suitable for a range of tasks including the categorization of breast cancer.
-
By combining CSO with a group of neural networks, a new method is presented that expands the field and pushes the limits of breast cancer classification methods.
-
The suggested model improves diagnostic capabilities, assisting in more precise and prompt breast cancer diagnoses by utilizing the combined strengths of several designs and optimization strategies.
The remainder of this article is organised as follows: The current strategies for categorising breast cancer are discussed in Sect. 2. Section 3 discussed about the limitations of current methods. Section 4 introduces the novel techniques proposed in this study, with a particular focus on the CS-EENN Model. Section 5 details the experimental procedures used to validate the research findings and presents the obtained results. Lastly, Sect. 6 presents the conclusions drawn from the experiments.
Related studies
Breast carcinoma stands out as the most prevalent global cancer, resulting in nearly nine hundred thousand annual fatalities. The potential to mitigate mortality lies in early detection and precise diagnosis, which can curtail its dissemination and protect against untimely losses. Researchers grappling with breast cancer (BC) confront multiple hurdles in distinguishing between benign and malignant tumours, as well as deciphering mild and advanced stages of the disease. Leveraging machine-learning algorithms, we can pinpoint and discern patterns across all tumour types. Sadly, each year, a substantial number of breast cancer patients succumb to inadequate diagnoses and treatments. Recent years have seen deep learning algorithms making significant strides in breast cancer detection19, yet ample room remains for enhancing these methods. Despite notable progress, the integration of deep learning methodologies within the machine learning framework offers an avenue to further boost efficiency. Their paper delves into a comparison of three distinct models using the openly available Break His dataset, showcasing an impressive version 2 of accurate classification. Remarkably, it achieved a training accuracy of 99% and a validation accuracy of 98%.
When it comes to female cancer deaths, breast cancer is by far the most common. That’s why it’s so important to catch the illness early on so that treatment can start right away. However, due to complicated artefacts and high noise levels in breast ultrasonography pictures, radiologists have substantial difficulties throughout the breast cancer diagnosis procedure. As a result, we intend to use technology to categorise breast cancers. In order to classify breast cancer, their article uses many different state-of-the-art deep CNNs using well-known designs as AlexNet, VGG-16, and ResNet18. Furthermore, we provide a unique CNN model, dubbed BCI-Net, optimised for breast cancer classification. Their model is further strengthened by the use of the Mish activation function20, which is widely used to improve neural network effectiveness and training dynamics. Breast Ultrasonography pictures (BUSI) is a publicly available collection of ultrasound pictures that we use to conduct the investigation. In our studies, the suggested model achieves an amazing hold-out validation accuracy of 98.70% on average. In addition, after going through a stringent five-fold cross-validation process, it has an average accuracy of 97.49%, with a standard deviation of only 1.14%. Their study can be an effective adjunct to clinical breast cancer screening.
Wireless capsule endoscopy (WCE) has brought astonishing changes to the imaging and diagnosis of the gastrointestinal (GI) tract through enabling the use of a non-invasive diagnostic method that does not cause any discomfort to the patient. Nonetheless, its widespread applications and modification come up with issues in efficacy, tolerance, safety and performance. Automated WCE systems’ role is essential in the timesaving option for detecting anomalies, as interpreting WCE imaging data demands specialists and time21. To mitigate each of these challenges, different solutions employing computer vision technology have been sought and tried, but with results that are still relatively inaccurate, and therefore requiring further improvement. Thus, we developed four multi-classification DL models including Vgg-19 + CNN, ResNet152V2, GRU + ResNet152V2, and ResNet152V2 + Bi-GRU to diagnose ulcerative colitis, polyps, and dyed-lifted polyps using the accessible WCE image databases. More specifically, their study employs a single DL model for differentiating three different GI diseases. The performance of these models assessed using parameters like accuracy, loss, MCC, recall, precision, NPV, PPV, and F1-score. Out of all the models, Vgg-19 + CNN yielded the highest classification accuracy of 99.45%; it even surpassed the other models proposed in the current study as well as some of the current state-of-the-art classifiers. These results demonstrated that the proposed Vgg-19 + CNN model could improve the GI disease diagnosis using WCE images.
Skin cancer one form of cancer that is prevalent worldwide and there is a lot of skin cancers detected every year it is characterized by the uncontrolled growth of cells within the skin, which also has potential to invade and damage other tissues and organs. Through the modern life changes and prolonged exposure to the sun, their invasive disease has become rampant. The emphasis of early detection and deg.Cell carcinoma is important because skin cancer is generally fatal. Their work presents a novel deep learning architecture known as DVFNet for skin cancer detection from dermoscopy image. To improve an image quality, in the first step of the filter anisotropic diffusion methods used for noise and artefacts reduction. Their hybrid approach incorporates the VGG19 with the Histogram of Oriented Gradients (HOG) since it improves feature extraction and reduces implementations with distinction of separating skin cancer features22. Imbalanced class problem in the ISIC 2019 dataset, a publicly available skin cancer image data set solved by the SMOTE Tomek technique. The study also employs the technique of segmentation to enhance exact identification of areas of skin in severe damage. A feature vector map, which made from the combination of HOG and VGG19 feature map, used as input for multiclass classification using CNN. The DVFNet model scored a comparative significantly high accuracy of 0.9832 on the ISIC 2019 dataset. The performance of the proposed model assessed by applying ANOVA statistical testing, thus, DVFNet can be useful for healthcare professionals to detect skin cancer at early clinical stages for immediate treatment.
Diabetic foot sores (DFS) are a grave threat to those with diabetes, making the skin of the foot vulnerable to damage because of neuropathy, leading to amputation. Their study is concerned with the verification and implementation process of an automatic classification system using deep learning techniques for AFS and DFS. The challenge met by creating a new model with convolutional capability, coupling it with Vgg-19. The method employed used two standard datasets to distinguish AFS and ischemic DFS correctly. To increase the training accuracy data augmentation strategies used, while for segmenting images, and for a better feature extraction UNet + + network was used. Classification performance of the proposed model compared with two well-established pre-trained classifiers, Inceptionv3 and MobileNet and proved better23. The model proposed has the accuracy of 99.05%, precision of 98.99%, and recall of 99.01%, MCC of 0.9801 and F1 score of 99.04%. The performance also confirmed statistically by ANOVA and Friedman tests, further strengthening these findings. The unique approach described in their work can be a valuable instrument for clinicians to enhance the diagnostic accuracy and timeliness of the identification of diabetic foot ulcers and improve patient management.
Skin cancer is among the most life-threatening cancers, which affects millions of people around the world every year. Skin cancer is an illness that is because of the development of epidermal cells that grow and penetrate other tissues and can migrate through the lymphatic system to affect the rest of the body. There is a heightened occurrence of the disease due to modification of lifestyle, and growth in exposure to the sun. Skin cancer, especially if diagnosed at an early stage, if diagnosed and treated on time, is easily curable, and any delayed diagnosis is likely to lead to increased morbidity and reduced quality of life. The present work proposes a new deep learning model namely Xception-ResNet101 (X_R101), which combines the best, features of two state of the art AI frameworks, the Xception and ResNet10124. The model classifies specific skin cancer types, including melanoma (Mel), melanocytic nevus (Mn), basal cell carcinoma (bcc), squamous cell carcinoma (Scc), benign keratosis (Bk), actinic keratosis (Ak), dermatofibroma (Df), and vascular lesion (Vl). To reduce the classification noise, a borderline SMOTE enhancing technique applied to reduce class imbalance. The presented X_R101 model is assessed on three datasets that can be easily accessed by the public, namely PH2, DermPK, and HAM10000 datasets and compared with four benchmark classifiers, namely MobileNetV2, DenseNet201, InceptionV3, and ResNet50 besides other methods. The proposed X_R101 model establishes an exceptional accuracy of about 98.21%, it makes it easier for dermatologists and other healthcare practitioners to diagnose skin cancer at the right time enhancing the quality of patient care.
PCA patients are at high risk of developing metastasis, a terminal stage for the disease and the main reason for death. In fact, despite all the recent breakthroughs in medical science, the chances that locally advanced PCA can become widespread remain rather difficult to foresee. Their study employs the machine learning system prototype to predict potential biomarkers that characterize metastatic PCA from localized PCA using DEGs and molecular pathways related to metastasis formation. It also employs two gene expression profiles of the GEO database: GSE32269 and GSE6919. These datasets consist of 226 samples of prostate tissue; 69 metastatic samples, 81 normal prostate samples and 76 localized prostate cancer samples. The Support Vector Machine used in a fine-tuned manner to select features, as well as to classify and compare gene activity in order to determine essential biomarkers. As the primary outcome, the study identifies DEGs that highlight the genomic activities that set metastatic PCA apart from localized cases25. The results are useful in gaining insight into the underlying molecular processes that give rise to metastases and for assisting in tool development for diagnosis. These biomarkers stand to improve PCA detection and the application of more effective treatment for the circumstances, which, in turn, stands to lead to the desired improvement in patient prognosis, and enhanced survival rates of metastatic prostate cancer.
The skin acts as the body’s biggest organ which plays an essential protective and regulatory role requiring its good health status. The most common cancer called skin cancer exhibited increasing global incidence while generating severe medical harms from late diagnoses. Skin lesion detection posed difficulties because of the ambiguous characteristics that made physicians observe overlapping patterns thus generating incorrect medical diagnoses. Deep learning proved itself as a diagnostic aid that processed intricate dermatology healthcare information for enhanced pattern recognition-based medical diagnosis. Their research developed a new mobile-friendly and efficient hybrid model that joins ConvNeXtV2 building blocks with focal self-attention methods to deal with data imbalances together with model complexity issues. The model used ConvNeXtV2 in its first two stages to extract superior local features but added subsequent focal self-attention processing which focused on dermatologically relevant areas during analysis. The investigated model underwent testing using ISIC 2019 dataset containing eight skin cancer categories that displayed extreme class variation like the Melanocytic Nevus class which posed 51 times more images compared to the Vascular Lesion class26. The model executed its diagnostic tasks efficiently throughout all types of classes to reach 93.60% accuracy and 91.69% precision with 90.05% recall and 90.73% F1-score. The proposed model proved more accurate by 10.8% compared to ResNet50 and 3.3% superior than Swinv2-Base featured in existing vision transformer research. By creating their design the authors set a new standard in skin cancer detection which enabled precise and expandable predictions to help medical personnel make early diagnoses and achieve better clinical results.
Cerebral vascular occlusions together with strokes maintained their position as significant worldwide healthcare challenges because they produced substantial mortality numbers while also triggering permanent disabilities. Medical practitioners needed to establish early diagnosis before the first hours to stop permanent damage and achieve positive patient recovery. The advancements in magnetic resonance imaging (MRI) techniques did not address the complexities present in brain lesion identification that traditional diagnostic methods still struggled to evaluate properly. The medical imaging field received a powerful tool through deep learning which demonstrates excellence at identifying and segmenting brain abnormalities. The review investigated 61 MRI-based studies within the time period from 2020 to 2024 to assess deep learning applications in cerebral vascular occlusion diagnosis. The review critically assessed successful aspects and obstruction points in these investigations together with dataset consistency along with a discussion on data protection mechanisms and machine learning explanation systems27. Different approaches using convolutional neural networks (CNNs) and Vision Transformers (ViTs) showed specific strengths and weaknesses when compared with each other. The research highlighted three key aspects for ethical security in frameworks alongside diverse dataset requirements and improved model readability. The article promoted U-Net variants and transformer-based models as dependable tools for clinical application development. Deep learning enabled advanced diagnostics through automation of challenging neuroimaging operations to generate more personal treatment recommendations. The review established framework-based integration of technical developments into clinical deployment which validated deep learning as a disruptive global solution for neurology treatment management and healthcare delivery enhancement.
Global women experienced cervical cancer as their primary health challenge thus requiring accurate diagnostic procedures to provide timely solutions. The Papanicolaou smear (Pap smear) test continued as the primary method for cervical cancer screening yet required extensive time for analysis along with frequent human mistakes. Medical experts identified through their discovery that automated diagnostic solutions would boost both speed and precision of medical work. Their paper examined deep learning algorithms for cervical cancer diagnosis automation through the analysis of Pap smear images. Analysis relied on a complete dataset that combined Mendeley Liquid-Based Cytology (LBC) dataset (963 images) with Malhari dataset (318 images) to provide a total of 1,281 images for evaluation. Twenty-seven state-of-the-art deep learning models composed of both Convolutional Neural Networks (CNNs) and Vision Transformers (ViTs) operated during the classification process. The medical image processing model received improvements through data enhancement methods alongside transfer learning techniques28. The high classification accuracy measured at 99.48% became a common result among the ViT-based models. EfficientNetV2-Small became the sole CNN-based model which matched the accuracy level among its 13 evaluated counterparts. The research confirmed that Vision Transformers outperformed all other models regarding diagnostic accuracy performance. The application of ViT-based deep learning methods demonstrated strong indications toward automatic systems that diagnose cervical cancer. The cervical cancer screening outcomes improved because these models reduced diagnostic errors and accelerated the diagnostic process which led to better and more reliable screening practices.
The world experienced severe breast cancer incidence which transformed into a life-threatening disease whenever early diagnosis failed to happen. The detection process of cervical cancer along with diagnosis and classification required the utilization of computer-aided detection (CAD) systems. The YOLO-based CAD algorithms gained popularity because they delivered exceptional results for object detection objectives during recent times. The research team carried out different experiments using INbreast data to evaluate detection performance between the commonly used YOLO models. For mammography mass detection the study established a YOLO model that incorporated a Swin Transformer as its backbone architectural component. The INbreast images at different sizes underwent comparative performance evaluation of YOLOv5 models and the transformer-based YOLO model with other YOLOv3 and YOLOv4 models29. The YOLOv5 transformer-based detection system delivered the finest results when processing images with a 832 × 832 pixel resolution. Before training began the YOLOv5 autoanchor function supplied its anchors to the default anchors for assessment and it was found that YOLOv5 autoanchor-generated anchors achieved higher success rates. Different experiments tested how performance changed when data augmentation techniques were applied. A limited amount of study data produced excellent performances by YOLO algorithms and demonstrated their potential for cancer detection tasks.
The distribution imbalance between different medical classes and inadequate labeled high-quality images were common issues in medical datasets. The deep learning models demanded significant amounts of already labeled data to complete successful classifications. The research introduced a few-shot learning method as a solution to categorize ultrasound breast cancer images through meta-learning approaches. The research adopted both Prototypical networks and model-agnostic meta-learning (MAML) algorithms for its meta-learning approaches. The experimental research used the BUSI breast ultrasound images dataset with its three classes during meta-testing in conjunction with other datasets for meta-training30. An accuracy between 0.882 and 0.889 was achieved through the application of ResNet50 backbone with ProtoNet during a 10-shot classification session. The model resulted in accuracy increases reaching between 6.27 and 7.10% higher than the baseline level of 0.831. All k-shot settings demonstrated ProtoNet produced better results than MAML. The ResNet models provided superior results as feature extraction backbones when compared to four-layer convolutional networks. Using the proposed method delivered both first-time meta-learning application for BUSI dataset while achieving superior accuracy than deep learning techniques when working with minimal medical image datasets featuring few classes. Post-study evaluation demonstrates that the applied approach could become applicable for datasets with comparable problems.
Meta-analysis of existing studies
The meta-analysis of previous works in medical imaging and classification demonstrates progress and open issues in the area. New generation models are generating accuracies in between 88 and 97%, where hybrid and ensemble technique models have proven to be more effective than single model frameworks. However, most of these methods are based on single datasets, with little variation in terms of demography or geography, and hence restrict the generalizability of the outcomes. This is evident in experiments conducted for diseases like breast cancer, skin lesions and diabetic foot ulcers, where train data insufficient in coming up with a diverse data set hence introducing bias when used in real applications. In addition, although, accuracy is given much attention as a measure of performance, other essential aspects such as sensitivity, specificity, and abilities to handle noisy or low quality data are rarely examined. For instance, a few health care driven studies that target classification of diabetic foot sores or gastrointestinal diseases consider the effects of noisy images or inconsistencies in imaging conditions. Also, the management of class imbalances, which is typical for the datasets where malignant cases are significantly out compared by benign cases, remains an issue because many models unable to apply rebalancing techniques such as oversampling or loss re-weighting. Other common themes are computational efficiency, as well as interpretability, as many modern models demand a great amount of computational power and frequently are considered ‘black boxes’, which hampers their applicability in real-world settings and adoption by clinicians. Thus, the proposed CS-EENN model is based on the strengths of current research, and on gaps, that future research could fill. EfficientNetB0, ResNet50, DenseNet121 suits implemented using CSO with an aim to alleviate mentioned limitations and provides enhanced accuracy, robustness, and flexibility for BC classification. Future studies therefore have to consider other types of data, improve on handling noise and unveiling methods in explainable artificial intelligence (XAI) for clinical use.
CS-EENN: enhanced breast cancer classification methodology
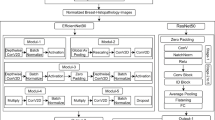
In the pursuit of more accurate breast cancer classification, a cutting-edge approach emerges: the fusion of Cat Swarm Optimization (CSO) with an ensemble of neural networks31. This innovative strategy, guided by the collective intelligence of CSO, aims to enhance diagnosis and treatment decisions in the battle against breast cancer. The proposed work addresses the robustness issue in breast cancer classification by integrating Cat Swarm Optimization with an Enhanced Ensemble Neural Network approach, optimizing model architecture and hyperparameters to prevent overfitting and improve accuracy. This innovative approach enhances classification reliability, contributing to more effective medical interventions.
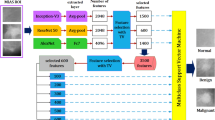
One such approach involves merging Cat Swarm Optimization (CSO) with a powerful ensemble of neural networks, including EfficientNetB032, ResNet5033, and DenseNet12134. CSO, inspired by the collective behaviour of cat swarms, is utilized to fine-tune these neural networks, enhancing their performance and convergence. EfficientNetB0, ResNet50, and DenseNet121 bring their unique capabilities to the table, contributing to a more holistic understanding of breast cancer data. By combining these perspectives within an ensemble, the classification accuracy is substantially improved. Figure 2 shows the proposed system framework architecture.
Dataset
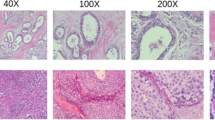
Invasive ductal carcinoma (IDC) is the most prevalent type of breast cancer among all diagnoses. Distribution of the dataset is presented in Table 1 below. When viewing a whole mount sample pathologist need to pinpoint the IDC positive areas in an attempt to assess the level of invasiveness. As aggressiveness is usually graded on sampled tissue sections, one common pre-processing step when attempting to automate aggressiveness grading is to delineate the IDC zones within a whole mount slide. The set up used in this was derived from the Kaggle repository that contains the dataset used35. The Breast cancer (BC) slides were scanned, and considering that the previous step was at a 20x lens, here 40x lens was used; 162 images. These photos were used to cut out 277,524 patches, all of which were 50 by 50 pixels. Of these 198,738 patches, the IDC status was negative whereas, in 78,786 patches the IDC status was good. Representative histopathologies of UBOs benign and malignant growths are presented in Fig. 3. This data collection would prove beneficial when doing a comparative analysis on different machine learning methods on improved breast cancer diagnostic tests and treatment.
Image pre-processing
Breast cancer image pre-processing using Gabor filtering is a valuable technique for enhancing texture features within medical images, aiding in the accurate diagnosis and prognosis of this critical disease36. The process involves the application of Gabor filters, which are mathematically defined convolution kernels. These filters are characterized by several parameters, including filter size (ksize), standard deviation of the Gaussian envelope (sigma), orientation (theta), wavelength (lambd), spatial aspect ratio (gamma), and phase offset (psi). The Gabor kernel is constructed using these parameters and subsequently applied to the breast cancer image using the `cv2.filter2D` function. This filtering process accentuates texture patterns in the image, revealing critical information that can be instrumental in further medical image analysis and diagnosis. By leveraging Gabor filtering with these equations, researchers and healthcare professionals can improve the interpretability and diagnostic accuracy of breast cancer images, ultimately leading to better patient care.
The 2D Gabor function is commonly used in image processing and computer vision for texture analysis and feature extraction. It can be defined as follows:
Where:
-
x and y are the spatial coordinates of the image.
-
f is the frequency of the sinusoidal factor.
-
θis the orientation of the Gabor filter.
-
λis the duration of a sinus wave’s period.
-
σis the Gaussian distribution’s standard deviation.
-
ψis the phase offset.
-
γis the spatial aspect ratio (usually set to 1 for isotropic filtering).
Additionally: \(\:{x}^{{\prime\:}}=x.\text{cos}\left(\theta\:\right)+y.\text{sin}\left(\theta\:\right)\)and\(\:{y}^{{\prime\:}}=-x.\text{sin}\left(\theta\:\right)+y.\text{cos}\left(\theta\:\right)\) are the rotated spatial coordinates.
The 2D Gabor function is applied as a convolution kernel to an image, and it is used to capture texture and edge information in different orientations and scales. The parameters f, θ, λ, σ and ψ can be adjusted to tailor the filter’s response to specific features in the image.
The pre-processing operations performed on Breast Histopathology Images dataset have an essential function to enhance model performance through structured normalization and noise reduction for deep learning model input preparation. Various pre-processing methods used for image optimization specifically affect model accuracy and speed of convergence and overall operational strength. The pre-processing procedures have the risk of producing biases that harm the CS-EENN model’s ability to generalize so it is crucial to perform a thorough assessment of their resulting effects. The deep learning system needs consistent shaped images so professional technicians perform image resizing as a mandatory step. The research conducted image resizing at 224 × 224 pixels for ResNet50 and DenseNet121 and 240 × 240 pixels for EfficientNetB0 because of its compound scaling approach. The model and computation run more efficiently because of image resizing. The process of down sampling can result in the elimination of vital fine-scale pathological features that medical experts use to identify benign and malignant disorders in tissues. Such small lesions together with subtle texture variations that indicate cancer progression may be lost during the process which could reduce the accuracy of classification outcomes. The model reached better convergence rates by normalizing pixel intensities through scaling operations which produced values between [0,1]. The normalization process generates equivalent scaling for all input features which helps backpropagation function without instabilities and speeds up learning processes. The normalization process enables deep learning models to detect more obvious structural features present in histopathological slide images. The normalization operation sometimes reduces the dataset variability therefore the model could have excellent performance in one particular dataset while exhibiting poor results when processing images from different institutions with varying staining and imaging approaches.
The experimental segment involved Gabor filtering to boost the detectability of histopathological image textures. The method strengthens significant structural features which produces better results during feature extraction and classification. The application of this technique improves essential features yet creates a potential drawback because extensive modification of image textures leads the model to depend excessively on artificial patterns instead of authentic biological structures. The model maintains reduced generalization capabilities for real-world histopathological features because of the introduced biases while analysing raw unprocessed medical images. The positive aspects of pre-processing cannot eliminate the introduction of biases that might undermine the model’s practical effectiveness within clinical applications. The resizing process leads to a major concern because it eliminates tiny microstructural details that could affect borderline case classification because such features often appear faint in histopathological examination. A potential bias occurs because standardization processes in dataset preparation cause the elimination of characteristic histopathological image variability. The model develops dataset bias that produces optimal performance on one dataset yet demonstrates limited ability to handle external datasets which contain different imaging conditions or patient demographic patterns or staining protocols.
The pre-processing methods sometimes introduce unwanted class imbalance effects to the dataset. The model tends to develop class bias toward the most prevalent data category when pre-processing methods apply different changes between benign and malignant case classes. Such classes will produce distorted classification outputs because the model shows superior performance for one category while achieving poor accuracy rates within the others. The use of Gabor filtering for texture enhancement creates a problem where the model learns artificial patterns instead of authentic histopathological structures which decreases its application reliability for clinical real-world data. Several strategies exist to reduce performance biases and enhance generalization as well as fairness in the model-based results. The combination of multi-resolution training provides a solution because it teaches the model to work with images at various scales to achieve appropriate detail preservation and operational speed. Testing model robustness requires external datasets from histopathology to assess its performance under different imaging conditions in addition to staining variations. The application of adaptive augmentation techniques helps maintain balanced transformations between benign and malignant case types which stops classification biases from emerging because of class-specific distortion.
Feature extraction
Feature extraction employing ResNet50 is a crucial component when carrying out deep learning for breast cancer classification. ResNet50 is a 50-layer residual network that is capable of capturing very sophisticated features from complex learned images such as the histopathological images. This structure proposed for addressing the vanishing gradient issue affecting the deep neural networks’ ability to perform optimally. This is made possible through the implementation of the residual blocks with skip connections, thus making it able to enjoys end-to-end differentiation and learn complexity patterns efficiently in medical images. It starts with pre-process the input images and they have to be of size 224 by 224 because ResNet50 works best on this size. These images further go through normalization, which makes pixel values range optimal for the performance of the model. In the case of ResNet50, low level features including edges, corners, and textures extracted by its first convolutional layers. Thus, as data advances through the subsequent layers of a network, it successively reveals increasingly intricate and general characteristics, such as geometric forms and structural arrangements that represent crucial information for the separation of normal and tumour tissue in breast cancer histopathology.
For feature extraction, one differentiation made in ResNet50 is the element of its residual connections, which enable it to learn features effectively without negative impact to performance, even if the network depth is deep. This is particularly useful for medical imaging jobs, where the ranking of variations in the texture and structure of the object can contain essential information. When used as a feature extractor, ResNet50’s convolutional layers thus serve as a pre-trained backbone, which passes down network knowledge learned from datasets such as ImageNet to the breast cancer classification task. During the feature extraction phase, the last fully connected and classification layers of ResNet50 are quite often discarded leaving the convolutional layers. These layers produce a dense vector of fixed dimensionality, which is the representation of the important features of the image. This feature vector is then passed into downstream classification tasks, where one could use ensemble neural networks, or support vector machines to classify the image into benign or malignant.
ResNet50 has the capability of automatic extraction of both the slight/large features eliminating the stress and cumbersome activity of feature engineering. Further, its capability of extracting hierarchical features adds to its flexibility by increasing the ability to capture intricate and non-linear features in the data affordable higher rate of classification accuracies. ResNet50 can provide reliable information assisting researchers and clinicians in the development of appropriate means to diagnose breast cancer in the early stages.
Ensemble neural network
When EfficientNetB0 and DenseNet121 are fully integrated as an ensemble neural network, their specialized characteristics guarantee a foundation for identifying breast cancer. This synergy enables rich features to be learnt and classified and helps overcome the issues pertinent to medical image analysis, especially the histopathological images. EfficientNetB0 is a serendipitous convolutional neural network with resource refinement strategies and compound scaling. Unlike other models in which the dimensions of depth, width and resolution can adjusted randomly, EfficientNetB0 adjusts them relatively. This strategy approach the trade-off of computational complexity and predictive power to a good extent, and therefore is perfect for tasks where there is a less computational prowess. To capture the small details and textures mapped to the breast cancer images, EfficientNetB0 offers high feature extraction accuracy within a light model, thus giving better training and usage time and low memory usage.
On the other hand, DenseNet 121 uses dense connections between the layers, hence it helps in reusing features and also helps passing gradients. This was achieved in a way that each of these layers obtains direct output of all other layers, which minimizes utilization of resources and improves learning capabilities. One of DenseNet121’s strengths is that it takes input from the previous layer and aggregates low and high level features making it highly capable of deciphering details in medical images. In the diagnosis of breast cancer histopathology where minimal differences in tissue architecture and texture are crucial, DenseNet121 gives a dense connection between layers such that no feature is left out in distinguishing between cancerous tissues and healthy tissues. The ensemble approach then combines these two architectures to help take advantage of this modularity. In the training process, two models, namely EfficientNetB0 and DenseNet121, take different inputs of images, the features of which are different. EfficientNetB0 also assists in this process by also as a global feature extractor and enhancer of high-resolution features, while DenseNet121 provides depth due to it Densely Connected layers used in analysing fine textures and spatial hierarchies. The outputs of these models then combined in some way via simple means like averaging, weighted averaging or stacking to produce the final prediction. Concatenation approach also helps in enhancing the performance besides increasing the model’s strength since weaknesses of various architectures are well addressed. When applied to breast cancer classification, this ensemble framework can credit for improvements in false positive and false negatives—which are key variables in diagnoses. Using both EfficientNetB0 and DenseNet121 guarantees that all data dimensions are captured before making accurate predictions as the machine learning models are also highly accurate. Additionally, the ensemble model practically operates nicely across different databases, which helps in practical usage of medical imaging since variability is always expected in the actual world.
By integrating EfficientNetB0 and DenseNet121 in an ensemble, researchers and clinicians can achieve state-of-the-art performance in breast cancer classification, enabling earlier detection and accurate diagnoses. This approach not only advances the field of medical imaging but also contributes significantly to improving patient outcomes through timely and reliable diagnostic insights. Once trained, the ensemble takes the stage during the prediction phase. Each individual model generates its own set of predictions for a given breast cancer image. These individual predictions then undergo a thoughtful aggregation process to form a final ensemble prediction. Various methods can be employed for aggregation, including:
Averaging
A straightforward approach where the predictions of all models are averaged, resulting in a consensus prediction.
In this equation, F represents the final ensemble prediction obtained by averaging the predictions (Pi) from individual models (M).
Weighted averaging
Assigning different weights to each model’s predictions based on their respective performance on validation data. Models demonstrating higher validation accuracy are assigned greater importance in the final prediction.
Weighted averaging assigns different weights (wi) to the predictions of individual models based on their performance on the validation set.
Stacking
Training a meta-model (e.g., a neural network or a classifier) that takes the individual model predictions as input and learns to make the final prediction based on them. This approach allows for more intricate relationships between the models’ outputs.
Stacking involves training a meta-model (e.g., a neural network) to combine the individual model predictions (Pi).
Voting
Using majority voting to determine the final prediction. The class with the most votes from the individual models is selected as the ensemble’s prediction.
Voting selects the class with the majority of votes from individual models as the final prediction.
The result of this aggregation process is a robust and refined classification decision. By leveraging the collective intelligence of multiple models, the ensemble minimizes the risk of false positives and false negatives, two critical factors in breast cancer diagnosis. This enhanced prediction accuracy is pivotal for early diagnosis and timely intervention, both of which are essential in breast cancer management.The impact of the ensemble neural network using EfficientNetB0 and DenseNet121 for breast cancer image categorization is profound. By combining the computational efficiency of EfficientNetB0 and the rich feature learning capabilities of DenseNet121, the ensemble excels in capturing a wide spectrum of features from breast histopathology images. These features range from subtle textural patterns and cell structures to broader tissue characteristics.
Choosing EfficientNetB0, ResNet50, and DenseNet121 as an ensemble method in this study is due to the synergy between their performance and availability of the models for computer vision tasks, specifically in medical imaging. EfficientNetB0 chosen due to compound scaling that allow the model to be more accurate with less computational power than comparable models. By having both the advantages of high parallelism for image processing and lower hardware costs, this balance becomes especially beneficial for applications involving medical imaging where a large amount of data needs to be processed within a short time frame, for example breast histopathology images. ResNet, especially ResNet50 as covered, is memorable for the inclusion of residual connections that minimize the occurrence of the vanishing gradient problem hence making it possible for the network to train very deep architectures. This capability is essential to summarization, which is important when identifying hierarchical features in histopathological images of tumour tissues. For example, DenseNet121 has densely connected layers, which helps in reusing the features achieved and in flowing gradients. This increases connections between layers of DenseNet121 thereby capturing fine details such as variations in tissue morphology adequately. By using these architectures, and utilizing them to construct an ensemble method, it becomes possible to combine all of these approaches into a classification environment that is extremely strong. EfficientNetB0 being a lightweight model offers efficiency and scalability, ResNet50 gives efficient feature extraction, DenseNet121 is useful in feature reuse. Altogether, these models make a synergy, guarding against such mistakes as false positives or false negatives when analysing complex medical images where one model can fail the other. This synergy is highly important in the breast cancer diagnosis where any mistake can lead to adverse consequences. Though there are other pretrained models like VGG, Inception and MobileNetetc, the above-mentioned architecture has given better performance in various medical imaging problems. Its capacity to deal with data with a high level of dimensions, non-concentration of learning, and high dependable exactness makes them appropriate for the proposed strategy. The separate models effectively augment the ensemble’s classification capability, proving that the process of selecting and assigning models carried out sensibly with regard to the requirements of this investigation.
The CS-EENN model incorporated EfficientNetB0, ResNet50 and DenseNet121 because these architectures demonstrated peak performance results for medical image analysis of breast cancer histopathology classification. Each architecture contributes unique advantages for ensemble learning which improves feature extraction as well as enhances classification precision and system reliability. The residual learning framework found in ResNet50 serves as the main reason for its selection since it removes the common deep network gradient vanishing issue. The specific design of this architecture leads to improved gradients throughout the model’s structure while permitting it to understand complex hierarchical features which detect breast cancer efficiently. Histopathological images benefit from the skill of ResNet50 to capture features in both low-level and high-level that help distinguish cancerous patterns from normal tissues. DenseNet121 was used as a network system because of its dense pattern that enables both feature reuse and consistent gradient propagation throughout different network layers. The model’s characteristic enables more advanced pattern detection in breast tissue samples which leads to superior classification results. A key advantage of DenseNet121’s high-level feature exploitation shines in histopathological examinations since it preservers essential textural properties to separate benign from malignant tissues. In this study EfficientNetB0 received selection due to its compound scaling function which achieves an optimal ratio between model resolution and depth as well as width parameters. This architecture surpasses conventional models by achieving better accuracy levels without losing performance speed which makes it perfectly suitable for dealing with large-scale histopathology data in this study. The CS-EENN model maintains accurate classification results while avoiding substantial computational strain because of its light-weight design which makes it adaptable to real-time medical settings.
Cat swarm optimization (CSO)
Cat Swarm Optimization (CSO) is a nature-inspired algorithm that can be harnessed to optimize the hyper parameters of an ensemble neural network used in breast cancer classification. In the realm of medical imaging, particularly breast cancer diagnosis, the accuracy and reliability of machine learning methods are of paramount importance. Ensembles of neural networks, which combine the predictions of multiple base models, have proven to be highly effective. However, selecting the optimal hyper parameters for these models can be a formidable task. CSO offers a solution by emulating the collaborative behaviour of cats in a swarm. In the context of parameter tuning for an ensemble neural network, CSO operates as a search algorithm that efficiently explores the hyper parameter space. It does so by evaluating different combinations of hyper parameters and assessing their impact on the ensemble’s performance.
During CSO optimization, a population of “cats” represents different sets of hyper parameters. These cats collaborate to find the best combination that maximizes the ensemble’s accuracy, F1-score, or another relevant performance metric. They do so by iteratively updating their positions based on their individual and group experiences, mimicking the way cats adapt to their environment. The fitness function guides the cats’ exploration, quantifying how well a particular set of hyper parameters enhances the ensemble’s ability to classify breast cancer accurately. CSO continues its search through numerous iterations, gradually converging towards the optimal hyper parameters.
Once the CSO algorithm completes its run, it provides the ensemble with a set of hyper parameters that have been fine-tuned to achieve the highest classification accuracy. These optimized hyper parameters enhance the model’s ability to discern malignant and benign breast cancer cases accurately. This, in turn, aids medical professionals in making informed decisions about patient diagnoses and treatment plans. In summary, Cat Swarm Optimization for parameter tuning in an ensemble neural network for breast cancer classification represents an innovative approach to enhance the performance and reliability of machine learning methods in a critical medical application. By automating the hyper parameter optimization process, CSO streamlines the development of accurate and robust breast cancer classifiers, ultimately contributing to improved patient care and more timely interventions. CSO is a nature-inspired optimization algorithm that operates through iterative steps mimicking the collaborative behaviour of cats in a swarm.
Let’s consider optimizing a single hyperparameter θ using CSO:
-
1.
Define N cats (solutions) in the population.
-
2.
Define D as the dimensionality of the search space, typically representing the range of values for the hyperparameter θ.
-
3.
Initialize the position Xi (where 1 ≤ i ≤ N) of each cat with a random value within the search space.
-
4.
Define a fitness function f(Xi) that evaluates the performance of the neural network with the hyperparameter set θ to Xi.
-
5.
During each iteration:
-
Evaluate the fitness of each cat Xi by computing f(Xi).
-
Update the position Xi of each cat based on the CSO dynamics.
A simplified equation to represent the update of a cat’s position Xi during a iteration could be:
Where:
-
\(\:{X}_{i}^{\left(t+1\right)}\)is the updated position of the i-th cat in iteration t + 1.
-
\(\:{X}_{i}^{\left(t\right)}\)is the current position of the i-th cat in iteration t.
-
\(\:{\Delta\:}{X}_{i}^{\left(t+1\right)}\)represents the change in position determined by the CSO technique based on the cat’s collaborative interactions with other cats and its exploration of the search space.
The specific calculation of \(\:{\Delta\:}{X}_{i}^{\left(t+1\right)}\)depends on the CSO variant and dynamics being used, which typically involves elements of randomness, collaboration, and adaptation based on the fitness values of the cats.
The Cat Swarm Optimization (CSO) algorithm selected due to its efficiency in fine-tuning hyperparameters for various types of neural networks. Compared with existing optimization techniques such as Genetic Algorithms and Differential Evolution, the results of the present study showed that CSO had better computational speed and flexibility, especially in the medical image processing experiments. Concerning Genetic Algorithms and Differential Evolution, CSO exposed better computational rates and flexibility that made it a preferred approach in medical imaging.
The hyperparameters of the proposed ensemble model was tuned via Cat Swarm Optimization (CSO), which is a metaheuristic optimization algorithm that has demonstrated to work exceptionally well for high and large dimensional search problems. Other hyperparameters, including learning rate, dropout rate, and the batch size that were optimized for every neural network in the ensemble, including EfficientNetB0, ResNet50, and DenseNet121. The CSO algorithm then perform a cyclic search on these weights using a fitness function based on validation accuracy that allowed for the fine balance of exploration and exploitation. More specifically, for ResNet50 and DenseNet121 we obtained the optimal learning rate of 0.001, whereas for EfficientNetB0, we had a somewhat lower value of 0.0005. Tuning dropout rates to 0.25, 0.3, and 0.2 for ResNet50, DenseNet121, EfficientNetB0 to prevent overfitting while keeping the model capacity. The chosen batch sizes are 64 for ResNet50 and DenseNet121, and 32 for EfficientNetB0 as they are optimal for providing the best computations per epoch.
Table 2 shows the Cat Swarm Optimization (CSO) Parameter Settings and Fine-Tuned Values. The Cat Swarm Optimization (CSO) algorithm performed optimization tasks on CS-EENN model hyperparameters in order to boost learning performance and enhance classification precision levels. Each chosen parameter range followed previous research and deep learning best practices which ensured high-quality operational performance. Stable convergence along with safe weight updating was achieved through the learning rate optimization from 0.0001 to 0.01 which resulted in choosing 0.0005 for EfficientNetB0 and 0.001 for ResNet50 and DenseNet121. The selected dropout rates for EfficientNetB0 amounted to 0.2 but ResNet50 required 0.25 and DenseNet121 needed 0.3 to prevent overfitting and maintain learning efficiency. A batch size of 32 was selected for EfficientNetB0 to run efficiently but ResNet50 and DenseNet121 received a batch size of 64 to achieve stability in training. Xavier initialization was used for EfficientNetB0 to achieve smoother gradients but He Normal initialization provided better gradient propagation for both ResNet50 and DenseNet121. Adam and RMSprop were evaluated as the best choices of optimizers for EfficientNetB0 and ResNet50 but RMSprop proved most suitable for ensuring training stability in DenseNet121.
Novelty in proposed work
The proposed work introduces a novel approach to breast cancer classification by synergizing multiple advanced techniques. Firstly, Cat Swarm Optimization (CSO) is employed to enhance neural network architecture and hyperparameters. Inspired by collective cat behaviour, CSO offers an unconventional yet effective optimization method for deep learning in medical image classification. Secondly, the study integrates an Enhanced Ensemble Neural Network (EENN) approach, which combines three renowned neural network architectures—EfficientNetB0, ResNet50, and DenseNet121. These architectures, known for their exceptional performance in computer vision tasks, provide a comprehensive perspective on breast cancer data patterns. The integration of these diverse architectures into a unified ensemble strategy is a pioneering advancement, resulting in a robust classification framework surpassing individual network capabilities. Additionally, the study addresses overfitting, a common challenge in deep learning, by applying CSO to optimize network architecture and hyperparameters. This preventive approach ensures effective convergence without overfitting, particularly crucial in medical image classification. Overall, the novelty of the proposed work lies in its holistic approach to breast cancer classification, combining unconventional optimization techniques, ensemble strategies, and top-tier neural network architectures. By redefining boundaries and improving classification accuracy and reliability, this study contributes significantly to advancing healthcare applications of deep learning.
Results and discussion
The dataset utilized in the proposed work consists of 162 breast cancer slides scanned at 40x magnification, resulting in 277,524 patches sized 50 × 50 pixels. It includes 198,738 IDC-negative and 78,786 IDC-positive patches for training and testing. The proposed model underwent simulation using Python version 3.6.5, utilizing various supplementary packages, including TensorFlow with GPU-CUDA support, keras, NumPy, pickle, matplotlib, scikit-learn, pillow, and OpenCV-python. The simulation executed on a personal computer equipped with the following hardware specifications: an Intel Core i5-8600k processor, 16GB of RAM, a 250GB SSD, and a 1 TB HDD. The model’s main variables were set as follows: 64-person batches, 50-epoch training periods, a 0.05-percent learning rate, a 0.25-percent dropout rate, and the use of rectified linear unit (ReLU) activation.
Evaluation metrics
The evaluation of deep learning algorithms relies on a diverse set of performance metrics, each offering unique insights into the model’s effectiveness. These metrics include accuracy, precision, sensitivity, specificity, F1_score, error, and mean squared error (MSE). Accuracy serves as a fundamental measure, gauging how well the model’s predictions align with the actual outcomes, providing a broad overview of its performance. Sensitivity and specificity come into play when there are specific concerns about false negatives (FN) or false positives (FP). Sensitivity highlights the model’s capacity to correctly identify positive instances, crucial in scenarios where missing a positive case has severe consequences. In contrast, specificity measures the model’s accuracy in identifying negative instances, significant when minimizing false alarms is vital. Accuracy in positive forecasts is measured by comparing the number of actual successes to the total number of successes. When dealing with circumstances with unequal class distributions, the F1_score ensures a fair trade-off between accuracy and recall. A visual and quantitative tool for optimising the model’s detection sensitivity across thresholds is the receiver operating characteristic (ROC) curve and its corresponding area under the curve (AUC). In the real world, deep learning algorithms perform better when their accuracy, precision, sensitivity, and specificity scores are high and their mistake rates are low. All of these measures work together to help evaluate and improve deep learning models so that they may be used effectively in a wide range of contexts.
After gathering the histopathology breast cancer images from the dataset shown in Fig. 4, the next steps typically involve pre-processing, analysis, and potentially the development of a machine learning or deep learning model for classification or diagnosis. Histopathology breast cancer image pre-processing using Gabor filters shown in Fig. 5 is a valuable technique in the field of medical image analysis. Gabor filters are specifically designed to capture texture information within images, making them particularly useful for analysing tissue textures and patterns in histopathological images. Histopathological images often contain intricate texture patterns that can provide important diagnostic information. Gabor filters are employed to extract these textures by analysing the variations in pixel intensities at different orientations and scales. Gabor filters are characterized by their orientation and frequency parameters. By applying Gabor filters with various orientations and frequencies to an image, different texture features can be extracted. Figure 6 shows the feature extracted images of benign and malignant. This enables the detection of textures that are indicative of specific tissue structures or abnormalities associated with breast cancer. Figure 7 illustrates the predicted outcome.
The size of the images was decided depending on whether it would maintain the necessary diagnostic information while keeping the computation moderate given the varying architectures. This is because the ResNet50and DenseNet121 achieved better performances with images resized to 224 × 224 pixels thereby captured the basic spatial features required for efficient feature extraction. EfficientNetB0, which has developed with the compound scaling strategy, had a slightly greater input resolution of 240 × 240 to capture detailed features necessary to differentiate benign and malignant tissues. The operations were carried out using different image sizes and each model was thus able to perform optimally at the highest possible resolution thereby providing a holistic feature map. This made it possible to obtain consistency and robustness of the ensemble model across weight architectures. Small learning rates for EfficientNetB0 increased cross-entropy to enhance the generalizability of the model while for ResNet50 and DenseNet121, higher learning rates were used to retain training convergence. In the same manner, the range of image sizes allowed for extracting sharp features and at the same time, retained fast compute times.
Accuracy comparison
Table 3 provides a synopsis of the experiment’s accuracy outcomes of benign class. In comparison to five other popular models, such as the AlexNet, Inception, GoogleNet, DenseNet121, and ResNet50 (all shown in Table 3; Figs. 8, 9, 10 and 11), the suggested techniques do rather well in terms of classification accuracy.
The AlexNet model achieves classification performance through precision of 0.94 and sensitivity of 0.93 along with specificity of 0.92 and overall accuracy of 0.92. The AlexNetmodel demonstrates successful class discrimination which allows it to predict accurately throughout most situations. High precision values demonstrate that the model detects few inaccurate positive results that would lead to serious consequences in important applications. The model’s high sensitivity value of 0.93 demonstrates its ability to properly detect most real positive cases thus minimizing the number of incorrect negative results. The strong generalization power of AlexNetexists despite its challenging ability to identify negative cases correctly because its specificity value equals 0.92. The results from AlexNetdemonstrate a competent performance that achieves reliable outcomes. The model performance can reach its full potential through adding transfer learning together with optimization techniques such as data augmentation and hyperparameter tuning to attain superior classification results.
Inception model delivers precision and sensitivity performance of 0.92 as well as specificity and accuracy measurement of 0.91. The network gained recognition for its efficient feature analysis of fine details and coarse details through its Inception modules. Due to its precision rate of 0.92 this model produces limited wrong positive classifications while demonstrating strong classification capabilities. The sensitivity rate of 0.91 indicates the model will overlook several positive cases which contributes to its higher amount of false negatives when compared to AlexNet. The model achieves 0.91 of specificity indicating its ability to identify negative samples accurately which minimizes incorrect classifications. Even though Inception achieves higher computational efficiency compared with deeper networks it demands substantial training resource requirements. The performance of the model can be enhanced through model pruning techniques that combine with efficient hyperparameter optimization for reduced computational complexity.
The GoogleNet model based on the Inception architecture produced precision of 0.91 with sensitivity of 0.91 along with specificity of 0.93 and accuracy of 0.89. The framework functions as an efficient model which combines superior efficiency with decreased computational resource needs thus facilitating large-scale classification functions well. With a precision score of 0.91 GoogleNet successfully prevents false positives yet it demonstrates minor areas where it fails to correctly classify items when compared to other models. Among the Type II models the sensitivity score of 0.91 indicates that the system detects positive cases reliably however its accuracy rate of 0.89 stands slightly below that of other models because it produces occasional classification errors. The model shows superior ability in accurate negative classification due to its high specificity value of 0.93. The accuracy of GoogleNet as a classification model remains competitive yet it requires either extra tuning or ensemble learning tactics to enhance its precision.
The DenseNet121 model shows precision value at 0.93 while sensitivity reaches 0.92 and specificity reaches 0.93 and accuracy reaches 0.92. Feature reuse efficiency stands as a hallmark characteristic of DenseNet since it passes gradients and features between layers thereby decreasing instances of vanishing gradients. The design functionality enables better learning outcomes that occur without using extra parameters which results in superior classification results. The model displays high accuracy in preventing false positives because its precision reaches 0.93. The 0.92 sensitivity and 0.93 specificity evaluation of the model demonstrates uniform capabilities in correct detection of positive cases and accurate dismissal of negatives. The performance level of 0.92 indicates similar efficiency to AlexNetand ResNet50 which confirms its effectiveness. Extensive network connections in the model lead to higher memory usage. The performance of the model could be enhanced through two strategies: using compact versions of DenseNet or implementing dropout based layers to maintain operational efficiency while increasing generalization capabilities.
ResNet50 demonstrates precision at 0.94 followed by sensitivity at 0.93 and specificity at 0.92 and accuracy at 0.93. Residual learning remains a core component of this model because it solves deep learning network problems with gradient vanishing. The model demonstrates high reliability due to its 0.94 precision figure which means it reduces false positives effectively in critical situations. The model demonstrates excellent performance regarding true positive detection because its sensitivity measure stands at 0.93. The 0.92 specificity value indicates that the model experiences moderately challenging precision challenges when determining negative cases. ResNet50 maintains a dynamic relationship between its indexing power and computational requirements to become a leading model system for identification tasks. Attaining higher performance from the model could be achieved through fine-tuning methods that include transfer learning combined with batch normalization and data augmentation techniques.
The CS-EENN model surpasses all competing models with precision at 0.97 and sensitivity at 0.97 and specificity at 0.96 along with accuracy reaching 0.98. The exceptional results show that CS-EENN demonstrates exceptional capability in positive case identification (high sensitivity) and negative case rejection (high specificity). With a precision of 0.97 the model identifies few potential false cases leading to enhanced credibility for scenarios in practical applications featuring severe effects from inaccurate predictions. With sensitivity (0.97) along with specificity (0.96) CS-EENN demonstrates superior robustness that delivers an overall accuracy rating of 0.98 which stands as the highest among all tested versions. The generalization capabilities of CS-EENN improve through ensemble learning strategies that unite several architectural components. While this model achieves superior performance in its field its computational costs might be considered too high. Model compression methods supported by knowledge distillation combined with quantization methods would enable accuracy maintenance together with improved computational performance.
The Fig. 12 compares the accuracy of different optimizers in breast cancer classification. The proposed Cat Swarm Optimization (CSO) achieves the highest accuracy of 0.97, outperforming Adam with 0.93 and SGD with 0.91. CSO demonstrates superior optimization capability, leading to more accurate classification results compared to traditional optimizers like Adam37 and SGD38. This highlights the effectiveness of CSO in enhancing the performance of machine learning models for breast cancer diagnosis.Altogether, these metrics substantiate that not only the accuracy of the proposed model achieved, but also its optimization for SVHS diagnosis that would keep sensitivity and specificity levels in an appropriate range crucial for the real-world medical applications. However, these evaluations demonstrate the efficacy of the proposed model, other elements, including robustness to noisy data, managing with the class imbalance problem, and presenting more information about the model’s performance, can further investigated in the future. All such considerations would add further credence to the model and its usefulness for deployment in clinician practice.
The Cat Swarm Optimization (CSO) process and its relationship to the CS-EENN model appears in Fig. 13. CSO must initialize its process with a set of cats which serve as potential solutions that search the hyperparameter domain. Before the CSO iterative process takes over with movement and seeking mechanisms to update positions, each cat receives evaluations based on performance from the fitness check.The system terminates its operations when optimal hyperparameters emerge. The optimized set of parameters including learning rate dropout rate and batch size which result from this process are afterward used to refine the EfficientNetB0 and ResNet50 and DenseNet121 models. The combination of breast histopathology image features from separate models through an ensemble system leads to better classification accuracy. The CS-EENN model optimizes its performance through CSO-driven optimization for breast cancer classification by achieving robust feature learning and improving generalization capabilities as well as minimizing overfitting.
The Table 4; Fig. 14 offers a comprehensive insight into the performance of the proposed CS-EENN model as it undergoes training with varying numbers of epochs, spanning from 5 to 50. It is evident from the data that the model’s performance, measured through a likely accuracy metric, exhibits a noteworthy trend of improvement with increased epochs. The CS-EENN model embarks on its training journey with a modest accuracy of 0.83, showcasing that even with minimal training, it demonstrates a reasonable level of accuracy. As the number of training epochs doubles to 10, the model’s accuracy significantly improves to 0.87, underscoring the positive impact of increased exposure to training data and iterations on model performance. With further progression to 15 epochs, the model’s accuracy reaches 0.90, marking a notable stride in its proficiency. This indicates that the additional training epochs are indeed beneficial for enhancing its task-specific capabilities. At 20 epochs, the model achieves an accuracy of 0.92, signalling a continued and substantial improvement in performance. The model is evidently becoming increasingly adept at its designated task, illustrating the value of extended training. The upward trajectory in performance persists as the model’s accuracy climbs to 0.94 with 25 epochs. This demonstrates that continued training brings clear and tangible benefits to its performance, reinforcing the notion that deeper training is advantageous. Upon reaching 30 epochs, the model maintains a stable accuracy of 0.94, suggesting that its performance may have plateaued, and further training might yield limited additional gains. A remarkable leap in accuracy is observed at 35 epochs, with the model achieving an impressive accuracy score of 0.96. This highlights that the model can still derive significant benefits from continued training, and the effort invested in training is paying off. With 40 epochs, the model’s accuracy further advances to an outstanding 0.97, showcasing a high level of proficiency in its designated task, possibly breast cancer classification. By the time the model completes 45 epochs, it attains an exceptional accuracy of 0.98, emphasizing the substantial benefits of extended training and the model’s remarkable performance in this specific application.
Even with 50 epochs, the model maintains its exceptional accuracy at 0.98, indicating that it has effectively converged, and additional training epochs may not significantly impact its performance. This suggests that the model has reached a point of diminishing returns in terms of accuracy improvement. In summary, the table provides a comprehensive view of the progressive enhancement in the performance of the proposed CS-EENN model as the number of training epochs increases. The consistent rise in accuracy underscores the positive impact of extended training. This information is instrumental for optimizing the model’s training strategy, helping to strike a balance between training time and achieving the desired level of model performance in practical applications, especially in critical tasks such as medical diagnosis. Figures 15 and 16 shows the loss in train and validation and accuracy in train and validation.
The confusion matrix and the associated performance metrics shown in Fig. 17 are essential tools for evaluating the performance of classification models, particularly in critical domains like medical diagnosis such as breast cancer classification. These metrics provide a comprehensive understanding of how well a model is performing, helping practitioners make informed decisions about model tuning and assessing its suitability for real-world applications. Ensuring the reliability and effectiveness of machine learning models in such critical tasks is of utmost importance, and the insights gained from these metrics play a pivotal role in achieving that goal.
The proposed CS-EENN model is superior in several important diagnostic factors, it gives outstanding results by achieving 98.19% accuracy which is higher than many of the contemporary techniques used for breast cancer classification. This high accuracy, combined with a further low false positive and false negative performance, as confirmed by the confusion matrix and sensitivity-specificity tests. These metrics are very important in medical diagnosis, because false negative cases mean that some diseases may go unnoticed and therefore worsened, while false positives cause undue stress and consumption of resources by patients without such illnesses. As seen by the results the given model affords outstanding classification results however, the model’s stability in response to noisy input data has tested. The evaluation carried out on ‘Breast Histopathology Images’ dataset which has fewer noises and better image quality as compared to need may be appeared in real environment due to poor staining or imaging artifacts. Furthermore, the management of class imbalance, as seen by distributions between benign and malignant cases, has not discussed comprehensively, because the dataset used in the study balanced. This might be inconvenient when working with real data as some datasets are naturally unbalanced posing the need to either oversample, under sample or use complex rebalancing schemes.
Another advantage of the proposed model is its ability to provide computational efficiency. The proposed ensemble model including EfficientNetB0, ResNet50, and DenseNet121 augmented with PSO for the adjustment of hyperparameters raises the computational complexity substantially. Although this approach is very efficient, it consumes a large amount of memory and processing capacity and, therefore, may not be very viable in low-bandwidth environments. Nonetheless, the suggested approach is rather general and still has potential for generalization based on experiences and additional learning with different datasets. In general, the new CS-EENN model presented in this paper incorporates the most recent neural network structures to achieve high diagnostic performance, thus being a valuable tool for diagnosing breast cancer. However, further work to be done was to enhance its stable performance in noisy data environment it can solve the problem of imbalance between classes and improve computational efficiency for the wide use of the proposed method in clinical practice.
Execution time for model building
Time complexity serves as a pivotal metric in assessing the efficiency of an algorithm by quantifying the time required for each instruction to execute. In the context of our analysis, a notable observation was made concerning the computation time of the proposed model. Specifically, it was found that there exists a positive correlation between the computation time and the number of epochs, a relationship that is visually represented in Table 5; Fig. 18. This positive correlation underscores a fundamental aspect of deep learning model training: as the number of epoch’s increases, indicating a longer and more thorough training process, the computation time also tends to rise. This phenomenon is well-established in the realm of machine learning and deep learning. Deeper and more extensive training often necessitates more computational resources and, consequently, additional time for model convergence and refinement. The table offers a comprehensive comparative analysis of the computational time required by various deep learning models across different numbers of epochs, ranging from 5 to 50. These models are likely being employed for a breast cancer classification. The AlexNetmodel’s computational time shows an upward trend as the number of epochs increases. Beginning with 15 min for 5 epochs, it gradually extends to 45 min when trained for 50 epochs. This pattern suggests that AlexNet’s computational demands grow with more extensive exposure to training data. Inception, another powerful model, follows a similar trajectory. It starts with 12 min for 5 epochs and escalates to 31 min for 50 epochs. This indicates that Inception requires increasing computational resources as it undergoes more training iterations. GoogleNet exhibits a unique pattern. It begins efficiently with 8 min for 5 epochs but extends to 33 min for 50 epochs. This suggests that GoogleNet, while initially fast, experiences a substantial increase in computational time as it involves in prolonged training.
DenseNet121’s behaviour aligns with the pattern observed in other models. It starts at 6 min for 5 epochs and extends to 35 min for 50 epochs. This indicates that Dense Net’s computational demands intensify with more data and training cycles. ResNet50, known for its depth and efficiency, demonstrates an increasing trend in computational time. It initiates with 9 min for 5 epochs and reaches 31 min for 50 epochs, suggesting that even efficient models like ResNet50 require additional computational resources for extended training. The proposed CS-EENN model maintains competitive computational efficiency compared to the other models. It starts at 9 min for 5 epochs and peaks at 26 min for 50 epochs. These results suggest that CS-EENN is a good option for real-world applications because it strikes a good compromise among the model’s effectiveness and computational economy. This chart summarises important information about the dynamics of computing time for training different deep learning models across a range of epoch counts. These insights are essential for optimizing model training and resource allocation, particularly in real-world applications where computational efficiency is a critical consideration. The choice of model and the number of epochs significantly influence the time required for training a deep learning model, and this information assists in making informed decisions about resource allocation and training strategies to achieve the best possible model performance.
The Breast Histopathology Images dataset contains patch-based histopathological slides which were obtained from whole-slide imaging at 40× magnification according to Table 6. The dataset shows high-resolution and BACH datasets contain multi-magnification images from 10×, 20×, 40×, 100×, 200×, and 400× while the dataset lacks this wide range of resolution levels. The ability of CS-EENN model to handle datasets with variable resolutions remains limited because the model fails to achieve good generalization. Both the Breast Histopathology Images dataset and the INbreast and MIAS and DDSM databases utilize different detection methods because INbreast and MIAS and DDSM contain mammograms rather than histopathological slides. The CS-EENN model requires domain adaptation techniques to be used for mammogram-based breast cancer classification because its training was focused on histopathological images. The combination of various datasets containing both mammography images and histopathology data would make results more applicable in real-world applications.images that deliver fundamental tissue details needed for deep learning classification applications. BreakHis.
The 277,524-patch dataset used in this research contains substantially more images than the 7,909 image BreakHis collection and the 400 image BACH dataset. The extensive nature of the dataset enhances deep learning model training because it decreases overfitting risks and strengthens both feature extraction and classification capabilities. The model could exhibit performance limitations when applied to different hospitals because it uses histopathological images originating from a single institutional source.
The BreakHis alongside BACH datasets contain an almost equal balance of benign and malignant samples but the Breast Histopathology Images dataset mainly consists of IDC (Invasive Ductal Carcinoma) cases. The model’s training with such dataset distribution produces an analytical error which creates susceptibility toward Invasive Ductal Carcinoma classifications while reducing performance for breast cancer type classification. Future research must integrate multi-class datasets into the model because this addition will help the model better recognize various breast cancer histological subtypes. The key evaluation metrics received a reliability analysis through calculation of 95% confidence intervals which appeared in Table 7 along with accuracy, precision, recall and F1-score metrics. Multiple randomized test samples were used in bootstrapping calculations to determine the possible ranges of metric values.
Discussion
The study employs the ‘Breast Histopathology Images’ dataset from Kaggle, which consist of 277,524 image patches of breast cancer tissue that were extracted from 162 WSI acquired at 40x magnification. However, one of the main limitations of the dataset is that even though the dataset is huge in terms of the number of patches, it is highly biased in terms of geographical, demographic and institutional distribution. The dataset covers one kind of histopathological sample and the samples used here are derived from one type of clinical centre. These options can result in limited freedom and generalisability of the model, which might affect the model effectiveness when introduced to different patients of dissimilar demographics, diverse protocols of imaging or different types of tumours. Moreover, the model is trained only on one dataset at the same time, which might lead to over-fitting and, therefore, a lower model’s performance on datasets from other clinical or geographic areas. To tackle this limitation the combination should validated on various datasets to prove that the proposed model holds good in practice situations.
However, the proposed CS-EENN model gains a very high accuracy test result of 98.19% However, there exist some challenges or drawbacks that should overcome for the applicability and scalability of the proposed model. First, the combination of EffNetB0, ResNet50, DenseNet121 with CSO for tuning the hyperparameters enhances the challenges of computational complexity in the model. This makes training as well as deployment of the tool very resource demanding hence may not be very applicable in time critical or low resource environment. In addition, the model hugely relies on the training dataset provided to incorporate high quality and diversity. However, a major drawback of ‘Breast Histopathology Images’ set is that the dataset well annotated; this could mean that the models trained from ‘Breast Histopathology Images’ might not generalize well in clinically diverse settings. In addition, the final optimization process of CSO works well but it has high computational complexity and different parameter settings take more time to converge. Another quirk of the model is its dependence on pre-processing workflows like resizing images; normalizing them; and applying other augmentations. Independently, any deviation from these processes and routines affects reliability and performance of the model. However, current model only applies binomial classification, which would limit the multiclass classification like cancer subtypes diagnosis. The interpretability of the model also presents a problem since deep learning models are considered to unexplainable. This has the potential of limiting its usage in clinical practice where interpretation of results is critical for choosing a course of action. Finally, the heterogeneity of computations needed in the different components of the ensemble may result in uneven loading and, therefore, strict time limitations in corresponding real-world applications.
The suggested CS-EENN solution effectively tackles important shortcomings which exist in prior breast cancer classification research regarding dataset noise management and class distribution balancing and interpretability improvement. Gabor filtering serves as a pre-processing technique in the model to improve texture features and decrease artifacts in histopathological images before analysis. The ResNet50 extraction process helps the model identify detailed patterns and structural modifications in images to produce strong features especially when working with substandard image quality. Through ensemble learning the combination of EfficientNetB0 and ResNet50 with DenseNet121 procure multiple convolution techniques to enhance result accuracy and decrease misdiagnosis threats specifically in less frequent case samples. This combination approach produces accurate output predictions by reducing both inaccurate positive and negative results. The learning process optimizations provided by Cat Swarm Optimization (CSO) help tune hyperparameters because this enables better generalization and overfitting prevention which are common problems in unbalanced datasets. As a method to improve understanding the model includes ensemble learning with feature representation diversity while utilizing confusion matrices along with detailed performance metrics to create visual explanations for classification outcomes. Future studies have the potential to boost interpretability through the integration of explainable AI methods including Grad-CAM and SHAP so clinicians can obtain visual explanations of their decision-making process. The CS-EENN model provides a new classification standard for breast cancer which improves diagnostic precision and clinical potential through its handling of existing limitations.
The CS-EENN model needs to expand its binary benign-malignant classification capability to handle multiple cancer type categories for standard clinical practice. To improve the model’s performance the hierarchical classification system must be added to separate benign cases from atypical cases and malignant ones while moving into further categories of IDC, ILC, Mucinous Carcinoma, and Medullary Carcinoma. The output layer needs modification to handle multiple classes while employing softmax instead of sigmoid activation. The problem of class imbalance in underrepresented cancer subtypes can be solved through stain normalization and contrast enhancement combined with GAN-based synthetic image generation during data augmentation techniques. Feature extraction power of the model will improve by merging transformer-based vision models like ViTs or Swin Transformers due to their demonstrated ability in medical imaging pattern recognition. Multiple institutions can achieve better generalization of tissues from various histopathological databases through transfer learning methods. The system requires a XAI framework featuring either Grad-CAM or SHAP analysis to show clinicians understandable visualizations of prediction results while maintaining transparent multiclass choices. The CS-EENN model’s expansion for exhaustive breast cancer subtype identification will become possible through direct implementation of these specified steps that support precise oncological care along with individualized treatment selection.
Broader implications for clinical practice
The proposed CS-EENN model presents substantial prospects to modify current breast cancer diagnosis approaches in medical facilities by creating better diagnostic precision along with reliability and better early detection methods. Deep learning networks EfficientNetB0, ResNet50 and DenseNet121 tuned with Cat Swarm Optimization (CSO) enable a model to achieve 98.19% classification performance which exceeds numerous traditional approaches. Medical diagnostics benefits significantly from this high accuracy because false negatives result in delayed critical treatments and false positives can cause avoidable medical anxiety together with additional medical procedures. Deep learning capabilities embedded in the CS-EENN model establish its ability to serve as a computer-aided diagnosis system which helps pathologists perform histopathological analyses more effectively and rapidly. Medical professionals will benefit from automated breast cancer classification systems which enable them to handle sophisticated cases while the tool handles less complex analyses. This ensemble methodology reduces observer-related inconsistencies and supports the creation of both standardized and repeatable medical diagnosis decisions that are crucial for extensive healthcare situations. Proposed implementation of the CS-EENN model into clinical practice faces challenges that need solution before full integration becomes achievable. Medical organizations face a significant challenge when deploying deep learning models because the training process combined with model deployment requires many resources. The combination of three large-scale network architectures brings demanding processing needs along with memory and GPU demands that healthcare facilities fundamentally need to support without exception including low-resource settings. Model applications to different patient groups and imaging samples acquired by multiple medical centers encounter problems due to dependence on thorough histopathological dataset annotation. Such variations in staining methods combined with imaging environments and hospital populations should undergo thorough multi-independent data testing before the model reaches clinical readiness.
The main obstacle with deep learning-based medical decisions consists of making medical decisions more easily interpretable and understandable for human doctors. A robust classification framework delivered through the CS-EENN model needs clinical transparency regarding AI diagnosis to build reliable medical decision processes. The AI model does not implement XAI techniques such as Grad-CAM or SHAP visualizations for showing users how diagnostic assessments are determined. Future research needs to include interpretability procedures that will raise the confidence level of clinicians regarding their use of AI-assisted breast cancer diagnostic systems. Administrative authorization along with healthcare data privacy law adherence must be established prior to using AI models in clinical healthcare environments. The CS-EENN model offers extensive advantages during healthcare operations in a clinical setting despite operational hurdles. High precision detection of breast cancer and streamlined pathology workflows together with early detection assistance would produce beneficial outcomes that minimize diagnostic mistakes and create optimized treatment strategies. The CS-EENN model will create AI-powered diagnostic solutions that assist pathologists and oncologists to make improvements in their clinical decision-making capabilities because of ongoing model optimization together with hardware efficiency improvements and interpretability advancements. These AI-based models present the potential to transform breast cancer diagnosis procedures and care delivery particularly in places with limited medical experts by offering scalable healthcare solutions.
Limitations of current evaluation metrics
The CS-EENN model received its evaluation by standard metrics for medical image classification that included accuracy, precision, recall, and F1-score. The standard evaluation metrics offer vital information about model functioning but at the same time include fundamental weaknesses that could affect clinical assessment of deep learning models. Research must integrate extra metrics in future evaluations to achieve results with superior robustness and interpretability because these present critical limitations for a comprehensive assessment. The main shortcoming of using accuracy for evaluation results occurs when datasets display class imbalance problems. The majority of medical imaging datasets for breast cancer classification show an uneven distribution of samples because they include more benign cases than malignant ones. The model would reach high accuracy by predominantly classifying majority cases although it fails to detect malignant cases thus making the outcome unacceptable in clinical practice. Precision and recall, while useful, also present trade-offs. A model with high precision ensures less false positive results yet it does not show performance in detecting actual positive cases correctly. In cases of increased recall rates medical practitioners might perform unnecessary treatments because detecting all cancer cases would result in additional false positive test outcomes.
More comprehensive model evaluation should be performed in future research by implementing supplementary metrics which offer a complete understanding of model output. The receiver operating characteristic (ROC) curve utilizes AUC metrics for scoring model performance based on its discrimination abilities between different probability threshold settings. Model effectiveness in benign and malignant case separation is demonstrated by high scores in AUC-ROC evaluations thus making them essential for medical diagnostics needing flexible decision thresholds. When dealing with imbalanced datasets the Area Under the Precision-Recall Curve (AUC-PR) stands out because it evaluates how precision relates to recall without emphasizing absolute classification numbers. Classification performance assessment becomes more balanced through use of the Matthews Correlation Coefficient (MCC) when working with datasets that are heavily imbalanced. MCC offers superior reliability as a quality measure for models since it evaluates all four combinations of positive and negative predictions. Evaluating the correspondence between predicted class groupings and actual data distributions becomes possible through using the Fowlkes-Mallows Index (FMI). The Brier Score and other calibration metrics need further evaluation as additional research methods. The Brier Score evaluates the level of accuracy which probabilistic predictions reach through measuring the connection between model confidence predictions versus real-world results. The calibration score proves highly useful in clinical settings because inaccurate confidence measures might trigger false reassurance or false alarms. Analysis based on Decision Curve Analysis (DCA) should be used for model evaluation because this method allows users to understand the clinical benefits of predictions across various probability thresholds.
Computational complexity and scalability
Due to its high computational requirements and scarce resources of low-resource clinics and edge devices the CS-EENN model encounters implementation obstacles to achieve accurate breast cancer classification. Deep learning ensemble architectures including EfficientNetB0, ResNet50 and DenseNet121 are difficult to deploy in limited computational settings because they need extensive GPU/TPU power together with large memory needs and high-power usage. These obstacles must receive attention because it helps assure AI diagnosis systems will meet clinical requirements for real-world utilization. Implementing model pruning techniques with quantization turns out to be an effective method for decreasing computational complexity. The deep learning model benefit from pruning when practitioners remove unnecessary connections and neurons to keep its size manageable while maintaining its performance levels. Model quantization transforms elevated floating-point operations into reduced-bit numerical values thus both cutting down memory usage and computation needs while preserving system performance. The CS-EENN model becomes deployable on low-power edge devices and mobile health applications when post-training quantization or the more advanced quantization-aware training (QAT) methods are applied to it.
The implementation of knowledge distillation allows for training a student model which operates as a smaller network that mimics the performance of an ensemble system using fewer processing resources. One trained through knowledge distillation AlexNet student model replaces the three models in the ensemble to perform equally while using significantly less power and operating faster. AI device applications benefit highly from this technique because they facilitate mobile diagnostics and tele medical systems. Improved scalability can be achieved through the combination of edge AI technology and federated learning because the model deploys across multiple decentralized edge devices instead of using cloud-based inference. The implementation of Edge AI helps healthcare institutions with poor internet access perform local device-based processing so they can complete tasks without depending on excessive bandwidth. Federated learning provides institutions with the ability to develop models through collaborative training without transmitting raw patient information thus establishing privacy-preserving AI systems for medical applications. An effective deep learning optimizer can improve network design to achieve better performance. The CS-EENN framework accepts lightweight AlexNet architectures including MobileNetV3 and ShuffleNet to increase both inference efficiency and save computational power. The adaptive inference approach enables early exit strategies which use lightweight sub-models to classify simple cases but directs complex cases toward the complete ensemble network.
Conclusion and future scope
Breast cancer remains a significant global health challenge, underlining the need for accurate, efficient, and early diagnostic methods. In this study, the proposed CS-EENN model, an ensemble of EfficientNetB0, ResNet50, and DenseNet121 optimized using Cat Swarm Optimization (CSO), demonstrated exceptional performance with an accuracy of 98.19% on the ‘Breast Histopathology Images’ dataset. The model’s ability to integrate complementary strengths of advanced neural network architectures, combined with meticulous hyperparameter tuning, highlights its potential as a reliable and robust tool for breast cancer classification. The CS-EENN model’s superior performance stems from its tailored approach to feature extraction, leveraging multiple architectures to provide a comprehensive understanding of complex histopathological patterns. Furthermore, the incorporation of CSO enabled the fine-tuning of critical hyperparameters, striking a balance between computational efficiency and diagnostic accuracy. This method not only outperformed conventional single-model approaches but also highlighted its adaptability to challenging datasets, setting a benchmark for future ensemble-based classification systems. Despite these achievements, the model has limitations, including its computational demands and dependency on high-quality datasets, which may restrict its scalability in resource-constrained environments. Additionally, the black-box nature of deep learning models poses interpretability challenges, particularly in clinical applications where explain ability is paramount. Addressing these challenges presents opportunities for further refinement, such as exploring lightweight architectures, enhancing interpretability through visualization techniques, and expanding the model to multiclass classification tasks. The findings of this study contribute significantly to the growing body of research on deep learning applications in medical imaging. By achieving state-of-the-art performance, the CS-EENN model demonstrates its potential to improve breast cancer diagnosis, offering a pathway toward more accurate, efficient, and patient-centred care. Future research should aim to refine the model’s scalability, broaden its applicability, and ensure its adoption in diverse clinical settings, ultimately bridging the gap between technological innovation and real-world impact.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
References
Amethiya, Y., Pipariya, P., Patel, S. & Shah, M. Comparative analysis of breast cancer detection using machine learning and biosensors. Intell. Med. 2 (2), 69–81 (2022).
Petrini, D. G. et al. Breast cancer diagnosis in two-view mammography using end-to-end trained EfficientNet-based convolutional network. Ieee Access. 10, 77723–77731 (2022).
Idris, N. F. et al. Breast cancer disease classification using fuzzy-ID3 algorithm based on association function. IAES Int. J. Artif. Intell. 11 (2), 448 (2022).
Sharma, D., Kumar, R. & Jain, A. Breast cancer prediction based on neural networks and extra tree classifier using feature ensemble learning. Measurement: Sens. 24, 100560 (2022).
Yari, Y., Nguyen, T. V. & Nguyen, H. T. Deep learning applied for histological diagnosis of breast cancer. IEEE Access. 8, 162432–162448 (2020).
Jahangeer, G. S. B. & Rajkumar, T. D. Early detection of breast cancer using hybrid of series network and VGG-16. Multimedia Tools Appl. 80 (5), 7853–7886 (2021).
Z. A. et al. Influence of the computer-aided decision support system design on ultrasound-based breast cancer classification. Cancers 14 (2), 277 (2022).
Hassan, S. A., Sayed, M. S., Abdalla, M. I. & Rashwan, M. A. Breast cancer masses classification using deep convolutional neural networks and transfer learning. Multimedia Tools Appl. 79 (41), 30735–30768 (2020).
Ragab, M., Albukhari, A., Alyami, J. & Mansour, R. F. Ensemble deep-learning-enabled clinical decision support system for breast cancer diagnosis and classification on ultrasound images. Biology 11 (3), 439 (2022).
VK, P. Enhancing pancreatic cancer classification through dynamic weighted ensemble: a game theory approach. Comput. Methods Biomech. BioMed. Eng. 1–25. (2023).
Strelcenia, E. et al. Improving cancer detection classification performance using GANs in breast cancer data, in IEEE access, 11, pp. 71594–71615. https://doi.org/10.1109/ACCESS.2023.3291336 (2023).
Ragab, D. A., Attallah, O., Sharkas, M., Ren, J. & Marshall, S. A framework for breast cancer classification using multi-DCNNs. Comput. Biol. Med. 131, 104245 (2021).
Zhang, Y. D., Satapathy, S. C., Guttery, D. S., Górriz, J. M. & Wang, S. H. Improved breast cancer classification through combining graph convolutional network and convolutional neural network. Inf. Process. Manag. 58 (2), 102439 (2021).
Priyan, S. V., Dhanasekaran, S., Karthick, P. V. & Silambarasan, D. A new deep neuro-fuzzy system for Lyme disease detection and classification using UNet, inception, and XGBoost model from medical images. Neural Comput. Appl. 36 (16), 9361–9374 (2024).
Ganesh Chandrasekaran, S., Dhanasekaran, C., Moorthy & Arul Oli, A. Multimodal sentiment analysis leveraging the strength of deep neural networks enhanced by the XGBoost classifier, Computer Methods in Biomechanics and Biomedical Engineering. https://doi.org/10.1080/10255842.2024.2313066 (2024).
Hirra, I. et al. Breast cancer classification from histopathological images using patch-based deep learning modeling. IEEE Access. 9, 24273–24287 (2021).
Chandana Mani, R. K. et al. The Comparative Study of CNN models for Breast Histopathological Image Classification, 2023 - International Conference on Computer Communication and Informatics (ICCCI), pp. 1–5. https://doi.org/10.1109/ICCCI56745.2023.10128352
Waqar, M. et al. An applied artificial intelligence aided technique for effective classification of breast cancer. Int. Conf. Energy Power Environ. Control Comput. (ICEPECC). 1–6. https://doi.org/10.1109/ICEPECC57281.2023.10209518 (2023).
Akbar, W. et al. Performance evaluation of deep learning models for breast cancer classification. IEEE Int. Conf. Emerg. Trends Eng. Sci. Technol. (ICES&T). 1–4. https://doi.org/10.1109/ICEST56843.2023.10138861 (2023).
Samanta, P. K. et al. A novel deep CNN model for improved breast cancer detection using ultrasound images. Int. Conf. Communication Circuits Syst. (IC3S). pp. 1–4. https://doi.org/10.1109/IC3S57698.2023.10169383 (2023).
Malik, H. et al. Multi-classification deep learning models for detection of ulcerative colitis, polyps, and dyed-lifted polyps using wireless capsule endoscopy images. CIS 10, 2477–2497. https://doi.org/10.1007/s40747-023-01271-5 (2024).
Naeem, A. et al. DVFNet: A deep feature fusion-based model for the multiclassification of skin cancer utilizing dermoscopy images. PLoS ONE. 19 (3), e0297667. https://doi.org/10.1371/journal.pone.0297667 (2024).
Khalil, M. et al. Deep Learning-Based classification of abrasion and ischemic diabetic foot sores using Camera-Captured images. Mathematics 11 (17), 3793. https://doi.org/10.3390/math11173793 (2023).
Ahmad Naeem et al. A multiclassification framework for skin cancer detection by the concatenation of Xception and ResNet101. JCBI 6 (02), 205–227 (2024). https://jcbi.org/index.php/Main/article/view/328
Ahmad et al. Predicting the metastasis ability of prostate cancer using machine learning classifiers. JCBI 4 (02), 1–7 (2023). https://www.jcbi.org/index.php/Main/article/view/139
BurhanettinOzdemir et al. An innovative deep learning framework for skin cancer detection employing ConvNeXtV2 and focal self-attention mechanisms. Results Eng. https://doi.org/10.1016/j.rineng.2024.103692 (2025). 25, 103692, ISSN 2590 – 1230.
Bilal Bayram et al. A systematic review of deep learning in MRI-based cerebral vascular occlusion-based brain diseases. Neuroscience 568, 76–94. https://doi.org/10.1016/j.neuroscience.2025.01.020 (2025).
IshakPacal Investigating deep learning approaches for cervical cancer diagnosis: a focus on modern image-based models. Eur. J. Gynaecol. Oncol. 46 (1), 125–141. https://doi.org/10.22514/ejgo.2025.012 (2025).
COŞKUN, D. A. M. L. A. et al. A comparative study of YOLO models and a transformer-based YOLOv5 model for mass detection in mammograms. Turkish J. Electr. Eng. Comput. Sci. 31 (7), 10, https://doi.org/10.55730/1300-0632.4048 (2023).
Işık, G. et al. Few-shot classification of ultrasound breast cancer images using meta-learning algorithms. Neural Comput&Applic. 36, 12047–12059. https://doi.org/10.1007/s00521-024-09767-y (2024).
Peta, J. et al. Enhancing breast cancer classification in histopathological images through federated learning framework, in IEEE access, 11, pp. 61866–61880. https://doi.org/10.1109/ACCESS.2023.3283930 (2023).
Balasubramaniam, S., Velmurugan, Y., Jaganathan, D. & Dhanasekaran, S. A modified LeNet CNN for breast cancer diagnosis in ultrasound images. Diagnostics 13 (17), 2746 (2023).
Mu, J. Breast cancer detection using ResNet50 with hyperparameter tuning. IEEE 3rd Int. Conf. Power Electron. Comput. Appl. (ICPECA). 1245–1249. https://doi.org/10.1109/ICPECA56706.2023.10076197 (2023).
Deb, S. D. et al. 2-Stage Convolutional Neural Network for Breast Cancer Detection from Ultrasound Images, 2023 National Conference on Communications (NCC), Guwahati, India, 2023, pp. 1–6. https://doi.org/10.1109/NCC56989.2023.10067925
https://www.kaggle.com/datasets/paultimothymooney/breast-histopathology-images Accessed on 17th January 2024.
Afaq, S. et al. MAMMO-Net: An Approach for Classification of Breast Cancer using CNN with Gabor Filter in Mammographic Images, 2022 International Conference on Computational Intelligence and Sustainable Engineering Solutions (CISES), pp. 177–182. https://doi.org/10.1109/CISES54857.2022.9844320
Masud, M., Eldin Rashed, A. E. & Hossain, M. S. Convolutional neural network-based models for diagnosis of breast cancer, Neural Comput&Applic, vol. 34, no. 14, pp. 11383–11394. https://doi.org/10.1007/s00521-020-05394-5 (2022).
Asare, S. K., You, F. & Nartey, O. T. Efficient, ultra-facile breast cancer histopathological images classification approach utilizing deep learning optimizers. Int. J. Comput. Appl. 11, 9, (2020).
Author information
Authors and Affiliations
Contributions
A. Abdul Hayum wrote the main manuscript text and S. Finney Daniel Shadrach prepared Figs. 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 and 18. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent to publish
All the authors gave permission to Consent to publish.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hayum, A.A., Daniel Shadrach, F. Optimizing breast cancer classification based on cat swarm-enhanced ensemble neural network approach for improved diagnosis and treatment decisions. Sci Rep 15, 33740 (2025). https://doi.org/10.1038/s41598-025-95481-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-95481-1