Abstract
Antimicrobial peptides (AMPs), including Tilapia piscidin 4 (TP4), have emerged as a promising therapeutic approach for treating different types of cancers. These peptides specifically target tumor cells while minimizing harm to normal tissues. This study aims to investigate the cytotoxic effects of TP4 and elucidate its molecular mechanisms of apoptosis in MCF-7 human breast cancer cell line. MCF-7 and MCF-10 cell viability was assessed after treating the cells with various concentrations of TP4 for 24 h using the MTT assay. The MCF-7 cells were allocated into three groups including control cells (Cnt), cells treated with 0.5 IC50, and 0.25 IC50 of TP4. The results demonstrated the inhibitory effect of TP4 on MCF-7 cell proliferation after 24 h (IC50 = 50.11 µg/mL), while no cytotoxicity was observed in normal breast cells (MCF-10) at this concentration. The selectivity index (SI) value exceeded 2, indicating TP4’s high specificity for cancer cells. Treatment with 25% and 50% IC50TP4 induced apoptosis, DNA fragmentation, and mitochondrial membrane potential changes (JC-1 staining) in MCF-7 cells. This induction was accompanied by increased expression of apoptotic genes (Bax, caspase3, and p53), decreased expression of the anti-apoptotic gene Bcl2, elevated levels of intracellular reactive oxygen species (ROS) and malondialdehyde (MDA) content, and reduced activity of the superoxide dismutase (SOD) and catalase (CAT) enzymes when compared to the control group (P < 0.05). Based on the data, we can infer that TP4 induces apoptosis in breast cancer cells via a ROS-dependent pathway.
Similar content being viewed by others
Introduction
Breast cancer exhibits significant global variations in incidence rates, yet it remains the most prevalent and deadliest form of cancer among women worldwide1. In 2020, breast cancer accounted for 11.6% of all new cancer cases, surpassing lung cancer as the most prevalent global cancer. This translated to 2.26 million reported new cases of breast cancer2. Breast cancer can be classified into invasive and non-invasive types, with further classification based on the specific origin site within the breast, such as the ducts, lobules, or stromal tissue3. The molecular characteristics of tumor cells have recently been utilized to assess treatment response and as criteria for stratifying breast cancer4. Despite significant progress in classical cancer treatments, such as chemotherapy, the limitations of these approaches include notable side effects and nonselective targeting of most currently employed anticancer drugs, which hinder their therapeutic efficacy5. In recent years, targeted therapy has sought natural and safe alternatives to mitigate the side effects associated with conventional anticancer drugs6.
Antimicrobial peptides (AMPs) hold promise as a potential alternative to cancer therapy. AMPs are naturally occurring peptides that are synthesized from specific genetic loci across a wide range of life forms7. While their primary synthesis occurs through transcription and translation, there are rare instances where AMPs can be produced independent of ribosomes. Extensive biochemical studies have elucidated their specialized roles in immune defense mechanisms, leading to their alternative designation as host defense peptides (HDPs)8,9. AMPs share several common structural features. First, they typically have a molecular weight of less than 10 kDa, with a length ranging from 5 to 50 amino acids. Second, they exhibit cationic properties at neutral pH. Third, AMPs possess a significant proportion of hydrophobic residues. Finally, they adopt an amphipathic structure10. The structural amphipathic nature of AMPs facilitates their electrostatic interactions with anionic molecules present on the plasma membrane of microbial or cancer cells11. In contrast to conventional chemotherapeutic drugs that harm healthy cells, AMPs exhibit selective targeting of cancer cells, resulting in lower toxicity towards non-cancerous cell types11,12. In addition to cancer, AMPs have various clinical applications including antimicrobial activity against yeast, fungi, influenza and HIV viruses, immune-modulatory activity (e.g., LL-37, Hbd-2, 3, 4), infections (e.g., cathelicidins, defensins, E6), wound healing (e.g., PXL150), anti-biofilm (e.g., LL-37), anti-microbial activity (e.g., nisin, gramicidin, polymyxins, daptomycin and melittin), diabetes (e.g., CRAMP)13,14.
Some previous studies evaluated the effect of AMPs on the breast cancer. Avand et al. (2018) reported that Lactococcus lactisantimicrobial peptide (Nisin) has the potential to be used against the breast cancer cells15. Previous study showed that pleurocidin-family cationic antimicrobial peptides (CAPs), NRC-03 and/or NRC-07, can be used alone or in combination with conventional chemotherapeutic drugs for the treatment of breast cancer16. It was shown that Piscidin-1 inhibits the migration of HT1080 cells in a dose-dependent manner17. Among AMPs, TP4 has a special place. TP4, a marine antimicrobial peptide from Nile Tilapia, has antibacterial, immune-modulatory, and wound-healing properties18,19. Furthermore, TP4 has garnered attention for its potent anticancer effects demonstrated in glioblastoma cells20, triple-negative breast cancer cells11, and human non-small-cell lung cancer cells21. In spite of these studies, there is no study to focus on the effect of TP4 on the MCF-7 cells and assess its selectivity and toxicity for this cancer cell. Moreover, the specific cellular mechanisms underlying the anti-tumor effects of TP4 with cationic properties, its potential interaction with cancer cell membranes, and downstream apoptotic signaling remain unknown. Gaining a comprehensive understanding of the molecular and cellular impacts of TP4 on cancer cells is crucial for advancing its pharmacological application. This study aimed to investigate the anticancer potential of TP4 against MCF-7 cells and elucidate the underlying mechanisms by examining the apoptotic pathway, and antioxidant status.
Materials and methods
Reagents and peptide
TP4 (with the amino acid sequence of FIHHIIGGLFSAGKAIHRLIRRRRR, the molecular weight of 2.981 kDa, and purity > 95%), received as a gift from the Antimicrobial Resistance Research Center, Mashhad University of Medical Sciences (Mashhad, Iran) was synthesized following the procedure outlined in Nashani et al. (2018)22 and stored at − 20 °C and protected from light. Phosphate-buffered saline (PBS), RPMI-1640 medium L-Glutamine, and the antibiotic penicillin/streptomycin were procured from Biowest (Maine et Loire, France). The fetal bovine serum (FBS) was purchased from Seralab (UK), Trypsin, Dimethyl sulfoxide (DMSO), and 3-(4,5)-dimethylthiazol(-z-y1)−3,5-diphenyltetrazolium bromide (MTT) reagent was obtained from Sigma-Aldrich Corporation (St. Louis, MO, USA).
Cell culture
The MCF-7 human breast cancer cell line was obtained from the Pasteur Institute of Iran located in Tehran. The cells were cultured in RPMI-1640 medium supplemented with l-Glutamine, 10% FBS (fetal bovine serum), and 1% antibiotic. They were incubated at 37 °C in a humidified atmosphere containing 5% CO2 for growth. Sub-culturing of the cells was performed every 2–3 days, and periodic examination for contamination was conducted using an inverted microscope.
Cell viability assay
MCF-7 and MCF-10 cell viability was assessed after treating the cells with various concentrations of TP4 for 24 h using the MTT assay. Cells were seeded in triplicate in 96-Well Microplates at a density of 5 × 10³ cells per well. After overnight incubation, the cells were treated with TP4 at concentrations of 0 (0.5% DMSO), 0.01, 0.1, 1, 10, 20, 50, 100, and 200 µg for 24 h. The MTT solution was then added to each well and the plates were incubated at 37 °C for 4 h to allow the conversion of MTT to insoluble formazan by viable cell dehydrogenases. The supernatant was removed, and the intracellular formazan was solubilized in 100 µL/well of DMSO for 15 min at room temperature. The absorbance at 570 nm was measured using a microplate reader (Dynatech Laboratories, Chantilly, VA, USA). Cell viability was assessed using the formula: % cell viability = (Mean absorbance in test wells/Mean absorbance in control wells) × 100. Subsequently, survival curves were plotted for each cell line based on the correlation between TP4 concentrations and the number of surviving cells23. To ascertain the 50% inhibitory concentration (IC50), it is necessary to construct concentration-response curves for each dosage. The cytotoxic selectivity of TP4 was evaluated by employing the selectivity index (SI), which can be calculated using the equation: SI = IC50 (non-cancer cells)/IC50(cancer cells). A selectivity index greater than 2 indicates a high level of selectivity24.
Experimental design
The MCF-7 cells were allocated into three groups. The untreated MCF-7 cells were considered control cells, while in the other two groups, cells were treated with 0.5 IC50 and 0.25 IC50 Treatments were applied in triplicate at 70–80% confluence. Cells were incubated in a CO2 incubator for 24 h at 37 °C and 95% humidity.
Estimation of intracellular reactive oxygen species
Intracellular reactive oxygen species (ROS) were quantified using a previously described method involving the fluorescent probe, 20,70-dichlorofluorescein diacetate (DCFDA)25. Cells were treated with TP4 and a separate set of cells was exposed to 25 µM H2O2 post-TP4 treatment for 2 h as a positive control to confirm assay sensitivity. After washing with PBS, DCFDA (5 µM) was added to the cells, which were then incubated in the dark at 37 °C for 30 min. Following another round of PBS washing, the fluorescence intensity of DCFDA was measured using a fluorescence microplate reader. The intracellular ROS levels were expressed as a percentage of the control.
Antioxidant enzymes activity and MDA contents
Superoxide dismutase (SOD) and catalase (CAT) activities and malondialdehyde contents (MDA) in MCF-7 cells were assessed using ELISA methods and commercially available kits provided by ZellBio GmbH, Germany. The manufacturer’s protocol was followed for the assays26.
Comet assay and JC-1 staining
The DNA damage in MCF-7 cells following treatment was assessed using the Comet assay kit (Cell Biolabs, STA-355, San Diego, CA, USA), following the manufacturer’s instructions. The quantification of tail DNA percentage was conducted following the previous study27. The CellProfiler software was used for the analysis, based on the comet morphology (counting pixel per comet). The software measures parameters such as the area and perimeter in the comet tail as objects based on the pipeline (fluorescent comet).
JC-1 staining was employed to visualize mitochondrial membrane potential changes and carried out according to previous study28. After culturing and treating MCF-7 cells, the medium was removed and MCF-7 cells from the control and treatment groups were incubated with JC-1 probe for 30 min. Then, cells were rinsed with phosphate-buffered saline (PBS) and the stained cells were observed with a fluorescent microscope.
Gene expression analysis
Total RNA was extracted from the cells using Trizol reagent (Qiagen RNA extraction method), followed by RT-PCR analysis. Briefly, after homogenizing the cells in Trizol reagent and adding chloroform, centrifugation (12000×g, 10 min, 4˚C) were carried out. Then, isopropanol was added and the centrifugation was repeated. The RNA pellet was washed with ethanol and centrifuged (75000×g, 5 min, 4˚C). DNase treatment was performed to eliminate genomic DNA contamination RNA concentration and integrity were determined using Nanodrop and 1% gel electrophoresis, respectively. The expression levels of Bax, Bcl-2, P53, and caspase 3 genes were assessed through reverse transcription with RevertAid H Minus Reverse Transcriptase, followed by qPCR using 2X SYBR Green Master Mix and specific primers (Table 1). All qPCR kits were obtained from Thermo Scientific, Waltham, MA, USA. The qPCR mixture (25 µL) consisted of 2 µL cDNA, 1 µL of each primer, and 12.5 µL of Maxima SYBR Green Master Mix. Thermal cycling involved an initial step at 95 °C for 10 min, followed by 45 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. The relative expression of the target genes was normalized to the housekeeping gene GAPDH and calculated using the 2 (−ΔΔCt)method29.
Statistical analysis
Statistical analysis was conducted using one-way analysis of variance (ANOVA) through the JAMOVI program (version 2.2.3.0). Tukey’s test at a 95% confidence level (p < 0.05) was employed to compare the means. Before conducting the ANOVA, the normality of the data was assessed using the Shapiro-Wilk test, while Levene’s test was utilized to examine variance. The statistical significance of the difference in IC50 values between MCF-7 and MCF-10 cells was evaluated using an independent samples t-test. All data were presented as mean ± standard deviation (SD) of replicates from independent experiments.
Results
Cytotoxicity
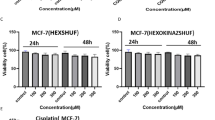
An MTT assay was performed on MCF-7 and MCF10 cells to evaluate the effects of TP4 at various concentrations after a 24-hour incubation period (Fig. 1). The resulting cell viabilities (%) for each cell line are depicted in Fig. 1 (P < 0.05). The data obtained indicates that the TP4 peptide exhibited a dose-dependent cytotoxic effect on the tested cell lines. At 24 h of treatment, the IC50 values of TP4 for MCF-7, and MCF-10 cells were determined to be 50.11 µg/mL, and 199.81 µg/mL, respectively. An independent samples t-test in IC50 values between MCF-7 and MCF-10 cells revealed a statistically significant difference (p < 0.05), confirming the high selectivity of TP4 for cancer cells. The SI value of TP4 for MCF-7 cells after 24 h of treatment was 3.98, demonstrating its high selectivity towards the MCF-7 cell line.
Cytotoxic activity of Tilapia piscidin 4 (TP4) against MCF-7 (a), and MCF-10 (b) cells after 24 h incubation. Each value represents the Mean ± SD of three replicates. Different letters show significant difference between groups at p < 0.05 level. Asterisks (*) indicates significant difference between control (DMSO) and treatment groups at p < 0.01 level. Control group was not shown due to no significant difference between DMSO and control group.
Intracellular ROS, MDA content, and antioxidant status
Treatment with sub-lethal doses of TP4 resulted in increased intracellular levels of ROS and MDA contents in MCF-7 cells, as compared to the control group (P < 0.05)(Fig. 2). Additionally, the activity of antioxidant SOD and CAT, exhibited a significant decrease when compared to the control group (P < 0.05) (Fig. 2). Notably, the most pronounced alterations were observed at a dose 25 µg/ml of TP4, which corresponds to 50% the IC50 value (Fig. 2).
Effect of Tilapia piscidin 4 (TP4) treatment at 25% and 50% concentrations on intracellular reactive oxygen species (ROS) levels (a), malondialdehyde content (MDA) (b), and the activities of superoxide dismutase (SOD) (c) and catalase (CAT) (d) in MCF-7 cells. Data are presented as mean ± standard deviation (SD). Each experiment was performed in triplicate, with three independent experiments conducted (n = 6, technical replicate). Different letters show significant difference between groups at p < 0.05 level. Asterisks (*) indicates significant difference between control and treatment groups at p < 0.01 level.
Comet assay
Treatment with 25% and 50% IC50TP4 resulted in elevated DNA breakage and increased tail length in comets, surpassing the control group (P < 0.05) (Fig. 3). Notably, the treatment with 50% IC50TP4 exhibited the most substantial increase in comet tail length (Fig. 3).
Comet assay results depicting the impact of Tilapia piscidin 4 (TP4) treatment on MCF-7 cells at concentrations of 25% and 50% for 24 h. The lower panel illustrates the quantification of DNA tail percentage in 100 cells. Mean ± standard deviation (SD). Different letters show significant difference between groups at p < 0.05 level. Asterisks (*) indicates significant difference between control and treatment groups at p < 0.01 level.
Results of JC-1 staining was shown in Fig. 4. According to this figure, normal cells have red color and cells with mitochondrial membrane potential changes have green color, indicating TP4 treatment led to the apoptosis of MCF-7 cells.
Expression of apoptosis-related genes
The analysis of gene expression revealed that treatment with 25% and 50% IC50 TP4 resulted in increased expression of apoptotic genes (Bax, casp3, and p53) and decreased expression of the anti-apoptotic gene (Bcl2) in MCF-7 cells compared to the control group (P < 0.05)(Fig. 4). The group treated with 50% IC50TP4 exhibited the most significant changes in gene expression among the aforementioned genes (P < 0.05) (Fig. 5).
Expression levels of Bcl2, Bax, p53, and caspase3 genes in MCF-7 cells were assessed following treatment with Tilapia piscidin 4 (TP4) at concentrations of 25% and 50%. The data were normalized to the housekeeping gene GAPDH and presented as the mean fold change ± SD. Each experiment was performed in triplicate, with a total of three independent experiments (n = 6, technical replicate). Different letters show significant difference between groups at p < 0.05 level. Asterisks (*) indicates significant difference between control and treatment groups at p < 0.01 level.
Discussion
The Tilapia piscidin 4 (TP4) is an antimicrobial peptide (AMP) with potential therapeutic efficacy in breast cancer treatment. This study evaluated the effect and molecular mode of action of TP4 on breast cancer cells. TP4 demonstrated a dose-dependent inhibition of cell growth in the MCF-7 cell line. The study by Ting et al. (2018) reported the cytotoxic effects of TP4 treatment on A549 lung cancer cells21, aligning with the findings of our present study. The IC50value for these cells was significantly lower compared to normal cells (MCF-10), indicating increased sensitivity. Additionally, the IS value exceeded 2, indicating the high selectivity of TP4 for cancer cells. AMPs exhibit selectivity due to the elevated negative charge present at the outer cellular surface of cancer cells compared to noncancer mammalian cells, a widely recognized phenomenon30.
Apoptosis induction through the use of cytotoxic natural substances is considered a pivotal approach in cancer treatment and the development of novel drugs31. Treatment of MCF-7 cells with 25% and 50% IC50TP4 resulted in upregulated expression of pro-apoptotic genes (Bax, P53, caspase) and downregulated expression of the anti-apoptotic gene (Bcl2). Additionally, the comet length was significantly greater in this experimental group compared to the control group. Conversely, assessment of oxidative stress indicators revealed enhanced intracellular levels of ROS and MDA content and reduced activity of antioxidant enzymes. TP4 has been found to effectively reduce the viability of MCF-7 cells by inducing excessive production of reactive oxygen species (ROS) in mitochondria. This, in turn, triggers caspase-mediated intrinsic apoptosis. Through real-time PCR analysis, we have demonstrated that TP4 exposure to MCF-7 cells triggers the initiation of the apoptosis pathway by perturbing the levels of Bax and Bcl-2. Bax, a proapoptotic gene, has been documented to be upregulated in p53-mediated apoptosis across various systems32. The increased level of p53 induced by TP4 in MCF-7 cells demonstrates its role in activating mitochondrial apoptosis. This activation is facilitated by the disturbance in Bax and Bcl-2 levels, leading to mitochondrial dysfunction and subsequent caspase-3 activation33. Based on our gene expression analysis, we observed a significant up-regulation of Bax mRNA expression in TP4-treated cells compared to the control group, indicating that TP4 influences the Bax/Bcl2 ratio, at least in part, through transcriptional regulation of Bax. While we observed a decrease in Bcl2mRNA expression, we cannot rule out the possibility that TP4 may also influence Bcl-2 activity through post-translational modifications. Caspase-3, a family of proteases, plays a central role in apoptotic development and serves as a critical component in DNA fragmentation, chromatin condensation, and other apoptotic processes34. Consistent with our findings, Su et al. (2019) reported that TP4 induces mitochondrial hyperpolarization and elevates mitochondrial ROS in human synovial sarcoma cells. Additionally, TP4 downregulates antioxidant enzyme levels, potentially increasing cellular susceptibility to ROS-induced damage in these cells. While the precise mechanisms require further investigation, ROS can activate caspase-3 through several potential routes. Firstly, ROS can directly oxidize cardiolipin, a phospholipid exclusively located in the inner mitochondrial membrane. Cardiolipin oxidation disrupts its interaction with cytochrome c, leading to the release of cytochrome c into the cytosol35. Secondly, ROS can indirectly modulate the Bcl-2 family of proteins and promote the activation of pro-apoptotic proteins like Bax, which are key regulators of mitochondrial outer membrane permeabilization (MOMP)36,37. The release of cytochrome c into the cytosol triggers the formation of the apoptosome, which in turn activates caspase-9, and subsequently caspase-3, leading to the execution of apoptosis38. The study also revealed that TP4 treatment induced elevated DNA breakage and elongated comet tails in U87MG and U251 cells, as observed in the comet assay20.
In addition to ROS, TP4 may also activate other apoptotic pathways, such as endoplasmic reticulum (ER) stress and autophagy, which could contribute to its anticancer effects. Previous studies reported that TP4 has the potential to target endoplasmic reticulum (ER), disrupt calcium homeostasis, activate the stress-induced transcription factor (FOSB), and finally leads to the necrosis of cancer cells11,39. In contrast to ER stress, a previous study showed that the autophagy is not activated by TP4 in the brain cancer cells40. Further investigations are needed to fully elucidate the relative contributions of these pathways in TP4-mediated apoptosis. The cytotoxicity induced by TP4 varies depending on the cancer cell type, transcriptome, and gene regulation. In MDA-MB-231 and MCF-7 cells, TP4 activates full-length FOSB (FosB Proto-Oncogene, AP-1 Transcription Factor Subunit), whereas, in MB453 cells, it mainly induces the truncated form (FOSDB)11. Overexpression of either FOSB or FOSDB leads to cell death in all tested breast cancer cells, and knockdown of FOSB attenuates TP4-mediated cytotoxicity11. TP4-induced activation of FOSB (or FOSDB) relies on Ca2+signaling and mitochondrial dysfunction, resulting in necrotic death11, indicating the involvement of Ca2+-dependent FOSB signaling in TP4-induced cytotoxicity. Additionally, TP4 induces apoptosis in an osteosarcoma cell line, through the activation of extrinsic Fas/FasL- and intrinsic mitochondria-mediated pathways41. Pre-treatment of these cells with caspase-8 inhibitor (Z-IETD-FMK) or caspase-9 inhibitor (Z-LEHD-FMK) significantly reduces caspase-3 activation and prevents apoptosis41. In non-small-cell lung cancer cells, TP4-induced necrotic death was observed, rather than apoptotic death21. These findings demonstrate that TP4 stimulates distinct cytotoxic pathways across different cancer types. Several factors could contribute to this difference. Firstly, differences in receptor profiles between MCF-7 and NSCLC cells may play a role. MCF-7 cells are estrogen receptor-positive, while NSCLC cells exhibit diverse receptor profiles42. It’s conceivable that estrogen receptor signaling pathways influence the cellular response to TP4. Secondly, variations in basal antioxidant enzyme levels could affect the mode of cell death. MCF-7 cells might have a lower capacity to handle TP4-induced ROS production compared to NSCLC cells, pushing them towards apoptosis43. Conversely, NSCLC cells might be better equipped to neutralize ROS, leading to necrosis due to other mechanisms. Finally, differences in the expression levels of pro-apoptotic and anti-apoptotic proteins or variations in cellular metabolic activity could also influence the cell fate decision following TP4 treatment.
In addition to TP4, previous studies showed that other AMPs also has the cytotoxic potential against breast cancer. Teleb et al. (2022) found that scorpion venom antimicrobial peptide smp43 has the IC50value of 11.9 µg/ml and SI value of 7.89 for MCF-744. Hepcidin (TH1-5) isolated from the freshwater fish Oreochromis mossambicus was another AMP with IC50of 20,000 µg/ml for MCF-7 but no significant effect against NIH/3 T345. Shi et al. (2022) reported that IC50 of AMP isolated from Brucea javanica(plant) globulin fraction for MCF-7 was 0.12 µg/ml. However, they did not report SI value for this AMP46. Based on these reports and the present study, it seems that AMPs can be a good candidate against MCF-7 cells with high selectivity and low toxicity.
While our in vitro findings provide valuable insights into the mechanism by which TP4 exerts its anti-cancer effects in MCF-7 cells, this study has limitations. These results are derived from cell culture models, which may not fully recapitulate the complex tumor microenvironment and systemic factors present in vivo. Therefore, future studies are warranted to validate these findings in more physiologically relevant models (In particular, in xenograft mouse models).
Conclusion
In this study, we have found that the marine antimicrobial peptide TP4 may have therapeutic advantages in treating human breast cancer based on our in vitro data. TP4 induces DNA fragmentation and apoptosis in cells by triggering oxidative stress.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Harwansh, R. K. & Deshmukh, R. Breast cancer: an insight into its inflammatory, molecular, pathological and targeted facets with update on investigational drugs. Crit. Rev. Oncol. Hematol. 154, 103070 (2020).
Wu, B. et al. Temporal trends of breast cancer burden in the Western Pacific region from 1990 to 2044: implications from the global burden of disease study 2019. J. Adv. Res. 59, 189–199 (2024).
Feng, Y. et al. Breast cancer development and progression: risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 5(2), 77–106 (2018).
Dai, X. et al. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 5(10), 2929–2943 (2015).
Abu Gazia, M. & El-Magd, M. A. Ameliorative effect of cardamom aqueous extract on doxorubicin-induced cardiotoxicity in rats. J. Cells Tissues Organs. 206(1–2), 62–72 (2019).
Attia, A. M. et al. New 2-oxopyridine/2-thiopyridine derivatives tethered to a benzotriazole with cytotoxicity on MCF7 cell lines and with antiviral activities. J. Lett. Drug Des. Discovery. 17(2), 124–137 (2020).
Mahlapuu, M. et al. Antimicrobial peptides: an emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 6, 194 (2016).
Divyashree, M. et al. Clinical applications of antimicrobial peptides (AMPs): where do we stand now? Protein Pept. Lett. 27(2), 120–134 (2020).
Aghamiri, S. et al. Antimicrobial peptides as potential therapeutics for breast cancer. J. Pharmacol. Res. 171, 105777 (2021).
Zhang, G. & Sunkara, L. T. Avian antimicrobial host defense peptides: from biology to therapeutic applications. Pharmaceuticals (Basel). 7(3), 220–247 (2014).
Ting, C. H. et al. Targeting FOSB with a cationic antimicrobial peptide, TP4, for treatment of triple-negative breast cancer. Oncotarget 7(26), 40329–40347 (2016).
Ting, C. H. et al. The mechanisms by which pardaxin, a natural cationic antimicrobial peptide, targets the Endoplasmic reticulum and induces c-FOS. J. Biomaterials. 35(11), 3627–3640 (2014).
Zhang, C. & Yang, M. The role and potential application of antimicrobial peptides in autoimmune diseases. Front. Immunol. 11, 859 (2020).
Guaní-Guerra, E. et al. Antimicrobial peptides: general overview and clinical implications in human health and disease. Clin. Immunol. 135(1), 1–11 (2010).
Avand, A., Akbari, V. & Shafizadegan, S. In vitro cytotoxic activity of a Lactococcus lactis antimicrobial peptide against breast cancer cells. Iran. J. Biotechnol., 16(3), 213–220 (2018).
Hilchie, A. L. et al. Pleurocidin-family cationic antimicrobial peptides are cytolytic for breast carcinoma cells and prevent growth of tumor xenografts. Breast Cancer Res. 13, 1–16 (2011).
Lin, H. J. et al. Piscidin-1, an antimicrobial peptide from fish (hybrid striped bass Morone saxatilis X M. chrysops), induces apoptotic and necrotic activity in HT1080 cells. Zoolog. Sci. 29(5), 327–332 (2012).
Hazam, P. K. & Chen, J. Y. Therapeutic utility of the antimicrobial peptide Tilapia Piscidin 4 (TP4). Aquaculture Reports, 17: p. 100409. (2020).
Pan, C. Y. et al. Study of the antimicrobial activity of tilapia piscidin 3 (TP3) and TP4 and their effects on immune functions in hybrid tilapia (Oreochromis spp). J. PloS One. 12(1), e0169678 (2017).
Su, B. C., Pan, C. Y. & Chen, J. Y. Antimicrobial peptide TP4 induces ROS-Mediated necrosis by triggering mitochondrial dysfunction in Wild-Type and mutant p53 glioblastoma cells. Cancers (Basel). 11(2), 171 (2019).
Ting, C. H. & Chen, J. Y. Nile tilapia derived TP4 shows broad cytotoxicity toward to non-small-cell lung cancer cells. Marin Drug. 16(12), 506 (2018).
Neshani, A. et al. Extended-Spectrum antimicrobial activity of the low cost produced tilapia piscidin 4 (TP4) marine antimicrobial peptide. J. Res. Med. Dent. Sci. 6(5), 327–334 (2018).
Kameyama, Y. et al. Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15 + C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics. 15(7), 513–522 (2005).
Rashidi, M. et al. Selective cytotoxicity and apoptosis-induction of Cyrtopodion scabrum extract against digestive cancer cell lines. Int. J. Cancer Manage., 10(5), 1–7 (2017).
El-Magd, M. A. et al. Incensole acetate prevents beta-amyloid-induced neurotoxicity in human olfactory bulb neural stem cells. Biomed. Pharmacother. 105, 813–823 (2018).
Niazvand, F. et al. Effects of Quercetin-Loaded nanoparticles on MCF-7 human breast cancer cells. Med. (Kaunas). 55(4), 114 (2019).
Lee, K. et al. Cyclo(phenylalanine-proline) induces DNA damage in mammalian cells via reactive oxygen species. J. Cell. Mol. Med. 19(12), 2851–2864 (2015).
Tang, Z. H. et al. Platycodin D from platycodonis radix enhances the anti-proliferative effects of doxorubicin on breast cancer MCF-7 and MDA-MB-231 cells. Chin. Med. 9, 1–7 (2014).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 – ∆∆CT method. J. Methods. 25(4), 402–408 (2001).
Schweizer, F. Cationic amphiphilic peptides with cancer-selective toxicity. Eur. J. Pharmacol. 625(1–3), 190–194 (2009).
Lee, C. C. et al. Spine venom of crown-of-thorns starfish (Acanthaster planci) induces antiproliferation and apoptosis of human melanoma cells (A375.S2). Toxicon 91, 126–134 (2014).
Farooq, Z. et al. p53 Tumor Suppressor: Functional Regulation and Role in Gene Therapy. p53: A Guardian of the Genome Beyond, : p. 57. (2022).
Lin, S. Y. et al. Emodin induces apoptosis of human tongue squamous cancer SCC-4 cells through reactive oxygen species and mitochondria-dependent pathways. J. Anticancer Res. 29(1), 327–335 (2009).
Su, T. R. et al. Induction of apoptosis by 11-dehydrosinulariolide via mitochondrial dysregulation and ER stress pathways in human melanoma cells. Mar. Drugs. 10(8), 1883–1898 (2012).
Fiorucci, L. et al. Cytochrome C interaction with Cardiolipin plays a key role in cell apoptosis: implications for human diseases. Symmetry 14(4), 767 (2022).
Liu, D. et al. MOMP: A critical event in cell death regulation and anticancer treatment. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer, : p. 189280. (2025).
Yapryntseva, M. A., Zhivotovsky, B. & Gogvadze, V. Permeabilization of the outer mitochondrial membrane: Mechanisms and consequences. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease, : p. 167317. (2024).
Zhou, Z. et al. Diverse functions of cytochrome C in cell death and disease. Cell. Death Differ. 31(4), 387–404 (2024).
Bhat, T. A. et al. Endoplasmic reticulum-mediated unfolded protein response and mitochondrial apoptosis in cancer. Biochimica et biophysica acta (BBA)-Reviews on cancer, 1867(1), 58–66 (2017).
Su, B. C., Pan, C. Y. & Chen, J. Y. Antimicrobial peptide TP4 induces ROS-mediated necrosis by triggering mitochondrial dysfunction in wild-type and mutant p53 glioblastoma cells. Cancers 11(2), 171 (2019).
Kuo, H. M. et al. MSP-4, an antimicrobial peptide, induces apoptosis via activation of extrinsic Fas/FasL- and intrinsic Mitochondria-Mediated pathways in one osteosarcoma cell line. Mar. Drugs. 16(1), 8 (2018).
Siegfried, J. M., Hershberger, P. A. & Stabile, L. P. Estrogen Receptor Signaling In Lung Cancer. In Seminars In Oncology (Elsevier, 2009).
Mitra, S. et al. Impact of ROS generated by chemical, physical, and plasma techniques on cancer Attenuation. Cancers 11(7), 1030 (2019).
Teleb, W. K. et al. Cytotoxicity and molecular alterations induced by Scorpion venom antimicrobial peptide Smp43 in breast cancer cell lines MDA-MB-231 and MCF-7. Int. J. Pept. Res. Ther. 29(1), 8 (2022).
Hassan, M. A. et al. Cytotoxic effect of Hepcidin (TH1-5) on human breast cancer cell line (MCF7). Jurnal Teknologi, 77(3), 73–79 (2015).
Shi, H. et al. Antitumor potential of peptides isolated from Brucea Javanica Globulin fraction on MCF-7 cells. Pharmacognosy Magazine, 18(80), 1129–1136 (2022).
Acknowledgements
We would like to thank Antimicrobial Resistance Research Center, Mashhad University of Medical Sciences (Mashhad, Iran) and the Pasteur Institute of Iran located in Tehran for providing the TP4 and cancer cell line.
Funding
There is no funding.
Author information
Authors and Affiliations
Contributions
Rashid Alijani Ardeshir designed the study and wrote the manuscript. Kosar Moarefvand performed the experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ardeshir, R.A., Moarefvand, K. Selective apoptosis induction by antimicrobial peptide TP4 in MCF-7 breast cancer cells. Sci Rep 15, 15061 (2025). https://doi.org/10.1038/s41598-025-97906-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-97906-3
Keywords
This article is cited by
-
Antimicrobial peptides for anticancer and antiviral therapy: last promising update
Discover Oncology (2025)