Abstract
Pathological complete response (pCR) serves as a critical measure of the success of neoadjuvant chemotherapy (NAC) in breast cancer, directly influencing subsequent therapeutic decisions. With the continuous advancement of artificial intelligence, methods for early and accurate prediction of pCR are being extensively explored. In this study, we propose a cross-modal multi-pathway automated prediction model that integrates temporal and spatial information. This model fuses digital pathology images from biopsy specimens and multi-temporal ultrasound (US) images to predict pCR status early in NAC. The model demonstrates exceptional predictive efficacy. Our findings lay the foundation for developing personalized treatment paradigms based on individual responses. This approach has the potential to become a critical auxiliary tool for the early prediction of NAC response in breast cancer patients.
Similar content being viewed by others
Introduction
Breast cancer (BC) is currently the most prevalent cancer among women, surpassing lung cancer1. Neoadjuvant chemotherapy (NAC), previously limited to locally advanced BC, is now being used more extensively2,3. This treatment not only reduces the stage of BC and improves the rate of breast-conserving treatment (BCT), but also minimizes the need for axillary surgery4. Pathologic complete response (pCR) serves as a standard for measuring the efficacy of NAC. Early identification of pCR is crucial to avoid unnecessary surgery and chemotherapy cycles. This can help to prevent toxic side effects and psychological impacts on patients who do not respond to NAC5. Therefore, obtaining reliable methods for early prediction of pCR is of utmost importance.
Based on existing research, there are variations in pathological responses to NAC among patients with different tumor subtypes6. Additionally, individual responses to chemotherapy may also vary due to tumor heterogeneity7,8. Several histopathology-based studies have confirmed that certain stromal parameters, such as tumor-infiltrating lymphocytes (TILs) and tumor-stroma ratio9,10,11, demonstrate predictive capabilities for pCR, in addition to the predictive information hidden in the tumor epithelium12. Therefore, extracting heterogeneous information implicit in tumor and associated stroma from histopathological images and integrating it effectively is crucial for accurately identifying pCR13,14,15. However, due to individual variations in responses to chemotherapy drugs, relying solely on pre-treatment tumor information, while valuable for predicting pCR, may not provide sufficient accuracy to serve as a basis for clinical decision-making.
The changes in tumors during treatment are important predictive features. To improve the accuracy of pre-treatment assessment for pCR, researchers have recognized the significance of combining pre-treatment tumor information with imaging changes during treatment. This integration provides a better representation of the temporal heterogeneity of tumors and their response to chemotherapy drugs, thereby uncovering crucial information for predicting pCR. An increasing corpus of research supports the idea that incorporating changes in tumors and related regions during the treatment process is crucial in predicting pCR for patients undergoing NAC. This approach has the potential to enhance predictive capabilities16,17,18.
The findings of a study on predicting the efficacy of NAC in advanced colorectal cancer suggest that a predictive model, which combines Whole Slide Images (WSIs) with radiomic features, outperforms a unimodal model in terms of predictive accuracy19. Wang et al. developed a nomogram model that integrates deep learning (DL) pathomic, radiomic and immune scores to predict postoperative overall survival (OS) and disease-free survival (DFS) in patients with lung metastasis of colorectal cancer20. However, there is limited research reported on combining histopathology and radiomics to construct predictive models of post-NAC pCR in BC patients. Further investigation is needed to determine whether integrating both modalities can provide additional predictive information related to pCR status, thereby improving the predictive performance for pCR.
In the field of oncology, the advancement of artificial intelligence (AI) has brought about a transformative era. DL networks have outpaced conventional machine learning (ML) methodologies and are renowned for their precision and user-friendly attributes. They have been continuously applied in recent years to various medical applications, including the segmentation of breast tumors21,22,23, benign-malignant classification24,25,26, assessment of hormone receptor status27, tumor subtyping28,29, prediction of drug efficacy30,31 and disease prognosis32. The DL models based on multi-time point images have demonstrated performance advantages in evaluating the effectiveness of NAC33,34,35. Nonetheless, the presently prevalent application of Convolutional Neural Networks (CNNs) tends to prioritize regional details within images, exhibiting limited capability to capture global context and distant relationships between image elements, and requiring substantial computational cost. In 2020, Dosovitskiy et al. pioneered the application of the transformer, an innovative neural network, to image classification. The vision transformer (ViT) was introduced and has achieved commendable performance in this context36. ViT’s unique global self-attention mechanism and substantial receptive field range have led to outstanding classification performance, resulting in its increasing utilization for imaging tasks37,38. Additionally, it has found growing applications in medical imaging research, demonstrating superior performance compared to CNN, particularly in studies related to breast tumor analysis39,40,41,42.
In this study, we chose B-mode ultrasound (US) as a monitoring tool to capture tumor responses to NAC due to its economical, non-radiative, repeatable, and convenient nature. Additionally, we employed WSIs of histopathologic hematoxylin-eosin (HE) stained sections obtained by core needle biopsy (CNB) of primary BC tumors, providing visual insights into pathological characteristics. The study utilized DL methods to integrate WSIs from pathological analysis of pre-treatment CNB specimens of the primary tumor with initial US images. This integration allowed for the derivation of comprehensive tumor heterogeneity information. We proposed a double ultrasound coherence network (Dou-Ult Net) to capture pixel changes in US images before and after two cycles of NAC at any given location, which reflected drug responsiveness. Given the critical importance of inter-scale feature correlations, we have improved the architecture based on transformer by integrating the long-range and short-range attention mechanisms of the CrossFormer43 vision architecture. This enhancement aims to efficiently leverage multi-scale features across multimodal images and establish correlations between large-scale global features and small-scale fine-grained features. To achieve effective interaction between multi-time point US images and pathological WSIs at different scales and channels within input sequences, we embed a Squeeze and Excitation (SE) block44, which dynamically adjusts the channel weights of input feature maps to enhance the network’s perception of important features. We named this novel network architecture “Squeeze and Excitation CrossFormer” (SE-CrossT), which integrates channel attention modules within a transformer-based interaction network, thereby fully extracting the feature information of the images.
Results
Clinicopathological characteristics
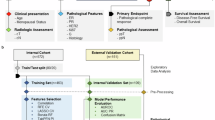
From July 2017 to July 2022, 596 patients were selected from a pool of 1835 candidates based on inclusion criteria at the Harbin Medical University Cancer Hospital, forming the final study group (Fig. 1). Overview of the study design is presented in Fig. 2. Clinicopathologic characteristics of patients in non-pCR and pCR cohorts are provided in Table 1. Among them, 491 patients with unifocal lesions were randomly assigned to the primary cohort, which was further divided into training, validation, and TC1. There were no statistically significant variances in clinical baseline features among the groups (P > 0.05). Additionally, 105 patients with multifocal lesions were used as TC2. Patient characteristics for each cohort are detailed in Supplementary Table 1.
a Diagnostic and treatment workflow for included breast cancer patients: All patients underwent ultrasound-guided core needle biopsy (CNB), followed by initiation of a standard NAC protocol within 1 week after the biopsy. Surgical intervention was conducted for all patients upon completion of the entire treatment course, and the achievement of pCR was determined through histopathological examination. b Obtaining pathology images: the process involved scanning HE-stained slides from CNB to create WSIs. The most representative region was selected as the ROI. c The process of obtaining ultrasound (US) images: collecting pre-NAC (labeled as US-0) and post two cycles of treatment (labeled as US-1) ultrasound images, followed by preprocessing. d Univariate and multivariate logistic regression analyses were performed to identify clinically meaningful features linked to pCR as baseline characteristics. These selected features were then used to construct a model integrating clinicopathological characteristics. e The breast cancer patients receiving NAC were divided into the training cohort (N = 391), validation cohort (N = 49), test cohort 1 (unifocal test cohort, N = 51), and test cohort 2 (multifocal test cohort, N = 105). Test cohorts were not involved in the training process. f Schematic diagram of the artificial intelligence model. g Performance results and visualization of the predictive model.
To evaluate the predictive abilities of established biomarkers and extract significant clinical characteristics, we applied univariate and multivariate analyses to investigate the relationships between existing histological and clinical imaging variables and NAC response. Univariate analysis revealed significant differences in several clinical factors between the pCR and non-pCR cohorts, encompassing ER, PR, HER2, Ki-67, clinical TNM staging, Breast Imaging Reporting and Data System (BI-RADS) classification, tumor subtype, chemotherapy regimen (P < 0.05). Figure 3 provides the landscape of PCR-related clinicopathological characteristics of BC. Multivariable analysis of pCR in relation to clinicopathological characteristics in the cohort is summarized in Table 2. It indicated that patients with ER-negative or ER-low positive status, high Ki-67 expression, non-HR+/HER2− subtype were more prone to achieve pCR following NAC. Simultaneously, the chemotherapy regimen is also a factor influencing pCR. This conclusion is consistent with previous research findings45,46,47.
Performance of DL models vs. other architectures in cross-modal multi-pathway tasks
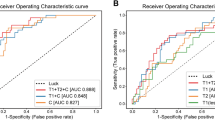
In this study, six DL methods were employed to establish a multimodal pCR prediction model based on pathology and US images. The results demonstrate that each DL model possesses the capability to predict pCR in the unifocal dataset. The cross-modal multi-pathway model based on the SE-CrossT network exhibited superior performance compared to other DL methods, with AUC values of 0.851 (95% CI: 0.793–0.869, 0.791–0.856) in both the validation cohort and TC1. ACC, SENS, SPEC, NPV, PPV values were 0.918, 0.896, 0.888, 0.866, 0.885 and 0.882, 0.878, 0.855,0.840, 0.864 (Fig. 4e, i). When comparing the SE-CrossT network with models established by five other DL methods in the validation cohort and TC1, the findings indicated that within the validation cohort, this model achieved the highest AUC, ACC, SENS, NPV, and PPV; In the TC1, it exhibited the highest AUC, ACC, NPV, and PPV values (Table 3). The ROC curves for the training, validation and test cohorts illustrate the comparative results of AUC for models based on six networks (Fig. 4a, e, i).
Muti ROC (a), (e) and (i) Cross-modal multi-pathway models based on six networks in the training, validation cohort and text cohort 1; Patho ROC (b), (f) and (j) Unimodal-pathology models based on six networks in the training, validation cohort and text cohort 1; Ult ROC (c), (g) and (k) Unimodal-ultrasound models based on six networks in the training, validation cohort and text cohort 1; Muti ROC (d), (h) and (l) Muti ROC ablation study for SE block in cross-modal multi-pathway model and fusion of clinicopathological characteristics into cross-modal multi-pathway model. SwinT Swin-Transformer, SE-CrossT based adding clinicopathological baseline characteristics.
Ablation study
To validate the predictive performance, the image set was divided, and six DL methods were employed to establish a unimodal pCR prediction model. Figure 4f, j showed that the SE-CrossT network had the highest performance for the unimodal-pathology model, with an AUC of 0.810 (95% CI: 0.777–0.819) in the validation cohort and 0.703 (95% CI: 0.649–0.745) in TC1. ACC, SENS, SPEC, PPV, NPV were 0.857, 0.730, 0.788, 0.751, 0.751 in the validation cohort and 0.776, 0.727, 0.738, 0.724, 0.729 in TC1 (Supplementary Table 2). Figure 4g, k shows that for the unimodal-US model targeting multiple time points, the SE-CrossT network achieved AUC values of 0.837 (95% CI: 0.806–0.855) and 0.817 (95% CI: 0.775–0.836) in the validation and TC1, respectively. ACC, SENS, SPEC, PPV, NPV were 0.878, 0.883, 0.882, 0.859, 0.879 in the validation cohort, and 0.823, 0.859, 0.867, 0.833, 0.855 in TC1 (Supplementary Table 3). Table 4 performance SE- CrossT models based on WSIs, US and Multimodal according to validation and test cohorts. The results indicate that cross-mode joint learning of breast tissue pathology and radiological images contributes to improved feature performance. The model’s performance surpasses that of unimodal models, demonstrating certain advantages in predicting pCR in the context of NAC. Furthermore, the results of the ablation experiment for the SE block also confirm the necessity of adding the SE block in training networks for cross-mode multi-pathway tasks (Supplementary Table 4), In order to visually depict the ablative experimental results of the SE module, we present the AUC curves without the addition of the SE module in each model’s respective cohort. Figure 4 shows the ablation study for the SE block.
Validation of model performance with multifocal cases as an independent test cohort
To assess the model’s robustness and generalization, we employed a multifocal lesion test cohort (TC2) that was not involved in training and exhibited greater tumor heterogeneity. The results revealed suboptimal performance for models based on CNN, with the best AUC reaching only 0.543 (95% CI: 0.496–0.553) (DenseNet121). Transformer-based models performed better, with SE-CrossT and Swin-transformer models exhibiting the highest performance, each achieving an AUC of 0.713 (95% CI: 0.668–0.714, 0.649–0.714, P = 0.931), the corresponding ACC, SENS, SPEC, PPV, and NPV values were 0.848, 0.794, 0.803, 0.815, 0.774, and 0.848, 0.793, 0.816, 0.786, 0.795, respectively (Supplementary Table 5). These results indicate that our cross-modal multi-pathway model based on SE-CrossT for breast tissue pathology and US images remains practical in predicting pCR for multifocal cases, expanding the model’s applicability.
Fusion of baseline features into cross-modal multi-pathway model
Four clinical variables associated with pCR, tumor subtype, ER status, Ki67 percentage, and chemotherapy regimen were integrated as features into the best-performing SE-CrossT-based cross-modal multi-pathway model. The AUC demonstrated improvement to 0.873 (95% CI: 0.834–0.898, P < 0.005) in the validation cohort, and in TC1, it exhibited an increase to 0.875 (95% CI: 0.835–0.890, P < 0.005) (Fig. 4h, l). Post-integration of clinicopathological variables, the model exhibited ACC, SENS, SPEC, NPV, and PPV values of 0.918, 0.883, 0.894, 0.861, 0.874, respectively (Table 5). Hence, integrating clinicopathological characteristics can enhance the performance of the cross-modal multi-pathway model while expanding its applicability. As an independent test cohort, multifocal cases exhibited an ACC of 0.886 post-clinicopathological characteristics fusion, with a significant increase in AUC to 0.819 (95% CI: 0.764–0.868, P < 0.001), Fig. 5 shows ROC curves for predicting pCR levels of cross-modal multi-pathway models based on the SE-CrossT network in the TC2. This also underscores the apparent importance of clinicopathological characteristics in multifocal cases.
Visual interpretation of the DL model
To improve the interpretability of predictions made by the DL model, we visualized response areas within the original lesion regions depicted in pathology and US modal images (Fig. 6). We created illustrative heatmaps by obtaining two distinct image sets: one featuring cases of pCR, and the other highlighting instances of non-pCR. The DL model demonstrated an impressive ability to produce accurate diagnostic results. The resulting heatmaps showed clear color patterns, with red regions indicating areas that provide valuable predictive information for pCR. After careful examination of all heatmaps, distinct patterns emerged that distinguished tumors with pCR from those without.
Instances of pathologic complete response non-pCR (a) and pCR (b) cases. The US-0 ultrasound images were acquired within the initial week preceding neoadjuvant chemotherapy (NAC), while the US-1 ultrasound images were captured during two cycles post-chemotherapy. In the domain of WSIs patches, emphasis is placed on regions hosting tumor-infiltrating lymphocytes (TILs), with invasive carcinoma securing the second position in terms of significance. Transitioning to ultrasound images, noteworthy regions often encompass a substantial portion of the primary tumor lesion area or are situated peripherally in the tumor’s periphery.
In the field of WSIs patch, the most valuable regions are those that contain TILs, with invasive carcinoma ranking second. In US images, valuable regions often include a significant portion of the primary tumor lesion area or are located in the tumor’s periphery. In US-0 images of tumors, valuable regions tended to coalesce within the internal facets of the tumor, regardless of the pCR status. In US-1 images, valuable regions that promote non-pCR tended to aggregate within the tumor, similar to the pattern observed in US-0 images. Additionally, in most US-1 images showing pCR tumors, the valuable regions are often concentrated along the tumor’s periphery. This explanation contributes to understanding the discriminative ability of the DL model, aligning with insights from previous clinical studies48,49.
Discussion
Nowadays, artificial intelligence (AI) has been extensively applied in medical imaging research due to its ability to uncover information that may escape human eyes. This has found widespread application in predicting the effectiveness of NAC for BC. For instance, studies using radiomics have discovered a correlation between posterior acoustic features in US images and the proliferative capacity and cell count of tumors. Tumors with rapid proliferation tendencies, it is found, are more likely to achieve pCR17. Tumors with no apparent change in size can be effectively analyzed for internal subtle changes through artificial intelligence. These changes often correlate with the loss of cell count post-NAC, thereby reflecting the responsiveness to NAC50. We have observed that while there is a considerable amount of research focusing on predicting pCR after NAC in BC, there is a limited emphasis on achieving optimal performance metrics during early-stage treatment. In the 2016 study by Noritake et al., a comparative analysis was conducted between 18F-FDG positron emission mammography (PEM) and whole-body 18F-FDG PET (WBPET) for predicting pCR in NAC51. The study revealed that WBPET exhibited superior predictive performance for pCR after the second cycle of NAC compared to PEM (AUC = 0.761). In 2022, in a large cohort study conducted by Wu et al., involving 801 patients, the results demonstrated that a radiomics model based on continuous US and clinical characteristics exhibited predictive performance for pCR after the second cycle of NAC, with AUC values of 0.834 and 0.829 in independent external test cohorts, respectively52. In 2021, a combined model based on siamese convolutional neural networks developed by Byra et al. predicted pCR after the second chemotherapy cycle, achieving an AUC of 0.84733. While these studies suggest the potential of imaging-based models relying on features at the second cycle of NAC to predict pCR, it is acknowledged that these insights may not be comprehensive. Li et al. utilized DL techniques to extract features from HE-stained WSIs of BC biopsy tissues53. They constructed a predictive model for the efficacy of neoadjuvant therapy, validating the spatial information of WSIs, such as tumor-infiltrating lymphocyte (TIL) density in the stroma, as being correlated with treatment response and prognosis. Additionally, the study confirmed the association of pCR status with histological features such as lymphocytes, vacuoles, mitosis, and nuclear pleomorphism. These findings also underscore that the initial pathological features of tumor tissues contain rich information predictive of treatment responsiveness, closely associated with post-NAC pCR. We observed complementary information in the predictive value for pCR between pre-treatment pathological information and imaging data during the treatment process. The effective integration of these two sources holds significant value for enhancing the accuracy of pCR predictions.
In this study, we propose a cross-modal multi-pathway DL model based on the novel vision architecture, SE-CrossT, for predicting pCR. This is the first model to jointly incorporate unsegmented HE-stained WSIs and multi-temporal US images into a network based on transformer, providing individualized predictions for post-treatment pCR in BC patients after the second cycle of NAC. Our model enables multi-temporal, multi-scale analysis of multimodal images. We compared our approach with other widely used methods in image classification including classical networks in CNNs such as VGG16, ResNet50, DenseNet121 and architectures in transformers like Vision Transformer (ViT), Swin-Transformer based on seven metrics: ACC, SENS, SPEC, PPV, NPV, and AUC. While CNNs are widely applied in medical image analysis, our results indicate that the performance of the CNN network is suboptimal. This may be attributed to the limited ability of CNNs, as a class of networks that primarily focus on perceiving local information, to capture the global receptive field. Similarly, although ResNet50 and DenseNet121 have added connectivity layers to enhance feature extraction and ensure information integrity, they still fail to address the issue of global information interaction. As a result, the model performance did not show a significant improvement. Due to various factors affecting the coherence of US image acquisition, it is challenging to achieve uniformity in the collected images. The performance of the model is consequently influenced by variations resulting from its autonomous learning of different imaging parameters. As a result, the performance of CNN based on absolute positional deviation further diminishes. Compared to CNNs, the self-attention mechanism in the transformer framework is more focused on the information relationships expressed by adjacent positions and pixels in the image domain, yielding a global receptive field. This aligns well with our need for integrating information across the image set. Our proposed SE-CrossT network demonstrating the optimal performance, the AUC values on the validation cohort and TC1 both were 0.851, significantly outperforming models established with other DL methods used for comparison in this study (Fig. 4e, i). In the multifactorial analysis of clinicopathological characteristics, we categorized ER and HER2 into three groups and introduced ER-low positive and HER2 low expression as categorical variables. The multifactorial analysis results indirectly support existing research findings on the subgroup characterized by ER-low positive, suggesting a biological behavior similar to ER-negative tumors54. The incorporation of clinicopathological characteristics further improved the model’s performance, with AUC values on the validation cohort and TC1 reaching 0.873 and 0.875 (Fig. 4h, l), respectively, both achieving ACC greater than 0.9. Our results confirm that the SE-CrossT architecture has successfully facilitated effective interaction between crucial regions in multimodal, multi-time point images for response prediction. This also validates that the predictive information contained in HE-stained WSIs from biopsy and US can complement each other. Our findings suggest that the multimodal DL model outperforms other networks in multimodal settings. Furthermore, ablation experiments also confirm that the multimodal model performs the best compared to the unimodal pathological model or US model.
Our model is trained through supervised learning. By deeply exploring the primary tumor and relevant lesion areas in multimodal images, it predicts based on the learned features. This approach can further reduce the varying impact of subjective factors among doctors on final clinical decisions. It achieves automatic acquisition and integration of hierarchical features from multi-scale ROI, avoiding limitations and biases seen in previous convolutional neural network feature fusion. This significantly increases the amount of information available for assessment, maximizing the utilization of existing image data. We translated the model’s image features into visual heatmaps to establish a link between input images and output labels, thereby overcoming the ‘black box’ nature of DL models to the fullest extent possible55,56. The results indicate that there are typically two valuable locations in US images for predicting pCR: the tumor boundary and the grayscale internal region of the tumor (Fig. 6). In the domain of WSIs patches, the regions considered most valuable are those containing TILs, followed by invasive carcinoma. This underscores the significant role these areas play in predicting pCR.
Incorporating clinicopathological characteristics related to pCR into our best model and validating the model’s generalizability using multifocal cases with stronger tumor heterogeneity, The results indicate that the model’s performance is enhanced when clinical information is included, with an AUC of 0.875 in the TC1 (Fig. 4l). Therefore, the addition of clinicopathological characteristics can improve the accuracy of predicting pCR when such information is available. However, in the multifocal lesions test cohort, we observed that the performance of the multifocal prediction model was lower than that of the unifocal cohort. This suggests that the spatiotemporal heterogeneity of multifocal tumors is stronger, affecting the accuracy of pCR prediction. Furthermore, the predictive performance combining clinicopathological characteristics is significantly better than the image-only model (AUC: 0.819 vs. 0.713), indicating that information from clinicopathological characteristics appears to be more crucial for multifocal tumors (Fig. 5). This also opens the possibility of attempting to predict pCR in multifocal cases.
This study has certain limitations. First, DL, especially transformer networks, requires extensive training datasets, and our data were obtained retrospectively from a single center, rendering the dataset insufficient. Future prospective studies with multi-center large datasets are needed to conduct more rigorous evaluations of DL methods and models. Second, the current framework still requires specialists to manually select tumor regions, and this simplistic approach makes the workflow cumbersome. Tackling this matter may entail the development of fully automated image segmentation techniques and investigating their effectiveness when incorporated into the NAC response prediction framework. Furthermore, with the continuous development of new drugs for NAC, certain medications such as pyrotinib, daparicyclin, and apatinib, which can enhance the pCR rate, have become potential treatment options for some patient groups57,58. In the future, additional chemotherapy regimens confirmed to improve pCR and disease prognosis can be incorporated into the model. However, due to limited data volume at present, they have not been included in the model training. Furthermore, it is well-known that different tumor subtypes exhibit distinct responses to chemotherapy drugs. While multifactorial analyses of baseline information before treatment have underscored the importance of these subgroups in predicting response, our study did not specifically delve into intra-subgroup investigations. Despite this, our model demonstrates greater versatility. It is worth noting that, in terms of model input selection, although WSIs are obtained through the invasive procedure of biopsy, this diagnostic process is an essential step in BC management. Additionally, US examinations do not impose additional physical or economic burdens on patients. Therefore, the model exhibits enhanced cost-effectiveness. Therefore, the proposed method in this study holds promise for potential application in clinical practice, providing robust evidence for clinical decision-making. Moreover, it may be applicable for analyzing multimodal data in other types of tumors. in subsequent investigations, we intend to incorporate more comprehensive clinical data and additional functional imaging information for analysis. This aims to acquire valuable data from multiple modalities early in treatment, enhancing the predictive efficiency of our model. Our goal is to provide a foundation for clinicians to implement early personalized precision treatment based on individual patient characteristics.
Methods
Patient recruitment and selection
The investigation followed the principles outlined in the Declaration of Helsinki and received approval from the Ethics Committee at Harbin Medical University Cancer Hospital (protocol code XJS2023-03, 12 April 2023). Given its retrospective design, the study was exempt from obtaining informed consent from patients. The study retrospectively collected data from 596 individuals diagnosed with invasive breast carcinoma during their hospitalization period from July 2017 to July 2022. The cohort was divided into two distinct groups: The pCR category (155 cases) and the non-pCR category (441 cases). The process of patient selection for model development is outlined in Fig. 1, which includes a subsequent set of criteria: (1) Biopsy specimens were initially acquired through CNB and confirmed by pathological examination as female patients with unilateral primary invasive BC who met the indications for NAC; (2) All patients underwent NAC treatment within 1 week after biopsy and did not receive any other treatments before chemotherapy; (3) US examinations were performed 1 week before biopsy and during the treatment process, with US images saved; (4) Surgical intervention was performed within 1 month after completing NAC, and tissue pathology results were obtained. The exclusion criteria for this study are: (1) a history of malignant tumors or concurrent other malignancies, severe infections, hematologic diseases, or autoimmune disorders; (2) failure to complete the prescribed NAC regimen or not receiving standard treatment, particularly for human epidermal growth factor receptor 2 (HER2)-positive tumors not treated with trastuzumab; (3) Poor US image quality, absence of tumor display, lack of visualization of tumor boundaries, or inconsistency between US findings and pathological descriptions may pose diagnostic challenges; (4) Lack of CNB pathology slides or poor quality of WSIs may lead to uncertain diagnoses; (5) In patients with unilateral multifocal tumors, where US findings and biopsy or postoperative pathological analysis remain uncertain, the diagnosis may be challenging; (6) Incomplete clinical medical records required for this study.
All patients received NAC treatment based on standardized protocols outlined by the National Comprehensive Cancer Network59 and the Chinese Anti-Cancer Association Breast Cancer Guidelines60 before surgery. The NAC regimens primarily included anthracycline with taxane, Taxane- and platinum-based, and other taxane-based regimens. Additionally, HER2-positive patients received chemotherapy based on taxanes in combination with anti-HER2 agents. Preceding NAC, data were extracted from medical records, encompassing comprehensive clinical information (including age, BMI, and menopausal status), US findings (tumor dimensions, breast imaging reporting and data system (BI-RADS) assessment) and detailed pathological characteristics (estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2) status, Ki67 status and pathological staging). Surgical resection was conducted for all patients within 2–3 weeks following the completion of the entire NAC regimen61, and postoperative pathological reports were utilized to determine the pCR classification (Fig. 2a, d).
US data and preoperative pathological WSIs collection
US examination: Breast US examinations were conducted using Siemens (ACUSON2000), Philips (EPIQ5), or Mindray (Resona7) US machines. Skilled radiologists with over 5 years of experience in breast US examinations acquired all US images. From the breast imaging database, we retrieved and analyzed US scans, selecting a pair of optimal static images from 15 to 20 captured during each B-mode US scan of the entire tumor at two time points: the first week before NAC (US-0) and two cycles after chemotherapy (US-1). These selected images, from the same plane, were saved. Using ImageJ software (version 1.48, National Institutes of Health, USA), irrelevant breast tissue information such as instrument model, scanning or imaging time, and patient information were removed from the images. Square regions of interest (ROI) were then selected on the remaining images, centered around the tumor and encompassing the surrounding area of the largest tumor. In cases of multifocal tumors, priority was given to selecting the US image that best represented the largest tumor. If the tumor became invisible after NAC treatment, the ROI images were cropped based on the tumor’s pre-NAC position (Fig. 2c). Two breast radiologists with 10 years of clinical experience independently reviewed the US images, and consensus was diligently reached in cases of disagreement.
WSIs acquisition: tissue samples were obtained via US-guided tumor CNB, and were formalin-fixed and paraffin-embedded (FFPE). Following standard hematoxylin and eosin (HE) staining, all processed sections were scanned at a ×20 magnification using a Leica digital pathology scanner. Two pathology experts, each with a decade of clinical experience, manually delineated rectangular blocks representing the most representative invasive cancer areas in the biopsy WSIs using ImageScope software (www.leicabiosystems.com) at a ×10 magnification (Fig. 2b). Additionally, two other experts with a decade of experience reviewed and confirmed the accuracy at a ×20 magnification.
Data preprocessing
Prior to training, a dataset comprising 491 patients with a total of 1473 images, including 491 WSIs from biopsy pathology and 982 US images at two time points for each unifocal lesion, was randomly partitioned into training (80%), validation (10%), and test (10%) cohorts, following an 8:1:1 data ratio. A total of 391 patients were utilized for training, with 49 and 51 patients assigned to the validation and test cohorts, respectively. Additionally, 105 patients with multifocal lesions were set aside as an independent test cohort. Considering the dataset’s pCR to non-pCR ratio of approximately 0.37, up-down and level-flipping techniques were applied to the pCR dataset in the training cohort to ensure a balanced ratio of approximately 1:1. Meanwhile, the original data distribution was preserved for the validation and test cohorts.
To prevent overfitting, we applied data augmentation techniques such as random rotations up to 10° and random scaling on various image classes. As a result, our training cohort has been expanded to 3005 samples, comprising 3005 pairs of US images and 3005 pathology images. Dealing with pathology images presented additional challenges during training due to the extensive information and large image scope inherent in WSIs. Therefore, we initially standardized the image size to 1536 × 1536. Subsequently, each image was divided into nine non-overlapping small image blocks with dimensions of 512 × 512 (following the approach by Byra et al., resizing images to 224 × 22462). Through this segmentation method, when training networks using patch-based pathology images, the quantity of images is sufficient for the network to adequately learn image features, thus avoiding the phenomenon of network overfitting. However, when training multi-time point US models, the smaller dataset size may lead to overfitting. To address this issue, we additionally augmented the ultrasound images, increasing their quantity fourfold, resulting in a final training cohort size of 12,020. The implementation of this strategy ensures consistency in reducing network overfitting between pathology and ultrasound images. Therefore, in the final training of the multimodal model based on pathology and US images, we applied one round of data augmentation to pathology images and two rounds of data augmentation to ultrasound images. Prior to inputting the data into the network, all images underwent standardization to expedite the network convergence process. To distinguish between test cohorts, the unifocal test cohort was labeled as test cohort 1 (TC1), and the multifocal test cohort was labeled as test cohort 2 (TC2) (Fig. 2e).
Model construction
In this investigation, we devised a novel DL vision architecture, SE-CrossT, to construct a multimodal model, as illustrated in Fig. 7. The architecture primarily comprises three components. (1) Input stage: The input image size is set to [3, 512, 512] by comparing the range of ROI, and the images are standardized to facilitate feature extraction and network convergence; (2) Feature extraction stage: Employing a multi-pathway CrossFormer, the pathological-ultrasound net (Path-Ult Net) is utilized to extract pathological features and baseline US features. The double ultrasound net (Dou-Ult Net) is employed to extract US features at two distinct time points; (3) Output stage: Utilizing an adaptive pooling algorithm, the pathological vector and the ultrasound vector are obtained at the same size. Consequently, the Path-Ult Net and the Dou-Ult Net each output an 8-dimensional vector. At the network’s output stage, baseline characteristics are incorporated, resulting in a 20-dimensional vector (8+8+4). The image features and baseline features are inputted into the classification network to determine the classification results.
Before inputting each modality image into the network, position encoding is applied. Then, the images are fed into a series of four-stage network modules. The Patho-Ult Net extracts pathology-ultrasound features, while the Dou-Ult Net incorporates a position-sharing module to capture time and position-related features. Finally, the pathological-ultrasound features derived from the Patho-Ult Net (purple dashed box) output and the dual-ultrasound features generated by the Dou-Ult Net (Red solid box) are input into the classification head.
The original hierarchical structure of CrossFormer consists of the following components: Cross-scale embedding layer (CEL), CrossFormer block, and classification head43. In the CEL layer, we introduced an SE block to comprehensively extract feature information from images (Supplementary Fig. 1). This module enhances channel features in the feature maps, directing the network to focus more on features related to pCR. The cross-scale embedding layer model based on the SE block is shown in Supplementary Fig. 2. With the SE-CEL module, strong correlation is added between features of different dimensions and global relevance of information is also ensured. In the Dou-Ult Net, we designed an US coherence network (Supplementary Fig. 3). This algorithm ensures positional coherence between network inputs while incorporating feature-sharing functionality. In contrast to existing feature fusion methods, the US coherence network shares features between the feature vectors of US at different time points. It effectively utilizes fusion when extracting low-level features, facilitating efficient training, and capturing key features, including changes in the pre- and post-lesion areas, which are crucial for pCR classification.
Experiments
Initially, we preprocessed the image data and baseline characteristics, splitting them into training, validation, and test cohorts. Using identical parameters, we compared our approach with five widely utilized methods in clinical image analysis: VGG16, ResNet50, DenseNet121, ViT and Swin-Transformer. Each of these methods was pretrained on ImageNet (http://www.imagenet.org/, accessed on 1 October 2022). In addition, we conducted ablation experiments building unimodal pathology or multi-time point US models for comparison. Simultaneously, ablation experiments were performed on the added SE block within the network to validate the necessity of incorporating SE blocks. All experiments were conducted on an RTX3090 graphics card using the Adam optimizer, with a fixed learning rate of 0.001 for all training networks. During the training process, the network was trained for 60 epochs using binary cross-entropy loss. At the 40th epoch, the model exhibiting the highest ACC over the preceding 20 epochs was chosen.
Evaluation of pathological results
pCR was evaluated based on the descriptions in the postoperative pathological reports. All reports underwent standard postoperative pathological examination and analysis following the guidelines of the American Joint Committee on Cancer. pCR63 was defined as the absence of invasive cancer in the primary breast lesion (with potential ductal carcinoma in situ) and negative regional lymph nodes, specifically classified as primary lesion MP5 grade64 and negative axillary lymph nodes. Two pathologists, each with over 8 years of experience, analyzed all cases and resolved discrepancies through mutual agreement.
Systematic evaluation
The data processing and statistical analysis were conducted using R software (version 4.1.0), Python software (version 3.8.0), and IBM SPSS Statistics 27 (Armonk, NY, USA). For inter-group comparisons, continuous variables were expressed as ranges and mean ± standard deviations (SDs) and analyzed using the Mann–Whitney U test or independent samples t-test. Categorical variables were represented as percentages and counts and analyzed using the chi-squared test or Fisher’s exact test. Univariate comparisons were applied to assess the relationship between each feature in the cases included in the study and pCR. Predictive variables correlated with pCR (P < 0.05) were selected. Subsequently, multivariate analysis was employed for ultimately filtering features for constructing a DL prediction model incorporating clinicopathological characteristics. Diverse DL methods were trained using the training cohort, and the model demonstrating optimal performance in the validation cohort was subsequently chosen for validation across the test cohorts. Matlab and GraphPad 9.0 software were used to calculate ROC curves for evaluating the diagnostic performance of the DL models. The AUC with a 95% confidence interval (CI) was computed and the DeLong test was used to compare the difference between AUCs. Accuracy (ACC), sensitivity (SENS), specificity (SPEC), positive predictive value (PPV), and negative predictive value (NPV) were compared. AUC curves were plotted to assess the accuracy of different models. All statistical analyses were two-sided, and P < 0.05 was considered statistically significant.
The gradient-weighted class activation mapping (Grad-CAM) method was employed to generate visual heatmaps65. This method furnishes detailed insights into the specific image regions that exert the greatest influence on the classification output for a given category, thereby enhancing the interpretability of network decisions. A meticulous visual examination of the produced heatmaps was undertaken to meticulously evaluate and compare the efficacy of attention mechanisms across a spectrum of networks.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data used in the study were collected in a real-world healthcare setting, and access to these data is restricted for privacy and proprietary reasons. Derived data supporting the findings of this study have been provided in the paper and its supplementary materials.
Code availability
The underlying code for this study (and training/validation datasets) is not publicly available but may be made available to qualified researchers on reasonable request from the corresponding author.
References
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Huber, S. et al. Locally advanced breast carcinoma: evaluation of mammography in the prediction of residual disease after induction chemotherapy. Anticancer Res. 20, 553–558 (2000).
Derks, M. G. M. & van de Velde, C. J. H. Neoadjuvant chemotherapy in breast cancer: more than just downsizing. Lancet Oncol. 19, 2–3 (2018).
Saleh, G. A. et al. Impact of imaging biomarkers and AI on breast cancer management: a brief review. Cancers 15, 5216 (2023).
Spanheimer, P. M. & Reeder-Hayes, K. E. Breast surgery after neoadjuvant chemotherapy: time for a change? Lancet Oncol. 23, 1477–1479 (2022).
An, Y. Y., Kim, S. H., Kang, B. J. & Lee, A. W. Treatment response evaluation of breast cancer after neoadjuvant chemotherapy and usefulness of the imaging parameters of MRI and PET/CT. J. Korean Med. Sci. 30, 808–815 (2015).
Teshome, M. & Hunt, K. K. Neoadjuvant therapy in the treatment of breast cancer. Surg. Oncol. Clin. N. Am. 23, 505–523 (2014).
Xiong, Q. et al. Multiparametric MRI-based radiomics analysis for prediction of breast cancers insensitive to neoadjuvant chemotherapy. Clin. Transl. Oncol. 22, 50–59 (2020).
Denkert, C. et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 19, 40–50 (2018).
Denkert, C. et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 28, 105–113 (2010).
Hagenaars, S. C. et al. Tumor-stroma ratio is associated with Miller-Payne score and pathological response to neoadjuvant chemotherapy in HER2-negative early breast cancer. Int. J. Cancer 149, 1181–1188 (2021).
Li, F. et al. Deep learning-based predictive biomarker of pathological complete response to neoadjuvant chemotherapy from histological images in breast cancer. J. Transl. Med. 19, 348 (2021).
Rouzier, R. et al. Nomograms to predict pathologic complete response and metastasis-free survival after preoperative chemotherapy for breast cancer. J. Clin. Oncol. 23, 8331–8339 (2005).
Cain, H. et al. Neoadjuvant therapy in early breast cancer: treatment considerations and common debates in practice. Clin. Oncol. 29, 642–652 (2017).
Meti, N. et al. Machine learning frameworks to predict neoadjuvant chemotherapy response in breast cancer using clinical and pathological features. JCO Clin. Cancer Inf. 5, 66–80 (2021).
Kim, S. Y. et al. Factors affecting pathologic complete response following neoadjuvant chemotherapy in breast cancer: development and validation of a predictive nomogram. Radiology 299, 290–300 (2021).
Cui, H. et al. Predicting pathological complete response after neoadjuvant chemotherapy in advanced breast cancer by ultrasound and clinicopathological features using a nomogram. Front. Oncol. 11, 718531 (2021).
Tadayyon, H. et al. Correction: Quantitative ultrasound assessment of breast tumor response to chemotherapy using a multi-parameter approach. Oncotarget 8, 35481 (2017).
Shao, L. et al. Multiparametric MRI and whole slide image-based pretreatment prediction of pathological response to neoadjuvant chemoradiotherapy in rectal cancer: a multicenter radiopathomic study. Ann. Surg. Oncol. 27, 4296–4306 (2020).
Wang, R. et al. Development of a novel combined nomogram model integrating deep learning-pathomics, radiomics and immunoscore to predict postoperative outcome of colorectal cancer lung metastasis patients. J. Hematol. Oncol. 15, 11 (2022).
Ma, Z. et al. ATFE-Net: axial transformer and feature enhancement-based CNN for ultrasound breast mass segmentation. Comput. Biol. Med. 153, 106533 (2023).
Xu, Y. et al. Medical breast ultrasound image segmentation by machine learning. Ultrasonics 91, 1–9 (2019).
Patil, S. M., Tong, L. & Wang, M. D. Generating region of interests for invasive breast cancer in histopathological whole-slide-image. Proc. COMPSAC 2020, 723–728 (2020).
Liu, M. Z. et al. Weakly supervised deep learning approach to breast MRI assessment. Acad. Radiol. 29, S166–S172 (2022).
Ye, H. et al. Automatic identification of triple negative breast cancer in ultrasonography using a deep convolutional neural network. Sci. Rep. 11, 20474 (2021).
Witowski, J. et al. Improving breast cancer diagnostics with deep learning for MRI. Sci. Transl. Med. 14, eabo4802 (2022).
Naik, N. et al. Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains. Nat. Commun. 11, 5727 (2020).
Niyas, S. et al. Automated molecular subtyping of breast carcinoma using deep learning techniques. IEEE J. Transl. Eng. Health Med. 11, 161–169 (2023).
Mathew, T., Niyas, S., Johnpaul, C. I., Kini, J. R. & Rajan, J. A novel deep classifier framework for automated molecular subtyping of breast carcinoma using immunohistochemistry image analysis. Biomed. Signal Process. Control 76, 103657 (2022).
Chiu, Y. C. et al. Predicting drug response of tumors from integrated genomic profiles by deep neural networks. BMC Med. Genomics 12, 18 (2019).
Dodington, D. W. et al. Analysis of tumor nuclear features using artificial intelligence to predict response to neoadjuvant chemotherapy in high-risk breast cancer patients. Breast Cancer Res. Treat. 186, 379–389 (2021).
Skrede, O. J. et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet 395, 350–360 (2020).
Byra, M., Dobruch-Sobczak, K., Klimonda, Z., Piotrzkowska-Wroblewska, H. & Litniewski, J. Early prediction of response to neoadjuvant chemotherapy in breast cancer sonography using Siamese convolutional neural networks. IEEE J. Biomed. Health Inf. 25, 797–805 (2021).
Xie, J. et al. Dual-branch convolutional neural network based on ultrasound imaging in the early prediction of neoadjuvant chemotherapy response in patients with locally advanced breast cancer. Front. Oncol. 12, 812463 (2022).
Qu, Y. H. et al. Prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer using a deep learning (DL) method. Thorac. Cancer 11, 651–658 (2020).
Dosovitskiy,A. et al. An image is worth 16x16 words: Transformers for image recognition at scale. In: Proceedings of the 9th International Conference on Learning Representations. Virtual Event, Austria: OpenReview.net, 2020.
Li, J. et al. DSMT-Net: dual self-supervised multi-operator transformation for multi-source endoscopic ultrasound diagnosis. IEEE Trans. Med. Imaging 43, 64–75 (2024).
Su, Y., Liu, Q., Xie, W. & Hu, P. YOLO-LOGO: a transformer-based YOLO segmentation model for breast mass detection and segmentation in digital mammograms. Comput. Methods Programs Biomed. 221, 106903 (2022).
Ayana, G. & Choe, S. W. BUViTNet: breast ultrasound detection via vision transformers. Diagnostics 12, 2654 (2022).
Ayana, G. et al. Vision-transformer-based transfer learning for mammogram classification. Diagnostics 13, 178 (2023).
Saednia, K., Tran, W. T. & Sadeghi-Naini, A. A hierarchical self-attention-guided deep learning framework to predict breast cancer response to chemotherapy using pre-treatment tumor biopsies. Med. Phys. 50, 7852–7864 (2023).
Clauwaert, J., Menschaert, G. & Waegeman, W. Explainability in transformer models for functional genomics. Brief Bioinform. 22, bbab060 (2021).
Wang, W. et al. Crossformer: a versatile vision transformer hinging on cross-scale attention. Preprint at https://arxiv.org/abs/2108.00154 (2021).
Hu, J., Shen, L., Albanie, S., Sun, G. & Wu, E. Squeeze-and-excitation networks. IEEE Trans. Pattern Anal. Mach. Intell. 42, 2011–2023 (2020).
Mason, S. R. et al. Platinum-based chemotherapy for early triple-negative breast cancer. Cochrane Database Syst. Rev. 9, CD014805 (2023).
Swain, S. M. et al. Event-free survival in patients with early HER2-positive breast cancer with a pathological complete response after HER2-targeted therapy: a pooled analysis. Cancers 14, 5051 (2022).
Tang, L. et al. Efficacy evaluation of neoadjuvant chemotherapy in patients with HER2-low expression breast cancer: a real-world retrospective study. Front. Oncol. 12, 999716 (2022).
Liu, Y. et al. Early prediction of treatment response to neoadjuvant chemotherapy based on longitudinal ultrasound images of HER2-positive breast cancer patients by Siamese multi-task network: a multicentre, retrospective cohort study. EClinicalMedicine 52, 101562 (2022).
Yu, F. H. et al. Pretreatment ultrasound-based deep learning radiomics model for the early prediction of pathologic response to neoadjuvant chemotherapy in breast cancer. Eur. Radiol. 33, 5634–5644 (2023).
Sui, L. et al. Ultrasound and clinicopathological characteristics-based model for prediction of pathologic response to neoadjuvant chemotherapy in HER2-positive breast cancer: a case-control study. Breast Cancer Res. Treat. 202, 45–55 (2023).
Noritake, M. et al. Evaluation of the response to breast cancer neoadjuvant chemotherapy using 18F-FDG positron emission mammography compared with whole-body 18F-FDG PET: a prospective observational study. Clin. Nucl. Med. 42, 169–175 (2017).
Wu, L. et al. An integrated deep learning model for the prediction of pathological complete response to neoadjuvant chemotherapy with serial ultrasonography in breast cancer patients: a multicentre, retrospective study. Breast Cancer Res. 24, 81 (2022).
Li, B. et al. Deep learning with biopsy whole slide images for pretreatment prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Breast 66, 183–190 (2022).
Yu, K. D., Cai, Y. W., Wu, S. Y., Shui, R. H. & Shao, Z. M. Estrogen receptor-low breast cancer: biology chaos and treatment paradox. Cancer Commun. 41, 968–980 (2021).
Price, W. N. Big data and black-box medical algorithms. Sci. Transl. Med. 10, eaao5333 (2018).
Zurowietz, M. & Nattkemper, T. W. An interactive visualization for feature localization in deep neural networks. Front. Artif. Intell. 3, 49 (2020).
Zhu, J. et al. Neoadjuvant efficacy of three targeted therapy strategies for HER2-positive breast cancer based on the same chemotherapy regimen. Cancers 14, 4508 (2022).
Niu, N. et al. A multicentre single arm phase 2 trial of neoadjuvant pyrotinib and letrozole plus dalpiciclib for triple-positive breast cancer. Nat. Commun. 13, 7043 (2022).
Gradishar, W. J. et al. NCCN Guidelines® Insights: Breast Cancer, Version 4.2021. J. Natl Compr. Canc. Netw. 19, 484–493 (2021).
Li, J. & Jiang, Z. Chinese Society of Clinical Oncology Breast Cancer (CSCO BC) guidelines in 2022: stratification and classification. Cancer Biol. Med. 19, 769–773 (2022).
Omarini, C. et al. Impact of time to surgery after neoadjuvant chemotherapy in operable breast cancer patients. Eur. J. Surg. Oncol. 43, 613–618 (2017).
Byra, M. et al. Breast mass segmentation in ultrasound with selective kernel U-Net convolutional neural network. Biomed. Signal Process. Control 61, 102027 (2020).
Giuliano, A. E. et al. Breast cancer—major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 67, 290–303 (2017).
Ogston, K. N. et al. A new histological grading system to assess response of breast cancers to primary chemotherapy: prognostic significance and survival. Breast 12, 320–327 (2003).
Selvaraju, R. R. et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 128, 336–359 (2019).
Acknowledgements
We thank all participating investigators who contributed to this study. This study was funded by the Wu Jieping Medical Foundation (grant number 320.6750.2023-11-10) and the Beijing Medical Award Foundation (grant number YXJL-2022-0561-0427). The funder played no role in the study design, data collection, analysis and interpretation of data, or the writing of this manuscript. Figure 2 was partly generated using Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 3.0, and partly sourced from Vecteezy.com under a free license.
Author information
Authors and Affiliations
Contributions
Conceptualization, Jianming Guo and Hongda Cao; Data curation, Baihui Chen and Youxue Zhang; Formal analysis, Jianming Guo and Baihui Chen; Funding acquisition, Dalin Li; Investigation, Jinfeng Zhang, Huanyu Zhang, Yuan Sui, Tianyu Chen, Dongxu Yang and Xue Gong; Methodology, Jianming Guo, Hongda Cao, Quan Dai and Ling Qin; Project administration, Dalin Li; Resources, Dalin Li, Jinfeng Zhang and Youxue Zhang; Software, Hongda Cao; Supervision, Dalin Li; Validation, Jianming Guo, Baihui Chen, Hongda Cao and Quan Dai; Visualization, Jianming Guo, Baihui Chen, Hongda Cao and Ling Qin; Writing—original draft, Jianming Guo; Writing—review & editing, Jianming Guo and Quan Dai. All authors will be informed about each step of manuscript processing, including submission, revision, revision reminder, etc., via emails from our system or assigned Assistant Editor.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Guo, J., Chen, B., Cao, H. et al. Cross-modal deep learning model for predicting pathologic complete response to neoadjuvant chemotherapy in breast cancer. npj Precis. Onc. 8, 189 (2024). https://doi.org/10.1038/s41698-024-00678-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41698-024-00678-8
This article is cited by
-
Fractal analysis of longitudinal MRI for predicting response to neoadjuvant chemotherapy in breast cancer
npj Precision Oncology (2025)
-
Artificial intelligence in digital pathology — time for a reality check
Nature Reviews Clinical Oncology (2025)
-
Multimodal deep learning for predicting neoadjuvant treatment outcomes in breast cancer: a systematic review
Biology Direct (2025)