Abstract
SLC25A19 is a protein that mediates the transport of thiamine pyrophosphate (TPP) across cell membranes, but its significance in cancer remains largely unknown. To address this knowledge gap, we conducted a pan-cancer analysis to investigate the role of SLC25A19 in cancer. To investigate the role of SLC25A19 in cancer, we obtained RNA-seq, survival, and clinicopathological data from 33 cancers in the TCGA database. We performed expression and survival analyses of SLC25A19 across these cancers. Additionally, we explored the diagnostic and prognostic value of SLC25A19 specifically in hepatocellular carcinoma (HCC). To understand the potential role of SLC25A19 in the immune microenvironment of HCC, we also analyzed its expression in relation to immune checkpoint genes. Then we identified the drug most strongly correlated with SLC25A19 expression in HCC. Finally, PCR and Western-Blotting experiments were performed on HCC and its paracancer tissues to verify the expression of SLC25A19. Our analysis revealed that SLC25A19 is overexpressed in hepatocellular carcinoma (HCC) and is associated with poor prognosis. We also found that high expression of SLC25A19 in HCC is associated with high expression of immune checkpoint genes, suggesting a potential role for SLC25A19 in immunotherapy. Furthermore, we identified drugs that are most strongly correlated with SLC25A19 expression in HCC, which could inform treatment decisions for HCC patients. PCR results showed that the expression of SLC25A19 in HCC was significantly up-regulated in 6 pairs of HCC and adjacent normal tissues. Our study can provide a reference for the diagnosis and treatment of HCC.
Similar content being viewed by others
Introduction
Metabolic reprogramming is a well-established hallmark of cancer that involves changes in glucose, amino acid, lactic acid, and nucleotide metabolism, among other processes1,2,3. Cancer cells require high metabolic activity to sustain their rapid replication and growth, making targeting cancer metabolism a promising approach to treatment4. In fact, efforts to inhibit cancer metabolism date back to the 1940s when methotrexate was discovered to inhibit nucleotide metabolism in cancer cells5. Since then, therapies targeting cancer metabolism, including 6-mercaptopurine, 5-fluorouracil, gemcitabine, and cytarabine, have shown promising clinical results6. As such, exploring cancer metabolism and identifying potential targets is a logical approach to developing new cancer therapies7.
The hypermetabolic state of cancer is dependent on the transmembrane transport of multiple substrates, with solute carrier (SLC) and ATP-binding cassette (ABC) transporters being the most important carriers6,7. While ABC transporters rely mainly on ATP hydrolysis for energy supply, SLC transporters primarily mediate secondary active transport and have a wide range of substrates, including small amino acids, lactic acid, ions, sugars, and vitamins8,9. SLC25A19 is a transporter that mediates the transmembrane transport of thiamine pyrophosphate, a vitamin that serves as a cofactor for several metabolic enzymes and is essential for homeostasis regulation10. Targeting SLC25A19 may therefore be a potential treatment for cancer. However, the role of SLC25A19 in cancer has not been comprehensively addressed in pan-cancer analyses11.
To investigate the role of SLC25A19 in cancer, we conducted a comprehensive bioinformatics analysis that included expression analysis, survival analysis, clinical correlation analysis, co-expression analysis, and immune microenvironment analysis across multiple cancer types. Our results suggest that SLC25A19 is overexpressed in hepatocellular carcinoma (HCC) and is associated with poor prognosis. High expression of SLC25A19 in HCC is also associated with high expression of immune checkpoint genes, suggesting a potential role for SLC25A19 in immunotherapy. Additionally, we identified drugs that are most strongly correlated with SLC25A19 expression in HCC. Overall, our study provides new insights into the significance of SLC25A19 in cancer and highlights its potential as a novel marker for cancer treatment.
Methods
Data acquisition
We obtained RNA-seq, survival, and clinicopathological data from the TCGA database using UCSC Xena (https://xena.ucsc.edu/). RNAseq data was converted by log2(x + 1). We analyzed data from 33 cancer types, including 374 hepatocellular carcinoma (HCC) samples and 50 normal liver controls, all of which had corresponding paired HCC tumor samples. To validate our findings, we obtained data sets GSE45436, GSE57957, and GSE121248 from the NCBI website (https://www.ncbi.nlm.nih.gov/geo/) to verify the expression level of SLC25A19 in liver cancer. We processed and mapped the data using R software (4.1.2). We performed expression analysis, survival analysis, clinical correlation analysis, co-expression analysis, and immune microenvironment analysis to evaluate the role of SLC25A19 in cancer.
Pan-cancer analysis of SLC25A19
We analyzed the expression of SLC25A19 in 33 types of cancer and normal samples using RNAseq data from the TCGA database. To compare expression differences between tumor and normal samples, we performed the Mann–Whitney U test. We then conducted survival correlation analysis, using the K–M curve to explore the relationship between SLC25A19 expression and patient survival. We considered P < 0.05 to be statistically significant.
Significance of SLC25A19 in hepatocellular carcinoma
We analyzed the expression of SLC25A19 in hepatocellular carcinoma (HCC) and normal controls in the TCGA database using the Mann–Whitney U test, with P < 0.05 considered statistically significant. We used the CCLE database (https://sites.broadinstitute.org/ccle/) to input the gene “SLC25A19” and then selected liver cancer-related cell lines to obtain the expression levels of this gene in various cell lines. We further analyzed SLC25A19 expression in 50 paired HCC and normal controls using the Shapiro–Wilk Normality test and paired t-test. To assess the accuracy of SLC25A19 expression in the TCGA database, we constructed a Receiver Operating Characteristic Curve (ROC) using the “pROC” R package and evaluated the area under the Curve (AUC). We also verified the expression of SLC25A19 in HCC using GEO data, again using the Mann–Whitney U test for analysis.
Additionally, we performed survival analysis using the K–M curve to explore the correlation between SLC25A19 expression and HCC survival, including overall survival (OS), disease-specific survival (DSS), disease-free survival (DFS), and progression-free survival (PFS), with P < 0.05 considered statistically significant. Finally, we investigated the correlation between SLC25A19 and clinical characteristics of HCC, including age, stage, grade, and T staging.
Co-expression analysis of SLC25A19 in HCC
We identified the top 50 genes with high co-expression correlation with SLC25A19 using the GEPIA website (http://gepia.cancer-pku.cn/). To investigate the function of these co-expressed genes, we performed a functional enrichment analysis using the Metascape website (https://metascape.org/gp/index.html). We then intersected the genes related to SLC25A19 obtained from the GEPIA website with the genes obtained from the Cbioportal website (http://www.cbioportal.org/) using Venn diagrams, resulting in the identification of the most significant co-expressed genes with SLC25A19. Finally, we used the “Pearson correlation analysis” method of the “Corrplot” R package to study the correlation between SLC25A19 and these intersection genes.
The role of SLC25A19 co-expressed genes in HCC
To investigate the role of co-expressed genes in HCC, we performed differential expression analysis, ROC curve analysis, and survival analysis. We extracted the expression data of co-expressed genes in HCC and used the Mann–Whitney U test to investigate the difference in expression between tumor and normal samples (*P < 0.05; **P < 0.01; ***P < 0.001; NS: P ≥ 0.05). We evaluated the prediction ability of co-expressed genes using the AUC value of the ROC curve and determined whether it had high accuracy. We also evaluated the prognostic value of co-expressed genes using the K–M curve, with P < 0.05 considered statistically significant.
Enrichment analysis and immune microenvironment analysis between high-SLC25A19 expression group and low -SLC25A19 expression group
We divided HCC samples in the TCGA database into two groups based on the expression level of SLC25A19: the high-SLC25A19 expression group and the low-SLC25A19 expression group. Using the “limma” R package with “fdrFilter” = 0.05 and “logFCfilter” = 1, we obtained all the differentially expressed genes between the two groups. Using the “ggplot2” package, a volcano plot was created to display the differentially expressed genes obtained from the differential analysis of SLC25A19. The top 5 genes with the most significant differences were highlighted. We then analyzed the function and pathway enrichment of these differentially expressed genes using gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment, with GO and KEGG enrichment analysis performed using the “ClusterProfiler” R package. Additionally, we analyzed differences in the immune microenvironment and immune checkpoint between the two groups by performing single-sample gene set enrichment analysis (ssGSEA) using the “GSVA” R package to obtain the abundance of different immune cells in HCC. We used the “limma” R package to analyze the difference in the level of immune cell infiltration between the high-SLC25A19 and low-SLC25A19 expression groups. Finally, we performed differential expression analysis of immune checkpoints by extracting the expression of immune checkpoint-related genes in HCC and analyzing the expression difference of immune checkpoint genes between the two groups using the “limma” R package.
Drug sensitivity analysis
We obtained gene expression data and drug correlation data from the CallMiner website (https://discover.nci.nih.gov/cellminer/home.do) and used the “limma” R package to analyze the correlation between SLC25A19 expression and drugs. We identified the top 4 drugs with the greatest association. The “pRRophetic” package was used to predict the sensitivity of relevant drugs based on the expression matrix of HCC patients. Patients were divided into high and low expression groups according to the median value of SLC25A17 expression. The rank-sum test method was then used to explore the distribution of IC50 of relevant drugs between the two groups.
PCR and WB to verify the expression of SLC25A19 in HCC
PCR experiments were conducted to compare gene expression levels between 6 pairs of HCC and Para-HCC tissues. The ethics committee of Fuyang Hospital of Anhui Medical University has approved this study (KY2021020). Total RNA was isolated from the tissues using the TRIzol reagent. cDNA was synthesized using the PrimeScript™ RT reagent kit. Real-time PCR was performed using the SYBR Green Real-time PCR Master Mix on an Applied Biosystems QuantStudio 7 Flex Real-time PCR System. The relative gene expression levels were calculated using the 2−ΔΔCt method, with GAPDH as the internal reference gene. The primer sequence of SLC25A19 is Forward: TCCAGGCCTCTAGGCAGATT; Reverse: ATAGACCTTGGGCTCACCCT. GAPDH Forward: AATGGGCAGCCGTTAGGAAA; Reverse: GCCCAATACGACCAAATCAGAG. All experiments were performed in triplicate, and the data were analyzed using GraphPad Prism software. To verify the expression of SLC25A19 at the protein level, Western blot (WB) analysis was conducted. Protein was extracted from the same 6 pairs of HCC and Para-HCC tissues using RIPA lysis buffer supplemented with protease and phosphatase inhibitors. Protein concentrations were determined using the BCA Protein Assay Kit. Equal amounts of protein (20 µg) were separated by 10% SDS-PAGE and transferred to PVDF membranes. The membranes were blocked with 5% non-fat milk in TBST (20 mM Tris-HCl, 150 mM NaCl, 0.1% Tween-20) for 1 h at room temperature and then incubated overnight at 4 °C with the following primary antibodies: anti-SLC25A19 (1:1000, Abcam, ab190228) and anti-β actin (1:5000, Abcam, ab8227) as a loading control. After washing with TBST, the membranes were incubated with HRP-conjugated secondary antibodies (1:3000, Cell Signaling Technology) for 1 h at room temperature. The protein bands were visualized using an ECL detection system (Tanon), and the intensity of the bands was quantified using ImageJ software. The relative expression levels of SLC25A19 were normalized to β actin and analyzed using GraphPad Prism software.
Cell culture
The human hepatocellular carcinoma (HCC) cell line Hep3B was cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, and maintained at 37 °C in a humidified incubator with 5% CO2.
Transwell migration and invasion assays
To investigate the functional role of SLC25A19, Hep3B cells were transfected with short hairpin RNA (shRNA) targeting SLC25A19 (sh-SLC25A19) or a non-targeting control shRNA (sh-NC) using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s protocol. Cell migration and invasion were assessed using Transwell chambers (8 μm pore size, Corning, USA). For the migration assay, 5 × 104 transfected Hep3B cells in serum-free DMEM were seeded into the upper chamber, while the lower chamber was filled with DMEM containing 10% FBS as a chemoattractant. After 24 h of incubation at 37 °C, cells that had migrated to the lower surface of the membrane were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and counted under a microscope. For the invasion assay, the procedure was similar, except the Transwell membrane was pre-coated with Matrigel (BD Biosciences, USA) to mimic the extracellular matrix. Experiments were performed in triplicate, and the average number of migrated or invaded cells was calculated.
Scratch wound healing assay
To evaluate cell migration capacity, Hep3B cells transfected with sh-NC or sh-SLC25A19 were seeded into 6-well plates and cultured to nearly 100% confluence. A uniform scratch was made across the cell monolayer using a sterile 200 µL pipette tip. The detached cells were removed by washing with phosphate-buffered saline (PBS), and the cells were incubated in serum-free DMEM. Images were captured at 0 and 24 h after scratching using a light microscope, and the relative migration distance was quantified using ImageJ software. Each experiment was performed in triplicate.
Statistical analysis
For differential expression analysis of unpaired samples, we used the Mann–Whitney U test. Paired samples were tested using the Shapiro–Wilk normality test and paired t-test. We evaluated the prediction ability of gene expression using the AUC value of the ROC curve, with AUC values ranging from 0.7 to 0.9 considered to have a certain accuracy and values above 0.9 considered to have high accuracy. We analyzed the relationship between gene expression level and patient survival using the K–M curve, with P < 0.05 considered statistically significant. We performed drug sensitivity analysis using Pearson correlation analysis, with P < 0.05 used as the statistical significance threshold.
Results
Pan-cancer analysis of SLC25A19
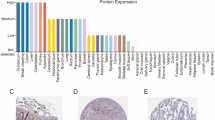
We analyzed the expression and prognostic value of SLC25A19 in pan-cancer. Figure 1A showed differential expression of SLC25A19 in 17 types of cancer in the TCGA database. We explored the relationship between SLC25A19 expression and tumor survival time using the K–M curve and found that SLC25A19 had prognostic significance in 9 types of cancer (P < 0.05) as shown in Fig. 1B–J. We also found that SLC25A19 was overexpressed in HCC and that the SLC25A19-overexpression group was associated with a poor prognosis (P < 0.05). Given this observation, we conducted a series of analyses to investigate the potential role of SLC25A19 in HCC.
Pan-cancer analysis of SLC25A19. (A) SLC25A19 was differentially expressed in 17 cancer types, including BLCA, BRCA, CESC, CHOL, COAD, ESCA, GBM, HNSC, KIRC, KIRP, LIHC, PCPG, PRAD, READ, STAD, THCA, and UCEC. (B) In ACC, high expression of SLC25A19 was associated with poor prognosis (P < 0.05). (C) In ESCA, high expression of SLC25A19 was associated with poor prognosis (P < 0.05). (D) In KICH, high expression of SLC25A19 was associated with poor prognosis (P < 0.05). (E) In KIRC, high expression of SLC25A19 was associated with poor prognosis (P < 0.05). (F) In LGG, high expression of SLC25A19 was associated with poor prognosis (P < 0.001). (G) In LIHC, high expression of SLC25A19 was associated with poor prognosis (P < 0.05). (H) In MESO, high expression of SLC25A19 was associated with good prognosis (P < 0.05). (I) In UCEC, high expression of SLC25A19 was associated with poor prognosis (P < 0.05). (J) In UVM, high expression of SLC25A19 was associated with poor prognosis (P < 0.05).
Significance of SLC25A19 in hepatocellular carcinoma
To verify the expression and clinical relevance of SLC25A19 in HCC, we performed TCGA and GEO expression analyses, survival analysis, and clinical relevance analysis. TCGA expression analysis showed that SLC25A19 was highly expressed in HCC (P < 0.001) as depicted in Fig. 2A. In addition, SLC25A19 expression was significantly up-regulated in HCC based on the differential expression analysis of 50 pairs of samples, as shown in Fig. 2B (***P < 0.001). The AUC value of the ROC curve of SLC25A9 was 0.904, indicating that SLC25A19 has strong diagnostic value for HCC (Fig. 2C). We then verified the expression of SLC25A19 using three datasets (GSE45436, GSE57957, and GSE121248) from the GEO database, which showed that SLC25A19 is highly expressed in HCC, consistent with the results from the TCGA database (Fig. 2D–F). Survival analysis demonstrated that high expression of SLC25A19 predicted poor OS (P = 0.004), DSS (P < 0.001), DFS (P = 0.009), and PFS (P < 0.001) in HCC patients, as depicted in Fig. 2G–J. Univariate and multivariate Cox analysis results presented in Table 1 indicated that stage and SLC25A19 expression levels can be used as independent prognostic factors of HCC. Table 2 showed the clinical data of the high-SLC25A19 expression group and the low-SLC25A19 expression group. Finally, we analyzed the clinical correlation, as shown in Fig. 2K–N. Figure 2K demonstrated that SLC25A19 expression was higher in HCC patients aged < = 65 years. Figure 2L showed that the expression level of SLC25A19 in HCC patients in the G3 phase was higher than that in G1 and G2 phases (P < 0.05). Figure 2M showed that the expression level of SLC25A19 in stage I HCC patients was lower than that in Stage II and Stage III (P < 0.05). Figure 2N showed that the expression level of SLC25A19 in stage T1 HCC patients was lower than that in stage T2 and T3 (P < 0.05). Furthermore, we established a nomogram for a patient with hepatocellular carcinoma based on SLC25A19 expression (Supplemental Fig. 1), and the results showed that the 1, 3, and 5-year mortality rates were 0.0876, 0.197, and 0.293, respectively.
Significance of SLC25A19 in hepatocellular carcinoma. (A) TCGA analysis. SLC25A19 was highly expressed in HCC (P < 0.001). (B) TCGA analysis. In 50 pairs of samples, SLC25A19 was significantly up-regulated in HCC (***P < 0.001). (C) TCGA analysis. The AUC value of the ROC curve of SLC25A9 was 0.904, indicating that SLC25A19 has a strong diagnostic value for HCC.
Co-expression analysis of SLC25A19 in HCC
To identify genes that act in concert with SLC25A19, we performed a co-expression analysis. The top 50 genes with high correlation to SLC25A19 analyzed by the GEPIA website are listed in Supplemental Table 1. Figure 3A shows the enriched functions and pathways among these genes, including the Regulation of TP53 Activity, which regulates tumor growth, and the Regulation of Cell Cycle, Mitotic, Cell division, and Cell Cycle process involved in Cell function. Additionally, we obtained genes with high correlation by intersecting the GEPIA (50 genes) and Cbioportal (20 genes) analyses (Fig. 3B), resulting in 10 genes: CENPA, CCDC137, WDR62, FANCI, KPNA2, MYBL2, PTTG1, KIF2C, TROAP, and CENPM. These genes are considered the most co-expressed genes associated with SLC25A19. Figure 3C shows the correlation between these 10 genes and SLC25A19, where red indicates a positive correlation, and “*” indicates a correlation P value less than 0.05.
Co-expression analysis of SLC25A19 in HCC. (A) The enriched functions or pathways of co-expressed genes. These include the regulation of TP53 Activity regulating tumor growth and the regulation of cell cycle, mitotic, cell division, and cell cycle process involved in cell function. (B) The intersection of GEPIA (50 co-expressed genes) and Cbioportal (20 co-expressed genes) analysis. 10 genes were obtained: CENPA, CCDC137, WDR62, FANCI, KPNA2, MYBL2, PTTG1, KIF2C, TROAP, and CENPM. (C) The correlation between these 10 co-expressed genes and SLC25A19.
The role of SLC25A19 co-expressed genes in HCC
To further analyze the 10 co-expressed genes associated with SLC25A19, we performed expression analysis, ROC, and survival analysis. Figure 4A demonstrates that these 10 genes are highly expressed in HCC (P < 0.001). ROC analysis depicted in Fig. 4B showed that eight of these genes had AUC values greater than 0.9, indicating strong diagnostic value for HCC. Finally, survival analysis indicated that high expression of these 10 genes predicted poor prognosis of HCC patients, as shown in Figs. 4C-L. Supplemental Fig. 2 reveals multivariate COX regression results of co-expressed genes, most of which are highly correlated with the prognosis of HCC patients. These findings suggest that SLC25A19 may synergistically promote the progression of liver cancer and affect the survival prognosis of patients with co-expressed genes.
The role of 10 co-expressed genes in HCC. (A) Expression analysis of 10 co-expressed genes in HCC. The results showed that all 10 genes were up-regulated in HCC (***P < 0.001). (B) ROC curves of 10 co-expressed genes in HCC. All of them have high diagnostic value for HCC. (C–L) Survival analyses of 10 co-expressed genes in HCC. All co-expressed genes were associated with poor prognosis of HCC (P < 0.05).
Enrichment analysis and immune microenvironment analysis between high-SLC25A19 expression group and low-SLC25A19 expression group
To investigate the role of SLC25A19 in HCC, we divided patients into high and low expression groups based on SLC25A19 expression level. Differentially expressed genes between the two groups were obtained (Fig. 5A). GO enrichment analysis showed that differentially expressed genes were associated with cell division, chromosomal region, and other cell functions, as shown in Fig. 5B. Similarly, KEGG pathway analysis revealed pathways such as cell circulation and DNA replication that are involved in tumorigenesis, as depicted in Fig. 5C.
Furthermore, we analyzed the difference in immune infiltration levels between the two expression groups. The high-SLC25A19 expression group showed lower infiltration of B cells, CD8+ T cells, DCs, Mast cells, neutrophils, NK cells, pDCs, T Helper cells, and TIL cells (Fig. 5D). Interestingly, we also found that many immune checkpoint genes were highly expressed in the SLC25A19 high expression group (Fig. 5E), including PDCD1, which can inhibit T cell activity. This finding is consistent with the role of SLC25A19 in promoting tumor progression.
Enrichment analysis and immune microenvironment analysis between high-SLC25A19 expression group and low-SLC25A19 expression group. (A) Volcano map of differentially expressed genes. (B) GO enrichment results of differentially expressed genes between the two groups. We found that differentially expressed genes were associated with cell division, chromosomal region, and many other cell functions. (C) KEGG pathway enriched by differentially expressed genes. These include pathways such as cell circulation and DNA replication that are involved in tumorigenesis. (D) The difference in immune infiltration level between the two groups with high and low-SLC25A19 expression. The high-SLC25A19 expression group had lower B cells, CD8+ T cells, DCs, Mast cells, neutrophils, NK cells, pDCs, T Helper cells, and TIL cell infiltration. (E) Many immune checkpoint genes were highly expressed in the SLC25A19 group.
Drug sensitivity analysis
Figure 6 shows the results of our drug sensitivity analysis for SLC25A19, which identified four drugs with a significant positive correlation (P < 0.001): Vorinostat (Cor = 0.488), Hydroxyurea (Cor = 0.486), Parthenolide (Cor = 0.454), and Cladribine (Cor = 0.451). Vorinostat induces cell growth arrest and apoptosis, while Hydroxyurea inhibits DNA synthesis by targeting ribonucleoside diphosphate reductase. Importantly, these mechanisms of drug action align with our previous enrichment analysis, supporting the notion that SLC25A19 could be a potential therapeutic target for HCC. In addition, we also conducted drug sensitivity analysis on the commonly used chemotherapy drugs for HCC, Imatinib and Lapatinib, and the results showed that HCC patients with low expression of SLC25A19 had lower IC50, that is, more sensitive to Imatinib and Lapatinib (Fig. 6B and C).
Drug sensitivity analysis. (A) Four drugs with the highest correlation with SLC25A19 were showed, including Vorinostat (Cor = 0.488), Hydroxyurea (Cor = 0.486), Parthenolide (Cor = 0.454), Cladribine (Cor = 0.451). The X-axis represents gene expression and the Y-axis represents correlation. (B) Drug sensitivity analysis of Imatinib. (C) Drug sensitivity analysis of Lapatinib.
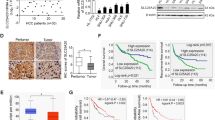
Validation of the expression of SLC25A19 in HCC patients
Firstly, PCR was performed to assess the expression of SLC25A19 in HCC and adjacent normal tissues. The results indicated that SLC25A19 expression was significantly upregulated in HCC tissues compared to adjacent normal tissues in six paired samples (Fig. 7A, P < 0.01). To further validate this observation, Western blotting was conducted on six paired samples (T group: HCC tissues, P group: para-normal tissues). Consistent with the PCR results, the protein expression levels of SLC25A19 were elevated in most HCC tissues compared to adjacent normal tissues (Fig. 7B). Additionally, Cox regression analysis was conducted on multiple datasets, including TCGA, ICGC, and GEO, to explore the prognostic significance of SLC25A19. The analysis revealed that SLC25A19 expression was significantly associated with overall survival (OS) and other clinical outcomes such as disease-free survival (DFS) and progression-free survival (PFS) in HCC patients, highlighting its potential prognostic value (Fig. 7C). Next, we performed a comprehensive analysis of SLC25A19 expression across multiple hepatocellular carcinoma cell lines using the CCLE database. The data demonstrated high levels of SLC25A19 expression in the majority of these cell lines, further supporting its upregulation in HCC (Fig. 7D). To examine the functional role of SLC25A19 in HCC, we conducted transwell migration and invasion assays. The results demonstrated that silencing SLC25A19 (sh-SLC25A19) significantly inhibited the migratory and invasive capabilities of HCC cells compared to the control group (sh-NC) (Fig. 7E, P < 0.01). Moreover, scratch wound healing assays further validated these findings, showing that knockdown of SLC25A19 markedly reduced the migration capacity of HCC cells after 24 h (Fig. 7F, P < 0.05).
Validation of SLC25A19 expression and its functional role in HCC. (A) RT-qPCR analysis revealed that SLC25A19 expression was significantly upregulated in six paired HCC and adjacent normal tissues (P < 0.01). (B) Western blotting further confirmed that the protein levels of SLC25A19 were elevated in HCC tissues (T group) compared to para-normal tissues (P group). (C) Cox regression analysis based on multiple datasets (TCGA, ICGC, and GEO) demonstrated the prognostic significance of SLC25A19 in overall survival (OS), disease-free survival (DFS), progression-free survival (PFS), and other clinical outcomes in HCC patients. (D) Analysis of SLC25A19 expression in hepatocellular carcinoma cell lines using the CCLE database showed consistently high expression levels. (E) Transwell assays revealed that silencing SLC25A19 (sh-SLC25A19) significantly reduced the migratory and invasive abilities of HCC cells compared to the control group (sh-NC) (P < 0.01). (F) Scratch wound healing assays confirmed that knockdown of SLC25A19 markedly inhibited the migration capacity of HCC cells at 24 h (P < 0.05).
Discussion
Cancer cell metabolism has long been recognized as a potential therapeutic target, often described as the “Achilles’ heel” of cancer12,13. Although the concept of targeting cancer metabolism predates Warburg’s discovery of aerobic glycolysis in cancer cells14, it was not until Sidney Farber demonstrated that targeting metabolic pathways—specifically folic acid metabolism with amethopterin—could effectively treat leukemia15, that this approach gained traction. Despite the development and clinical use of various metabolic inhibitors over the years, their effectiveness remains limited. In the current era of immunotherapy, exploring synergistic strategies involving the modulation of cancer metabolism, oncogene activity, and immune reactivation presents a promising research avenue16.
The SLC25 family, or mitochondrial carrier family, encompasses a vital group of transmembrane proteins that transport metabolites, nucleotides, and coenzymes across the mitochondrial membrane, crucial for maintaining cellular metabolic balance and energy production. Recent research underscores the relevance of this family in cancer, noting aberrant expression patterns of these transporters in various cancers, which are implicated in tumor development, progression, and resistance to therapy.
SLC25A19, in particular, has emerged as a critical member of this family. It encodes the mitochondrial deoxyribose-phosphate carrier, facilitating the transport of 5-phosphodeoxyribose from the mitochondria to the cytoplasm—a key step in DNA synthesis and repair. Aberrant expression of SLC25A19 is strongly associated with cancer prognosis. Elevated levels of SLC25A19 are linked to poorer outcomes in certain cancers, whereas its reduced expression is associated with increased tumor aggressiveness.
Our research extends these findings by focusing on the role of SLC25A19, a thiamine pyrophosphate (vitamin B1) transporter, in cancer. Through pan-cancer analysis, we identified differential expression patterns of SLC25A19 across various cancer types, with particularly adverse prognostic implications in hepatocellular carcinoma (HCC). Our survival analysis further validated high SLC25A19 expression as a robust predictor of poor outcomes. Moreover, our co-expression analysis highlighted ten genes closely associated with SLC25A19, suggesting their potential as novel biomarkers for HCC. We also explored the interaction between SLC25A19 expression and the immune microenvironment, identifying potential synergies with immunotherapeutic approaches.
Despite the promising therapeutic potential of existing tyrosine kinase inhibitors like sorafenib for advanced HCC, their overall efficacy is suboptimal17,18. The unique metabolic profile of the liver, being the largest metabolic organ, underscores the potential of targeting metabolic pathways in HCC treatment19,20,21. Our findings suggest that patients with high SLC25A19 expression might particularly benefit from immune checkpoint inhibitors, proposing a combination of immunotherapy with SLC25A19-targeted approaches as a future treatment strategy.
Furthermore, our study uncovered a co-expression network involving SLC25A19 that may regulate HCC progression and serve as a prognostic tool. Investigating the biological functions and interactions of these co-expressed genes offers valuable insights into their potential therapeutic roles.
While our study provides substantial evidence of the role of SLC25A19 in HCC, it is limited by the absence of a clinical cohort and relevant trials to confirm these findings. Future research will aim to address these limitations, enhancing our understanding and therapeutic targeting of HCC.
Conclusions
Our study provides a robust prognostic marker for hepatocellular carcinoma (HCC), SLC25A19. Our analysis of immune microenvironment can provide a reference for the immunotherapy of HCC.
Data availability
The datasets analyzed in this study were downloaded and accessed from TCGA database: https://gdc.cancer.gov/about-data/gdc-data-processing/resources-tcga-users.
Abbreviations
- LAML:
-
Acute myeloid leukemia
- ACC:
-
Adrenocortical carcinoma
- CHOL:
-
Cholangio carcinoma
- BLCA:
-
Bladder urothelial carcinoma
- BRCA:
-
Breast invasive carcinoma
- CESC:
-
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- COAD:
-
Colon adenocarcinoma
- UCEC:
-
Uterine corpus endometrial carcinoma
- ESCA:
-
Esophageal carcinoma
- GBM:
-
Glioblastoma multiforme
- HNSC:
-
Head and neck squamous cell carcinoma
- KICH:
-
Kidney chromophobe
- KIRC:
-
Kidney renal clear cell carcinoma
- KIRP:
-
Kidney renal papillary cell carcinoma
- DLBC:
-
Lymphoid neoplasm diffuse large B-cell lymphoma
- LIHC:
-
Liver hepatocellular carcinoma
- LGG:
-
Brain lower grade glioma
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- SKCM:
-
Skin cutaneous melanoma
- MESO:
-
Mesothelioma
- UVM:
-
Uveal melanoma
- OV:
-
Ovarian serous cystadenocarcinoma
- PAAD:
-
Pancreatic adenocarcinoma
- PCPG:
-
Pheochromocytoma and paraganglioma
- PAD:
-
Prostate adenocarcinoma
- READ:
-
Rectum adenocarcinoma
- SARC:
-
Sarcoma
- STAD:
-
Stomach adenocarcinoma
- TGCT:
-
Testicular germ cell tumors
- THYM:
-
Thymoma
- THCA:
-
Thyroid carcinoma
- USC:
-
Uterine carcinosarcoma
References
Park, J. H., Pyun, W. Y. & Park, H. W. Cancer metabolism: Phenotype, signaling and therapeutic targets. Cells 9 (10), 2308. https://doi.org/10.3390/cells9102308 (2020).
Bose, S. & Le, A. Glucose metabolism in cancer. Adv. Exp. Med. Biol. 1063, 3–12. https://doi.org/10.1007/978-3-319-77736-8_1 (2018).
Vazquez, A. et al. Cancer metabolism at a glance. J. Cell. Sci. 129 (18), 3367–3373. https://doi.org/10.1242/jcs.181016 (2016).
Xie, J. et al. Database mining analysis revealed the role of the putative H+/sugar transporter solute carrier family 45 in skin cutaneous melanoma. Channels (Austin). 15 (1), 496–506. https://doi.org/10.1080/19336950.2021.1956226 (2021).
Chen, L. et al. Pan-cancer analysis and single-cell analysis revealed the role of ABCC5 transporter in hepatocellular carcinoma. Channels (Austin). 15 (1), 541–554. https://doi.org/10.1080/19336950.2021.1968592 (2021).
Farhadi, P., Yarani, R., Dokaneheifard, S. & Mansouri, K. The emerging role of targeting cancer metabolism for cancer therapy. Tumour Biol. 42 (10), 1010428320965284. https://doi.org/10.1177/1010428320965284 (2020).
Miranti, E. H. et al. Low vitamin B12 increases risk of gastric cancer: A prospective study of one-carbon metabolism nutrients and risk of upper gastrointestinal tract cancer. Int. J. Cancer. 141 (6), 1120–1129. https://doi.org/10.1002/ijc.30809 (2017).
Morioka, S. et al. Efferocytosis induces a novel SLC program to promote glucose uptake and lactate release. Nature 563 (7733), 714–718. https://doi.org/10.1038/s41586-018-0735-5 (2018).
Xie, J. et al. Solute carrier transporter superfamily member SLC16A1 is a potential prognostic biomarker and associated with immune infiltration in skin cutaneous melanoma. Channels (Austin). 15 (1), 483–495. https://doi.org/10.1080/19336950.2021.1953322 (2021).
Nobili, S. et al. Role of ATP-binding cassette transporters in cancer initiation and progression. Semin. Cancer Biol. 60, 72–95. https://doi.org/10.1016/j.semcancer.2019.08.006 (2020).
Zastre, J. A. et al. Up-regulation of vitamin B1 homeostasis genes in breast cancer. J. Nutr. Biochem. 24 (9), 1616–1624. https://doi.org/10.1016/j.jnutbio.2013.02.002 (2013).
Kroemer, G. & Pouyssegur, J. Tumor cell metabolism: Cancer’s Achilles’ heel. Cancer Cell 13 (6), 472–482. https://doi.org/10.1016/j.ccr.2008.05.005 (2008).
Sancho, P., Barneda, D. & Heeschen, C. Hallmarks of cancer stem cell metabolism. Br. J. Cancer. 114 (12), 1305–1312. https://doi.org/10.1038/bjc.2016.152 (2016).
Koppenol, W. H., Bounds, P. L. & Dang, C. V. Otto Warburg’s contributions to current concepts of cancer metabolism [published correction appears in Nat Rev Cancer. ;11(8):618]. Nat. Rev. Cancer 2011, 11 (5), 325–337. https://doi.org/10.1038/nrc3038 (2011).
Farber, S. & Farber, S. Some observations on the effect of folic acid antagonists on acute leukemia and other forms of incurable cancer. Blood. 1949;4(2):160–167. Blood 127 (3), 271. https://doi.org/10.1182/blood-2015-12-685586 (2016).
Yang, Y. Cancer immunotherapy: Harnessing the immune system to battle cancer. J. Clin. Investig. 125 (9), 3335–3337. https://doi.org/10.1172/JCI83871 (2015).
Abdelgalil, A. A., Alkahtani, H. M. & Al-Jenoobi, F. I. Sorafenib. Profiles Drug Subst. Excip. Relat. Methodol. 44, 239–266. https://doi.org/10.1016/bs.podrm.2018.11.003 (2019).
Greten, T. F., Manns, M. P. & Korangy, F. Immunotherapy of HCC. Rev. Recent Clin. Trials. 3 (1), 31–39. https://doi.org/10.2174/157488708783330549 (2008).
Yang, C., Huang, X., Liu, Z., Qin, W. & Wang, C. Metabolism-associated molecular classification of hepatocellular carcinoma. Mol. Oncol. 14 (4), 896–913. https://doi.org/10.1002/1878-0261.12639 (2020).
Li, J. et al. CD147 reprograms fatty acid metabolism in hepatocellular carcinoma cells through Akt/mTOR/SREBP1c and P38/PPARɑ pathways. J. Hepatol. 63 (6), 1378–1389. https://doi.org/10.1016/j.jhep.2015.07.039 (2015).
Fang, G. et al. Inhibition of GSK-3β activity suppresses HCC malignant phenotype by inhibiting glycolysis via activating AMPK/mTOR signaling. Cancer Lett. 463, 11–26. https://doi.org/10.1016/j.canlet.2019.08.003 (2019).
Acknowledgements
We are very grateful for data provided by databases such as TCGA, GEO.
Funding
The work was funded by grants from the Key Discipline of Hepatobiliary and Pancreatic Surgery of Jiaxing City [2023-ZC-005], Translational therapy center for hepatobiliary pancreatic cancer [2021-YJZX-04], and the pancreatic cancer project [2022-QMX-009][2023-YB-006] and National Clinical Key Specialty Construction Project [2023-GJZK-001].
Author information
Authors and Affiliations
Contributions
Liang Chen designed the study. Liang Chen was involved in database search and statistical analyses. Liang Chen, Xiaomei Ying, Jiaheng Xie and Hong Liu were involved in the writing of manuscript and its critical revision. All authors were responsible for the submission of the final version of the paper. All authors approved the final version. All authors agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical standards
All procedures performed were in accordance with the declaration of the ethical standards of the institutional research committee and with the 1964 Helsinki 387 Declaration and its later amendments. The ethics committee of Fuyang Hospital of Anhui Medical University has approved this study(KY2021020). The ethics committee of Jiaxing First Hospital has approved this study(2023-LY-502).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, L., Ying, X., Xie, J. et al. SLC25A19 is a key prognostic marker for hepatocellular carcinoma. Sci Rep 15, 13435 (2025). https://doi.org/10.1038/s41598-025-98371-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-98371-8