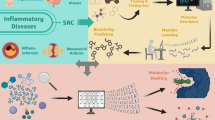
Abstract
The Gly2032Arg (G2032R) point mutation in proto-oncogene tyrosine-protein kinase 1 (ROS1) is one of the predominant factors of drug resistance to targeted therapies in patients with ROS1 fusion-positive non–small-cell lung cancer (NSCLC). This study aimed to identify novel inhibitors from a library of alkaloids (447 compounds) using computational approaches. Molecular docking-based virtual screening was performed to identify promising compounds, followed by ADMET property prediction and molecular dynamics simulations to assess their safety and stability. The top compounds identified were yibeinoside A and vomicine, which exhibited high binding affinities to the G2032R-mutant ROS1 protein. ADMET analysis indicated that yibeinoside A possessed better predicted pharmacokinetic profiles than vomicine and the positive control, lorlatinib. Molecular dynamics simulations demonstrated that yibeinoside A formed a highly stable complex with stable root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), and solvent accessible surface area (SASA) values. Molecular Mechanics Poisson–Boltzmann Surface Area (MM/PBSA) calculations further confirmed that yibeinoside A and vomicine had better binding free energies than lorlatinib. Collectively, these findings suggest that yibeinoside A, with its balanced binding interactions and favorable predicted pharmacokinetic profile, is a promising lead candidate for further development as a selective inhibitor against G2032R-mutant ROS1.
Similar content being viewed by others
Introduction
Cancer remains one of the most critical global health challenges, affecting millions of individuals worldwide1. In 2022 alone, an estimated 20 million new cases and 10 million cancer-related deaths were reported, underscoring the urgent need for more effective therapeutic strategies2. Over the past years, molecular oncology has advanced substantially. Improved understanding of tumorigenesis, together with the development of targeted therapies, has transformed cancer care by reducing adverse effects and improving patient survival3. Precision medicine, also referred to as personalized medicine, has further accelerated this progress by tailoring treatments based on individual genetic, lifestyle, and environmental factors4. Precision medicine has been widely adopted in cancer treatment, where genetic testing is used to identify specific mutations in cancer cells5. These mutated targets could provide valid information for targeted therapies and tailored treatments.
Proto-oncogene tyrosine-protein kinase 1 (ROS1) is a receptor tyrosine kinase that plays a critical role in cellular signaling, growth, and differentiation6. ROS1 becomes oncogenic through chromosomal rearrangements that generate ROS1 fusion proteins, which drive tumorigenesis by constitutively activating pathways that promote proliferation, survival, and migration7. ROS1 rearrangements occur across several cancer types, including glioblastoma, gastric cancer, and non–small cell lung cancer (NSCLC)8,9,10. NSCLC accounts for approximately 85% of all lung cancer cases worldwide and in Taiwan11,12. Approximately 39.7 and 22.4 cases per one million cancer patients in Taiwan and global, respectively11,12. ROS1 gene fusions have been identified in approximately 1–2% of patients with NSCLC13. Therefore, ROS1 has been proved as a key factor in NSCLC therapeutics.
ROS1 is a large and single-pass transmembrane protein, comprising 2347 amino acid residues14. Its N-terminal region contains an αC-helix, four β-sheets, and several flexible loop segments1,15, while its C-terminal region features a distinctive intracellular tyrosine kinase domain followed by a single transmembrane helix16. Crizotinib was approved by FDA in 2016 for the administration of ROS-1 rearranged NSCLC17. Crizotinib binds to the ATP-binding pocket and inhibits ROS1 kinase activity by competing with ATP6. However, the Gly2032Arg (G2032R) solvent-front mutation located at the entrance of the ATP-binding site is the most common mechanism of acquired resistance to crizotinib18. This substitution introduces a bulkier, positively charged arginine side chain that creates steric hindrance, reduces inhibitor accessibility, and markedly diminishes crizotinib efficacy19. Lorlatinib is a more potent, broader-spectrum ROS1 inhibitor than crizotinib and is often considered after crizotinib failure20. Lorlatinib is often used as a treatment option when crizotinib is no longer effective for patients with G2032R-mutant ROS1-fusion NSCLC21. However, the G2032R solvent-front substitution often reduces susceptibility to many kinase inhibitors, and lorlatinib shows limited or variable efficacy against this specific mutant16,22. Consequently, the development of potent and selective inhibitors capable of overcoming G2032R-mediated resistance is urgently needed to improve therapeutic outcomes in ROS1-positive NSCLC.
Alkaloids represent a diverse class of naturally occurring nitrogen-containing organic compounds produced by plants, as well as certain fungi, bacteria, and animals23. Alkaloids are characterized by their nitrogen-containing structures which construct with or without heterocyclic rings23. These compounds are predominantly basic in nature due to the presence of nitrogen atoms, which can act as proton acceptors in acid-base reactions24. Given their immense structural diversity, alkaloids are divided into many groups, encompassing pyrrolidine alkaloids, piperidine alkaloids, indole alkaloids, isoquinoline alkaloids, tropane alkaloids, steroidal alkaloids, purine alkaloids25. The diverse chemical structures of alkaloids allow them exhibiting an extraordinarily wide range of potent biological activities by interacting with multiple biological targets, including receptors, enzymes, and ion channels26. For instance, berberine, a natural compound isolated from several plants like Coptis chinensis, has been linked to anti-obesity and antidiabetic effects by inhibiting adipogenesis and decreasing PTP1B expression27. Quinine, extracted from the bark of the cinchona tree, is a renowned medication used to treat both babesiosis and malaria28,29. Vinblastine is clinically used to treat for various cancers, including leukemias, lymphomas, breast cancer, and testicular cancer30. Given their potent biological activities and pharmacologically privileged scaffolds, alkaloids represent a highly promising reservoir for anticancer drug discovery.
Molecular docking and molecular dynamics simulation, remarkable computational techniques in modern drug discovery31,32, provide invaluable insights into molecular interactions and significantly accelerate the drug development. In this study, virtual screening was applied to evaluate a diverse library of alkaloids against the G2032R-mutant ROS1 kinase, followed by molecular dynamics simulations to investigate the stability and binding behaviors of the resulting complexes. This integrated computational workflow aimed to identify structurally and energetically promising alkaloid candidates that may serve as potential therapeutic agents for overcoming G2032R-mutant ROS1-mediated resistance in NSCLC.
Materials and methods
Software preparation
AutoDock Vina v1.2.3-2.3, MGLT AutoDock tools v1.5.7, Open Babel v2.4.1, Discovery Studio Visualizer v25.1.0, Swiss-PDBViewer, PyMOL v3.1.1, GROMACS v2023.3, g_mmpbsa, admetSAR 3.0, and Grace v5.1.25 were used in this study Methods.
Alkaloid library preparation
A virtual library of alkaloids was collected from the PubChem database (447 compounds). Protonation states at physiological pH (7.4) were adjusted using Open Babel, followed by energy minimization with Universal Force Field (UFF). Subsequently, the minimized structures were converted to PDBQT format using MGL AutoDock tools.
Protein preparation
The X-ray crystallographic structure of the ROS1 kinase domain was obtained from the Protein Data Bank (PDB ID: 4UXL) with a resolution of 2.4 Å. The preparation of protein structure was conducted using software Discovery Studio Visualizer by removing co-crystallized ligands, water molecules, and ions not involved in active site interactions. The G2032R mutation of ROS1 was modified using the Build and Edit Protein tool of Discovery Studio Visualizer33. Then, the protein structure was validated, optimized, and minimized energy using the Swiss PDB Viewer. Polar hydrogens were added and Kollman charges were assigned by MGL AutoDock tools. Finally, the protein structure was saved in PDBQT format for further docking studies. Lorlatinib was used as the positive control in this study. Redocking validation was performed by computing the heavy-atom RMSD between the crystallographic lorlatinib and the redocked pose of lorlatinib. The resulting RMSD was 0.001 Å, indicating successful reproduction of the crystallographic binding mode.
Virtual screening analysis
Virtual screening analysis were executed using AutoDock Vina, which is fast and widely used for initial virtual screening and supports batch docking34. For grid preparation, the active site was defined based on the coordinates of the co-crystallized inhibitor (crizotinib) with a central reference point at coordinates of 42.32 for X, −19.55 for Y, and − 4.86 for Z. The grid box dimension was set as 20 Å × 20 Å × 20 Å to completely cover the binding pocket. Each alkaloid was docked into the active site of the G2032R mutation of ROS1 with the parameters (Exhaustiveness: 8, Number of output poses: 10 per compound compound). Docking scores were evaluated based on the binding free energy (kcal/mol). The top 2 highest negative value compounds for G2032R mutation of ROS1 were selected for downstream analysis. Lorlatinib was used as the positive control to analyze with alkaloids.
Post-docking analysis
Top-ranked 2 alkaloids were separated to carry out further analysis. The best binding pose of the selected ligands was analyzed using PyMOL and Discovery Studio Visualizer to assess their 2D and 3D structures and protein–ligand interactions, such as hydrogen bonding, salt bridges, and hydrophobic forces with the active site of G2032R ROS1.
ADMET property prediction
The ADMET (Adsorption, Distribution, Metabolism, Excretion, and Toxicity) properties were evaluated to assess the predicted pharmacokinetic and safety profiles of the top-ranking ligands after virtual screening analysis. Prediction platform admetSAR 3.0 was employed to predict the ADMET of the top 2 highest negative value compounds. The SMILES (Simplified Molecular Input Line Entry System) of the selected ligands was retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov) and submitted to admetSAR 3.0 server to analyze.
Molecular dynamics simulation
Molecular dynamics simulations were performed for the apo form of the G2032R mutant of ROS1 and the 2 top-ranked protein–ligand complexes obtained from molecular docking. All simulations were conducted using GROMACS with the CHARMM36 parameter set of the CHARMM force field. Ligand topology and parameter files were generated via the CGenFF server. Each system was solvated in a periodic cubic box with the transferable intermolecular potential with three points (TIP3P) water model, and Na⁺ and Cl⁻ ions were added to achieve electroneutrality and a physiological salt concentration of 0.1 M. Energy minimization was performed using the steepest descent algorithm until convergence. NVT equilibration 100 ps with V-rescale thermostat at 300 K, NPT equilibration 100 ps with Parrinello–Rahman barostat at 1 bar, production 50 ns, coordinates saved every 100 ps. The production molecular dynamics simulations were run for 50 ns with coordinates saved every 100 ps. Analysis included root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), solvent accessible surface area (SASA), and hydrogen-bond interactions for both apo and complex systems was performed to assess conformational stability by the software Grace (version 5.1.25). Each ligand–protein complex was simulated in three independent replicates.
MM/PBSA binding free energy calculations
The binding free energies of the protein–ligand complexes were estimated using the Molecular Mechanics Poisson–Boltzmann Surface Area (MM/PBSA) method. g_mmpbsa tool was used to calculate the binding free energy. The last 10 ns of molecular dynamic trajectories were used to calculate the binding free energy for the ligands complexed with the G2032R mutation of ROS1. The overall binding free energy is defined by the following equation:
Where Gcomplex, Gprotein, and Gligand represent the free energies of the protein–ligand complex, the protein alone, and the ligand alone, respectively.
Results and discussions
ROS1 is a proto-oncogene encoding a receptor tyrosine kinase that belongs to the insulin receptor family35. Owing to its high similarity of sequence and the kinase domain, anaplastic lymphoma kinase (ALK) inhibitors, such as crizotinib and lorlatinib, have been repurposed for the treatment of ROS1-rearranged non-small cell lung cancer (NSCLC)36. However, these drugs show limited efficacy against the G2032R ROS1 mutation, a well-known resistance mutation16,22. Alkaloids constitute a broad and diverse class of naturally occurring organic compounds, fundamentally characterized by the presence of at least one nitrogen atom in their molecular structure25. Alkaloids exhibit a wide range of pharmacological activities, making them valuable candidates in drug development37. The structure-based drug discovery has increasingly focused on natural product scaffolds as potent tyrosine kinase inhibitors. For example, lamellarins, a group of alkaloids have shown potential anticancer activity by inhibiting various protein kinases, including mutant EGFR38. Thus, alkaloids are considered a promising compound class for the development of anticancer agents. Computational methods represent modern approaches in drug discovery. They allow the virtual prediction of binding affinities and the analysis of molecular interactions and dynamics between proteins and ligands, enabling the efficient assessment of compounds for therapeutic development. In the current study, Computational methods analysis of alkaloids was conducted to identify potential inhibitors against G2032R-mutant ROS1.
Virtual screening
Structure-based molecular docking was performed to screen a database of 447 alkaloid compounds (Table S1). As summarized in Table 1, yibeinoside A and vomicine were identified as the two top-ranked ligands based on their predicted binding affinities. Both natural products yielded docking scores comparable to that of lorlatinib (− 11.05 kcal·mol⁻¹ by AutoDock Vina), a clinically relevant ROS1 inhibitor. In contrast, betaine, which exhibited the lowest docking score (− 3.05 kcal·mol⁻¹), failed to form strong interactions and served as a negative control (Table S1). The 2D and 3D structures of yibeinoside A, vomicine, and lorlatinib are shown in Fig. 1, The 2D and 3D structures of yibeinoside A, vomicine, and lorlatinib are shown in Fig. 1. Yibeinoside A is a steroidal saponin alkaloid isolated from the bulb of Fritillaria pallidiflora and contains a C21 steroidal core with a tertiary nitrogen and an appended sugar moiety39. Vomicine is an indole alkaloid from Strychnos nux-vomica characterized by a tetracyclic scaffold with a central nitrogen and multiple polar groups (hydroxyl and carbonyl)40. In the drug-likeness evaluation, vomicine complied with Lipinski’s Rule of Five, whereas yibeinoside A violated the molecular-weight criterion but satisfied other filters such as Veber and PAINS. These characteristics suggest that both compounds may possess pharmacological potential. Although the two ligands obtained favorable docking scores against the G2032R ROS1 mutant, AutoDock Vina scores are approximate and may be biased toward large, highly polar molecules *. To strengthen confidence in their predicted affinities, the top candidates and lorlatinib were therefore rescored using MM-PBSA.
ADMET property prediction
Following virtual screening, yibeinoside A and vomicine were further evaluated using in-silico ADMET prediction via the admetSAR 3.0 platform. Key pharmacokinetic and toxicity parameters are summarized in Table 2. Both compounds were predicted to have high human intestinal absorption; however, only vomicine was predicted to be Caco-2 permeable, indicating potentially more efficient transcellular uptake. Vomicine was also predicted to cross the blood–brain barrier (BBB), suggesting possible central-nervous-system exposure. Plasma protein binding (PPB) was predicted to be high for yibeinoside A, suggesting a lower free drug fraction, whereas vomicine showed low PPB, potentially resulting in higher unbound concentrations in plasma. Lorlatinib, used as the positive control, was predicted to inhibit CYP1A2. Both alkaloids were predicted to undergo relatively high plasma clearance (CLp). Toxicity predictions differed markedly between the two alkaloids. yibeinoside A was predicted to be non-mutagenic and non-carcinogenic in rodent models, whereas vomicine was predicted to be mutagenic (Ames-positive) and rodent-carcinogenic. This prediction indicated that yibeinoside A probably might be safer than vomicine. In contrast, vomicine was predicted to be hepatotoxic, mutagenic, and carcinogenic, raising concerns over its potential adverse effects. In addition, toxicity predictions indicated that lorlatinib exerted positive toxicity profiles on hepatotoxicity, mutagenicity, and carcinogenicity. The positive results of Ames mutagenicity and rodent carcinogenicity for vomicine and lorlatinib suggest they may have properties of high risk of DNA damage and cancer induction. Although alkaloids often possess diverse therapeutic activities, many are known to be toxic even at low doses23. Reported mechanisms include neurotoxicity, disruption of signaling pathways, interference with enzymatic processes, and inhibition of DNA synthesis and repair41. In this study, vomicine was predicted to exhibit multiple toxic liabilities, whereas yibeinoside A only showed a risk of drug-induced hepatotoxicity. The positive control, lorlatinib contains 6 nitrogen atoms also displayed positive results across all three toxicity assays in ADMET prediction. In addition, side effects, such as edema, difficulty breathing, and diarrhea are recorded by FDA42. Nevertheless, these computational ADMET estimations provide only preliminary insights and cannot substitute for experimental validation or clinical evaluation.
Binding modes and interactions
Virtual screening was performed to identify alkaloids capable of binding to the G2032R-mutant ROS1 kinase and to explore their potential binding poses within the active site. This computational analysis provided a structural basis for the predicted inhibitory activity of the ligands by revealing key molecular interactions stabilizing the protein–ligand complexes. As shown in Fig. 2, yibeinoside A was predicted to bind deeply within the active-site cleft of G2032R ROS1. The detailed 2D and 3D interaction diagrams revealed interactions with residues that likely contribute to its binding affinity. A strong conventional hydrogen bond was predicted between the ligand and the side chain of ASN2084 at a distance of 3.22 Å (3.22 Å), a type of interaction known to play an important role in anchoring ligands to protein active sites43. Weaker carbon-hydrogen bonds were also observed with LEU2086 and GLY2102 of G2032R ROS1 protein. Beyond hydrogen bonding, the complex was further stabilized by multiple hydrophobic interactions. The steroidal framework and nitrogen-containing functional groups of yibeinoside A enabled extensive hydrophobic contacts, including three Pi–alkyl interactions with LEU1951, ARG2032, and LEU2086, helping to secure the ligand within a hydrophobic binding pocket. Notably, the direct interaction with the mutant residue ARG2032 may enhance binding specificity and potency toward this drug-resistant kinase. As illustrated in Fig. 3, vomicine also occupied the active site of G2032R ROS1, driven by several stabilizing forces. The most prominent interaction in its predicted binding mode was a strong attractive charge interaction between a protonated nitrogen atom on vomicine and the negatively charged carboxylate of ASP2038. Additional hydrogen bonds and Pi-system interactions were also detected. Importantly, a predicted interaction with the mutant residue ARG2032 was observed, similar to the binding behavior of yibeinoside A. This suggests that vomicine may also serve as a promising scaffold for developing potent and selective inhibitors targeting the drug-resistant G2032R ROS1 kinase. For comparison, the third-generation ROS1/ALK inhibitor lorlatinib was docked as a positive control. As shown in Fig. 4, lorlatinib formed two halogen bonds through its fluorine atoms with CYS2085 (3.31 Å) and ASN2084 (3.67 Å). Additional hydrophobic and Pi-system interactions were also present, indicating that lorlatinib employs diverse noncovalent forces to stabilize its binding pose. However, unlike the top-ranking alkaloids, lorlatinib did not form a direct interaction with the mutant residue ARG2032 in the predicted pose. This difference suggests that the two newly identified ligands may interact with the mutant position more effectively than lorlatinib, potentially contributing to improved inhibition of the drug-resistant G2032R-mutant ROS1. Finally, the 2D interaction diagram indicated that betaine formed only a single interaction with G2032R ROS1 (Figure S1), consistent with its lowest docking affinity observed in the virtual screening analysis.
Molecular simulation
To evaluate the structural stability and flexibility of the G2032R-mutant ROS1 protein in complex with the top-scoring alkaloids from virtual screening, 50-ns molecular dynamics (MD) simulations were performed for each ligand-bound system, along with the apo form and the lorlatinib-bound complex as a positive control.
The RMSD of the protein backbone atoms absence or presence with ligands was computed for 50 nanoseconds, and the results illustrate in Fig. 5A. the apo structure (black line) exhibited fluctuations ranging from 0.25 to 0.42 nm, serving as a baseline for evaluating ligand-induced dynamic changes. The lorlatinib-bound complex (blue line) showed the lowest RMSD values, maintaining stability between 0.25 and 0.32 nm, indicating that lorlatinib formed a highly stable complex with the mutant ROS1 protein. Yibeinoside A (red line) maintained a stable RMSD profile ranging from 0.30 to 0.40 nm, closely resembling the apo and lorlatinib-bound systems. This suggests that yibeinoside A remained well accommodated within the binding pocket and preserved the structural integrity of the protein throughout the simulation. Vomicine (green line) displayed slightly higher RMSD values, reaching 0.45 nm, with a mild fluctuation observed between 20 and 35 ns. The complex stabilized after 35 ns, indicating that although vomicine induced somewhat greater deviation from the initial structure, the overall fluctuation remained within an acceptable range. Collectively, the RMSD results indicate that yibeinoside A formed a more stable complex with G2032R ROS1 than vomicine, with stability comparable to that of the apo protein and lorlatinib. Lower RMSD values typically reflect a favorable fit within the active site and minimal perturbation to the protein structure44. Overall, yibeinoside A demonstrated superior stability compared to vomicine, with low RMSD indicating favorable active-site fitting and minimal structural perturbation.
Molecular dynamics simulation analysis of protein stability over 50 ns. A Root Mean Square Deviation (RMSD) of the apo protein and its complexes with Yibeinoside A, Vomicine, and Lorlatinib. B Root Mean Square Fluctuation (RMSF) of the apo protein and its complexes with Yibeinoside A, Vomicine, and Lorlatinib.
The RMSF of the protein’s Cα atoms was calculated over the simulation to identify changes in flexibility at the residue level. The results are presented in the Fig. 5B. All systems exhibited similar overall fluctuation patterns, with major peaks and troughs occurring in the same regions, indicating that ligand binding did not induce significant structural disruption. For the apo protein, the highest fluctuations were observed in the loop region spanning residues ~ 2100–2120. As expected, the lorlatinib-bound complex displayed the most stable RMSF profile, consistent with tight and effective protein–ligand interactions. Both yibeinoside A and vomicine induced RMSF patterns similar to the apo protein and showed values closely aligned with lorlatinib. Although yibeinoside A exhibited slightly higher fluctuations in the 2100–2120 region, its overall RMSF profile remained more stable than that of vomicine. Since higher RMSF values correspond to increased flexibility and lower values reflect more rigid, stabilized regions45, these results suggest that yibeinoside A contributed to greater structural rigidity and stabilization of G2032R ROS1 compared with vomicine. This result suggests that yibeinoside A-bound complex formed a more stronger structural rigidity with G2032R ROS1 than the complex with vomicine-bound complex.
The Rg values over the 50-nanosecond simulation trajectory are presented in Fig. 6A. The apo protein and the protein in complex with lorlatinib exhibited the lowest and most stable Rg values, fluctuating tightly around 2.00–2.02 nm. The complex with yibeinoside A maintained a compact structure with an average Rg of approximately 2.05 nm, only slightly higher than the apo protein and the lorlatinib complex. The vomicine complex showed a slightly higher Rg of ~ 2.07 nm.
Analysis of protein compactness from the 50-ns molecular dynamics simulation. A Radius of Gyration (Rg) of the apo protein and its complexes with Yibeinoside A, Vomicine, and Lorlatinib. B Solvent Accessible Surface Area (SASA) of the apo protein and its complexes with Yibeinoside A, Vomicine, and Lorlatinib.
The Rg plot provides insights into the conformational stability of the protein. A stable and consistent Rg value indicates that the protein maintains a stable fold, whereas large fluctuations or an increasing trend suggest unfolding or structural expansion46. The relatively low Rg values observed for both yibeinoside A and vomicine complexes indicated that the protein structure remained stable throughout the simulation. Although both ligands slightly increased the Rg compared with the apo protein, these minor changes likely reflected subtle conformational adjustments upon ligand binding. Overall, the results suggest that yibeinoside A is the most favorable candidate for forming a stable complex with the G2032R ROS1 protein.
The SASA for each system over the 50 ns molecular dynamic simulation was performed. As illustrated in Fig. 6B, the lorlatinib complex exhibited the lowest and most stable SASA profile, with an average value of approximately 153 nm², even lower than that of the apo protein. The apo protein and the complex with yibeinoside A showed nearly identical SASA values, averaging around 158 nm², with the trajectory of yibeinoside A almost superimposable on that of the apo protein. The vomicine complex displayed a slightly higher average SASA (~ 164 nm²). The SASA plot reflects changes in the protein’s solvent-exposed surface during the simulation. An increase in SASA typically corresponds to a less compact or partially unfolded protein, whereas a lower and stable SASA indicates a more compact and well-folded structure47. Lorlatinib reduced the SASA below that of the apo protein, suggesting that its binding promoted a more compact structure. The SASA profile of the yibeinoside A complex indicated that its binding did not disrupt the global structure. Similar to the Rg results, the slightly increased SASA of the vomicine complex suggested a less compact structure than yibeinoside A.
Taken together, the RMSD, RMSF, Rg, and SASA analyses provided a comprehensive assessment of the ligand–protein complexes. Both yibeinoside A and vomicine formed stable interactions with G2032R ROS1. However, yibeinoside A emerged as the superior binder, as it consistently maintained the global structure (based on Rg and SASA) and exhibited lower RMSD and RMSF values, indicating a highly stable and minimally disruptive interaction with the G2032R ROS1 protein.
Hydrogen bonds between a ligand and the protein are a key force that maintains the stability of a complex. As shown in Fig. 7, both yibeinoside A and lorlatinib demonstrated the formation of a robust and stable hydrogen-bonding network. They consistently maintained multiple hydrogen bonds throughout the simulation, indicating strong and specific anchoring to the binding site of the G2032R ROS1 protein. In contrast, vomicine formed markedly fewer hydrogen bonds, maintaining only 0–1 hydrogen bond during the trajectory. The strong hydrogen bonding of yibeinoside A likely contributed to the enhanced structural stability observed in the RMSD, RMSF, Rg, and SASA analyses. These findings suggest that the stable complex formed by yibeinoside A with the G2032R ROS1 protein may be attributed to this robust hydrogen-bonding pattern. Because vomicine formed only weak hydrogen-bond interactions, the structural stability of the vomicine complex appeared to rely on alternative forces. Previous 2D and 3D interaction analyses showed that vomicine formed a strong salt bridge between its positively charged nitrogen and the negatively charged carboxylate of ASP2038. This observation indicates that the major stabilizing force in the vomicine complex is likely its salt bridge interaction rather than hydrogen bonding.
MMPBSA analysis
MM-PBSA calculations were performed to estimate the binding free energies of the ligand–protein complexes. The energy components were: van der Waals energy, electrostatic energy, polar solvation energy, and nonpolar solvation energy in Table 3. Vomicine showed the most favorable binding free energy (− 24.13 kcal·mol⁻¹), followed by yibeinoside A (− 21.51 kcal·mol⁻¹), and the positive control lorlatinib (− 18.13 kcal·mol⁻¹). All three ligands exhibited substantial van der Waals contributions, indicating significant hydrophobic interactions with the ATP-binding pocket of G2032R ROS1. Vomicine stood out for its exceptionally large electrostatic interaction energy (− 65.51 kcal·mol⁻¹), consistent with the strong salt bridge identified with ASP2038. However, vomicine also displayed the highest polar solvation energy, suggesting that desolvation penalized the net affinity. Yibeinoside A showed a more balanced interaction profile, characterized by moderate van der Waals energy (− 34.51 kcal·mol⁻¹), moderate electrostatic energy (− 12.10 kcal·mol⁻¹), and the lowest solvation penalty (+ 25.10 kcal·mol⁻¹). This balance resulted in a favorable ΔG_total of − 21.51 kcal·mol⁻¹, often considered advantageous for drug-like binding because affinity does not depend excessively on a single interaction type. Interestingly, lorlatinib exhibited a reasonable MM-PBSA binding free energy (− 18.13 kcal·mol⁻¹), primarily driven by van der Waals and moderate electrostatic interactions. Both yibeinoside A and vomicine displayed stronger predicted binding affinities than lorlatinib, suggesting that these two alkaloids may interact more tightly with the G2032R ROS1 mutant. However, MM-PBSA provides relative trends which does not include entropy. Thus, MM-PBSA should be interpreted with caution. Therefore, definitive binding free energies require more rigorous alchemical methods or experimental validation.
Our computational analyses nominate yibeinoside A as a promising lead alkaloid against the drug-resistant G2032R ROS1 variant. Across docking, MD-derived stability metrics (RMSD, RMSF, Rg, SASA), hydrogen-bond profiling, and MM-PBSA rescoring, yibeinoside A consistently displayed a balanced interaction profile, emcopassing moderate van der Waals and electrostatic contributions with a relatively small solvation penalty. In addition, a key distinction between yibeinoside A and the clinical inhibitor lorlatinib was their interactions with the ARG2032. The G2032R mutation causes a bulky and positively charged arginine into the solvent-front region, generating steric hindrance that disrupts the binding of many ATP-competitive inhibitors. Our docking analysis showed that lorlatinib failed to form direct stabilizing interactions with ARG2032, resulting in a less favorable accommodation of the mutant pocket. In contrast, yibeinoside A was predicted successfully adapting to this steric and electrostatic alteration. Its steroidal scaffold establishes a π–alkyl interaction with the side chain of ARG2032, enabling the ligand to not only avoid the steric clash but also exploit the mutation to enhance binding stability. Although arginine is formally positively charged at physiological pH, its side chain contains several methylene groups that can participate in van der Waals contacts and CH–π interactions with ring systems or steroidal scaffolds. In contrast, although vomicine yielded a competitive MM-PBSA score, its predicted toxicity profiles reduce its immediate development appeal. We emphasize that these conclusions are computational predictions. they provide a structural rationale and prioritize compounds for follow-up, but experimental validation is required to confirm activity and safety.
This work is an in silico prioritization that carries several methodological limitations. First, AutoDock Vina scores are approximate (typical uncertainty ≈ 2–3 kcal·mol⁻¹) and are sensitive to ligand size and polar-contact number; to reduce size bias we report ligand-efficiency and performed MM-PBSA rescoring, but small score differences should be interpreted cautiously. Thus, additional docking engine or alternative scoring function will be adapted in our future research. Second, the alkaloid library contains many large, glycosylated natural products that can bias docking results toward high-contact, high-molecular-weight molecules. Third, in silico ADMET predictions are only estimates and must be confirmed experimentally. Fourth, because kinase inhibitors can have clinically relevant off-target activities, we recommend cross-target profiling. Top computational hits should be cross-docked and rescored against a panel of clinically important kinases and signaling proteins (for example EGFR, FGFR, VEGFR, AKT, MEK, PD-L1, BIRC3, CYFRA21-1, and related kinases) and tested experimentally. Finally, performing comparative docking on both the wild-type ROS1 and the G2032R-mutant ROS1 would strengthen the study by validating the docking protocol and clarifying whether candidate ligands truly overcome solvent-front resistance or instead bind nonspecifically.
Conclusion
In this study, based on this comprehensive computational study, yibeinoside A is predicted as a highly promising lead candidate for the inhibition of the drug-resistant G2032R ROS1 kinase from the alkaloid library. In silico ADMET assessment further suggested that yibeinoside A has a more favorable predicted safety and pharmacokinetic profile compared with lorlatinib and vomicine, both of which exhibited multiple toxicity alerts. Collectively, these computational results prioritize yibeinoside A as a high-value lead for further investigation in G2032R-Mutant ROS1. Nevertheless, these findings remain computational predictions. Experimental validationis is essential to confirm the therapeutic potential of yibeinoside A and to determine whether it can serve as a viable next-generation candidate against drug-resistant ROS1-driven NSCLC.
Data availability
All data generated and analyzed during this study are included in this articl.
References
Rod, N. H. et al. Cancer burden among adolescents and young adults in relation to childhood adversity: a nationwide life-course cohort study of 1.2 million individuals. Lancet Reg. Health Eur 27 (2023).
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Min, H. Y. & Lee, H. Y. Molecular targeted therapy for anticancer treatment. Exp. Mol. Med. 54, 1670–1694 (2022).
Kosorok, M. R. & Laber, E. B. Precision medicine. Annu. Rev. Stat. Appl. 6, 263–286 (2019).
Farhat, J., Alzyoud, L., AlWahsh, M., Acharjee, A. & Al-Omari, B. Advancing precision medicine: the role of genetic testing and sequencing technologies in identifying biological markers for rare cancers. Cancer Med. 14, e70853 (2025).
Yang, X., Tang, Z., Li, J., Jiang, J. & Liu, Y. Progress of non-small-cell lung cancer with ROS1 rearrangement. Front. Mol. Biosci. 10, 1238093 (2023).
Bergethon, K. et al. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 30, 863–870 (2012).
Charest, A. et al. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial Del (6)(q21q21). Genes Chromosomes Cancer. 37, 58–71 (2003).
Lee, J. et al. Identification of ROS1 rearrangement in gastric adenocarcinoma. Cancer 119, 1627–1635 (2013).
Gendarme, S., Bylicki, O., Chouaid, C. & Guisier, F. ROS-1 fusions in non-small-cell lung cancer: evidence to date. Curr. Oncol. 29, 641–658 (2022).
Hsu, J. C. et al. Lung cancer survival and mortality in Taiwan following the initial launch of targeted therapies: an interrupted time series study. BMJ Open. 10, e033427. https://doi.org/10.1136/bmjopen-2019-033427 (2020).
Zhou, J. et al. Global burden of lung cancer in 2022 and projections to 2050: incidence and mortality estimates from GLOBOCAN. Cancer Epidemiol. 93, 102693 (2024).
Drilon, A. et al. Repotrectinib in ROS1 fusion–positive non–small-cell lung cancer. N Engl. J. Med. 390, 118–131 (2024).
Roskoski, R. Jr ROS1 protein-tyrosine kinase inhibitors in the treatment of ROS1 fusion protein-driven non-small cell lung cancers. Pharmacol. Res. 121, 202–212 (2017).
Vilachã, J. F., Wassenaar, T. A. & Marrink, S. J. Structural aspects of the ROS1 kinase domain and oncogenic mutations. Crystals 14, 106 (2024).
Bubendorf, L. et al. Testing for ROS1 in non-small cell lung cancer: a review with recommendations. Virchows Arch. 469, 489–503 (2016).
Zhong, E. & Huang, H. Crizotinib in ROS1 rearranged non-small cell lung cancer (NSCLC), from response to resistance. BMJ Case Rep. 2016, bcr2016217322. https://doi.org/10.1136/bcr-2016-217322 (2016).
Katayama, R. et al. The new-generation selective ROS1/NTRK inhibitor DS-6051b overcomes Crizotinib resistant ROS1-G2032R mutation in preclinical models. Nat. Commun. 10, 3604 (2019).
Thawani, R. et al. TKI type switching overcomes ROS1 L2086F in ROS1 fusion-positive cancers. NPJ Precis Oncol. 8, 175 (2024).
Girard, N. et al. Lorlatinib for advanced ROS1 + non-small-cell lung cancer: results of the IFCT-1803 LORLATU study. ESMO open. 7, 100418 (2022).
Jóri, B. et al. Acquired G2032R resistance mutation in ROS1 to lorlatinib therapy detected with liquid biopsy. Curr. Oncol. 29, 6628–6634 (2022).
Fabbri, L. et al. From development to place in therapy of lorlatinib for the treatment of ALK and ROS1 rearranged non-small cell lung cancer (NSCLC). Diagnostics 14, 48 (2023).
Adibah, K. Z. M. & Azzreena, M. A. Plant toxins: alkaloids and their toxicities. GSC biol. pharm. sci. 6, 21–29 (2019).
Qian, J. & Brouwer, A. M. Excited state proton transfer in the Cinchona alkaloid cupreidine. Phys. Chem. Chem. Phys. 12, 12562–12569 (2010).
Olofinsan, K., Abrahamse, H. & George, B. P. Therapeutic role of alkaloids and alkaloid derivatives in cancer management. Molecules 28, 5578 (2023).
Bribi, N. Pharmacological activity of alkaloids: a review. Asian J. Bot. 1, 1–6 (2018).
Cai, Y. et al. Efficacy and underlying mechanisms of Berberine against lipid metabolic diseases: a review. Front. Pharmacol. 14, 1283784 (2023).
Renard, I. & Ben Mamoun, C. Treatment of human babesiosis: then and now. Pathogens 10, 1120 (2021).
Shanks, G. D. Historical review: problematic malaria prophylaxis with quinine. Am. J. Trop. Med. 95, 269 (2016).
Banyal, A. et al. Vinca alkaloids as a potential cancer therapeutics: recent update and future challenges. 3 Biotech. 13, 211 (2023).
Agu, P. C. et al. Molecular Docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci. Rep. 13, 13398 (2023).
Wei, H. & McCammon, J. A. Structure and dynamics in drug discovery. NPJ Drug Discov. 1, 1 (2024).
Parate, S., Kumar, V., Hong, J. C. & Lee, K. W. Identification of flavonoids as putative ROS-1 kinase inhibitors using pharmacophore modeling for NSCLC therapeutics. Molecules 26, 2114 (2021).
Eberhardt, J., Santos-Martins, D., Tillack, A. F. & Forli, S. AutoDock Vina 1.2. 0: new Docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 61, 3891–3898 (2021).
Raez, L. E., Manca, P., Rolfo, C. & Singh, V. ROS-1 rearrangements in Circulating tumor cells. J. Thorac. Oncol. 13, e71–e72 (2018).
Shaw, A. T. et al. First-line lorlatinib or Crizotinib in advanced ALK-positive lung cancer. N Engl. J. Med. 383, 2018–2029 (2020).
Heinrich, M., Mah, J. & Amirkia, V. Alkaloids used as medicines: structural phytochemistry Meets biodiversity—An update and forward look. Molecules 26, 1836 (2021).
Baunbæk, D. et al. Anticancer alkaloid lamellarins inhibit protein kinases. Mar. Drugs. 6, 514–527 (2008).
Xu, D. et al. Isolation and identification of Yibeinoside A. Acta Pharm. Sin. 25, 795–797 (1990).
Bhati, R., Singh, A., Saharan, V. A., Ram, V. & Bhandari, A. Strychnos nux-vomica seeds: pharmacognostical standardization, extraction, and antidiabetic activity. J. Ayurveda Integr. Med. 3, 80 (2012).
Rajput, A., Sharma, R. & Bharti, R. Pharmacological activities and toxicities of alkaloids on human health. Mater. Today Proc. 48, 1407–1415 (2022).
Bauer, T. M. et al. Clinical management of adverse events associated with lorlatinib. Oncologist 24, 1103–1110 (2019).
Steiner, T. & Koellner, G. Hydrogen bonds with π-acceptors in proteins: frequencies and role in stabilizing local 3D structures. J. Mol. Biol. 305, 535–557 (2001).
Guterres, H. & Im, W. Improving protein-ligand Docking results with high-throughput molecular dynamics simulations. J. Chem. Inf. Model. 60, 2189–2198 (2020).
Fatriansyah, J. F. et al. Molecular dynamics simulation of ligands from Anredera cordifolia (Binahong) to the main protease (Mpro) of SARS-CoV-2. J. Trop. Med. 2022, 1178228 (2022).
Rampogu, S., Lee, G., Park, J. S., Lee, K. W. & Kim, M. O. Molecular Docking and molecular dynamics simulations discover Curcumin analogue as a plausible dual inhibitor for SARS-CoV-2. Int. J. Mol. Sci. 23, 1771 (2022).
Bagewadi, Z. K. et al. Molecular dynamics and simulation analysis against superoxide dismutase (SOD) target of micrococcus luteus with secondary metabolites from Bacillus licheniformis recognized by genome mining approach. Saudi J. Biol. Sci. 30, 103753 (2023).
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Shu-Chi Cho: Conceptualization, data curation, formal analysis, methodology, software, writing - original draft. Yi-Wen Wang: Formal analysis, methodology. Chien-An Chu: Data curation, software. Ming-Chih Huang: Conceptualization, software. Chung-Ta Lee: Conceptualization, funding acquisition, project administration, writing - review and editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cho, SC., Wang, YW., Chu, CA. et al. Virtual screening of novel alkaloids as potent inhibitors for G2032R-mutant ROS1 kinase in non-small-cell lung cancer. Sci Rep 16, 5342 (2026). https://doi.org/10.1038/s41598-026-36317-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-026-36317-4