Abstract
Basal-like breast cancers (BLBC) have limited targeted therapies and poor outcomes. We found that CREB5 is a transcription factor overexpressed in 15% of BLBCs and was upregulated in breast cancers that metastasize to the brain. In cell lines, CREB5 overexpression regulated cell phenotypes and transcriptional changes, including IL13RA2, a cell surface receptor that is currently druggable and represents a novel target in BLBC.
Similar content being viewed by others
Introduction
Breast cancers (BC) are the most frequently diagnosed cancers in women1 and treatments are guided by the expression of hormone receptors or HER2. Triple-negative breast cancers (TNBCs) lack these targets and represent a large portion of the transcriptional subtype basal-like breast cancers (BLBCs) as compared to the luminal A/B or HER2 subtypes. BLBCs represent 15% of all BCs and exhibit poor clinical outcomes due to the lack of targeted therapeutics2. Therefore, further identifying oncogenic drivers of BLBCs and evaluating new druggable targets could lead to promising developments of targeted therapeutics for these patients.
Several oncogenic regulators have been associated with BLBCs in the search of potential therapeutic targets. As one example, MET overexpression functions to promote stemness in several cancers and is also overexpressed in BLBCs3, suggesting similar functions. Transcription factors, while not druggable, have also been implicated in BLBC progression4. Recent pan-cancer studies determined that the transcription factor cyclic adenosine monophosphate (cAMP) responsive element binding protein 5 (CREB5) promotes cancer development and progression, including stemness5. CREB5 has developmental functions and is necessary for general cell survival. Several studies, including ours, have indicated that CREB5 is selectively overexpressed in metastatic tumors, including cancers of the prostate, colon, ovaries, brain and more5,6,7,8. In two in vitro BC studies, CREB5 promoted angiogenesis in BLBC and regulated proliferation in TNBC cell lines9,10. Regardless, few studies have examined the role of CREB5 in BC patients, and particularly BLBCs, which is the focus of this study.
We first evaluated CREB5 mRNA expression based on data from The Cancer Genome Atlas (TCGA, n = 1069) and METABRIC (n = 1608)11 across four BC molecular subtypes: BLBC, HER2, Luminal A/ and B subtypes. In both datasets, median CREB5 expression was the highest in BLBCs (Fig. 1A, B). In the same BC tumors, CREB5 was amplified or overexpressed in 15–16% of BLBC cases as compared to less than 5% in all other subtypes (Fig. 1C). High CREB5 expression in patients with BLBC was associated with worse overall survival (Fig. 1D, HR 1.88, 95% CI 1.07–3.31, p = 0.025). We next examined a cohort of 45 BC patients with biopsies collected from their primary tumor and their breast cancer brain metastasis (BCBM). Here the basal-like BCBMs exhibited the highest baseline CREB5 expression (Fig. 1E). Further, 9 out of 16 BLBCs exhibited increases in CREB5 in the secondary metastatic sample relative to the matched primary (Fig. 1F)12. Altogether, CREB5 expression was the most enriched in BLBCs and was increased in certain BCs that have poor outcomes due to brain metastasis.
Box plots showing CREB5 median expression across A TCGA and B METABRIC datasets with the boxes representing the ends of the first and third quartiles in breast cancer molecular subtypes. Red dotted line denotes the global median while blue dotted lines are global standard deviations. C Percent of tumors with CREB5 amplifications or overexpression in BLBC as compared to all other BC subtypes in the same two datasets. Overexpression is defined by a z-score >2 and amplifications are defined by cBioPortal parameters. D Kaplan–Meier plot comparing overall survival of CREB5 high (red line) versus low (black line) BLBC patients (n = 309) from KMPlot.com. E BCBMs acquired by Cosgrove et al.12 are stratified by BC subtypes including basal (n = 16), HER2 (n = 9), luminal A (n = 11), and luminal B (n = 8). Median CREB5 expression across subtypes are shown in a box plot with whiskers extending to minimum and maximum values. *p < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001, ns not significant. F Waterfall plot shows the percent change of CREB5 expression in each patient from the primary BC biopsy to the BCBM biopsy in the same dataset.
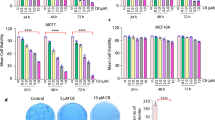
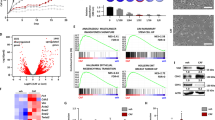
We next evaluated several tumorigenic phenotypes by overexpressing CREB5. We explored two BLBC cell lines, MDA-MB-231 and HCC1806, which have been characterized as distinct transcriptional subtypes by Lehmann et al. as mesenchymal stem-like and basal-like 2 subtypes, respectively13. CREB5 overexpression increased the proliferation rate of both cell lines based on median live cell counts (Fig. 2A, B, p < 0.05). Therefore, we sought to further evaluate the role of CREB5 on SCL phenotypes, which have been identified as a key feature of BCs with increased metastatic capacity14. We assessed tumor-like spheroid, or tumorsphere, formation efficiency based on spheroid counts and size through a 3D culture experiment that specifically selects for SCL cells15. This indicated significant increases in the MDA-MB-231 cells while exhibiting notable trends in HCC1806 cells (Fig. 2C–E). Furthermore, CREB5 overexpression led to substantial alterations in the transcriptomes in both 2D and 3D cultures (Fig. 2F). In 2D, CREB5 overexpression significantly (FDR adjusted p-value ≤ 0.05) up-regulated 4584 and 4899 genes in MDA-MB-231 and HCC1806 cells, respectively. In 3D, CREB5 overexpression up-regulated 6548 and 9376 genes in MDA-MB-231 and HCC1806 cells, respectively. While the two cell lines expectedly exhibited differences in their baseline transcriptome, Gene Set Enrichment Analysis (GSEA)16 based on C6 oncogenic signatures from the MSigDB database17 indicated that KRAS associated signatures were consistently perturbed in both cell lines in 3D culture (Fig. 2G). Altogether, CREB5 overexpression promoted several tumorigenic phenotypes in 2D and 3D cultures, and this was accompanied by significant changes in their transcriptomes.
Proliferation assay showing median live cell count over 10 days for A MDA-MB-231 and B HCC1806 cell lines with either CREB5 or luciferase (LUC) overexpressed. C Tumorsphere formation assay results for MDA-MB-231 (top row) and HCC1806 (bottom row) cell lines with CREB5 (left panels) or luciferase overexpressed. Scale bar shown is 200 μm. Quantitative analysis of median D tumorsphere formation efficiency and E tumorsphere size (μm²). F Volcano plots illustrating changes in gene expression from RNA sequencing in MDA-MB-231 (left panels) and HCC1806 (right panels) cell lines, in 2D (top row) and 3D (bottom row) culture. Each dot represents a gene that either passes the significant thresholds for both the p-value and log-fold changes (red), for only the p-value (blue), or neither (gray). G Net enrichment scores (NES) from GSEA based on C6 oncogenic signatures depicted across cell lines in 3D culture. Red circles: significantly upregulated both cell lines. Orange circles: only significantly upregulated in the MDA-MB-231 cells. Purple circles: only significantly upregulated in the HCC1806 cells. Significance is based on FDR < 0.05.
To consider accessible cell surface targets for CREB5-high BLBC, we evaluated changes in the surfaceome induced by CREB5 overexpression. In each BLBC cell line, we found that CREB5 overexpression led to a 1 to ≥100-fold increase in expression of 198 and 159 cell surface proteins in MDA-MB-231 and HCC1806 cells, respectively (Fig. 3A). Further, the expression of 22 and 19 proteins was increased by 10-fold or greater in CREB5 overexpressed MDA-MB-231 and HCC1806 cells, respectively. Three proteins that increased by greater than 10-fold were present in both cell lines: IL13RA2, NTM, and THSD1 (Fig. 3B). As IL13RA2 is currently an early phase CAR-T cell target for glioblastomas18 and a gene previously implicated in BCBMs19, we further examined its significance in our BLBC models, and found that the IL13RA2 protein expression was supported by 14 unique peptides that increased upon CREB5 overexpression, (Supplemental Table 1). In reviewing the RNA-seq data, IL13RA2 mRNA expression was also significantly increased by CREB5 overexpression. In HCC1806 cells, CREB5 overexpression increased median IL13RA2 expression by roughly 30-fold in 2D culture and 3-fold in 3D culture. Similarly, in MDA-MB-231 cells, IL13RA2 expression was increased by 100-fold in 2D culture and 230-fold in 3D culture (Fig. 3C, D).
A Bar chart depicts the counts of unique cell surface proteins increased by CREB5 overexpression in MDA-MB-231 and HCC1806 cell lines, as categorized by set ranges of abundance ratios. B Venn diagram compares cell surface proteins with greater than 10- fold changes in MDA-MB-231 and HCC1806 cell lines. The overlapping proteins (n = 3) are detailed in the table beneath. IL13RA2 mRNA expression in C MDA-MB-231 and D HCC1806 cell lines with CREB5 or luciferase overexpressed in 2D and 3D cultures. Box plots display median expression levels with whiskers extending to minimum and maximum values. p ** < 0.01, **** < 0.0001. E Log2 fold changes in IL13RA2 expression with respect to primary BC biopsies from patients with BCBMs, as organized by BC subtypes from Cosgrove et al.12. Each circle represents the change in expression of a paired sample (n = 44). The dotted line at 0 indicates no change in expression. F Percentage of BCBM samples with IL13RA2 expression increased from the primary BC sample by 0–100%, ≥100%, or ≤0%. Change in IL13RA2 expression is shown across BC subtypes including basal (n = 16), HER2 (n = 9), luminal A (n = 11), and luminal B (n = 8). G Bar graph displays the frequency of IL13RA2 genetic alterations including high mRNA expression, amplification, and homozygous deletions, across multiple cancer types from TCGA.
Having observed the relationship between CREB5 and IL13RA2 in our cell models, we next sought to examine the characteristics of IL13RA2 expression in patients with BC. In these BCBM samples12, IL13RA2 expression exhibited the greatest relative increase in the suite of primary BCs characterized as BLBCs (Fig. 3E). When instead observing the proportion of tumors with increased IL13RA2 expression, BCBMs that developed from BLBC primary tumors, as compared to other subtypes, exhibited the highest proportion of metastatic tumors with increased IL13RA2 (Fig. 3F). Here >62% of patients with both primary and BCBM samples exhibited increased IL13RA2 expression in the BCBM compared to the paired primary tumor. 70% of the basal-like BCBMs that had increases in IL13RA2 expression increased by greater than 100%. Furthermore, we found that IL13RA2 exhibits similar behavior to many epithelial-mesenchymal transition (EMT) transcription factors (Fig. S1); this may explain the association between increased IL13RA2 and BCBMs, which have increased metastatic potential to the brain. To further investigate the potential of IL13RA2 as a general drug target, we evaluated IL13RA2 expression at a pan-cancer level in the TCGA dataset (n = 10,967). Across cancers, IL13RA2 was notably amplified or exhibited elevated mRNA expression in several cancer types, such as brain lower grade gliomas (Fig. 3G). Altogether we have observed in our cell line models of BLBC that CREB5 regulates the mRNA and cell surface expression of IL13RA2, a current immunotherapy target and that in patients with cancer, IL13RA2 is frequently overexpressed.
There are no current efforts to directly inhibit CREB5 or to perturb its downstream transcriptional activity. Of importance to the development of therapies, we previously determined that CREB5 was functionally distinct from other canonical CREB family members that are regulated by Protein Kinase A20. Therefore, the development of strategies to block oncogenic CREB5 activity requires a distinct effort, either to directly drug CREB5 activity, or to target robust downstream cell surface targets of CREB5 specifically, such as IL13RA2. As a cell surface receptor that binds with high affinity to IL13, IL13RA2 was initially identified as a driver of glioma18,21. It has also been implicated in BCBMs19,22, which represent a population of BC patients with exceptionally aggressive disease. In previous studies, IL13RA2 expression was functionally necessary for metastasis of BC cells that are hormone receptor + , HER2 + , or BLBCs19,22. Interestingly, among the TNBC patients that developed brain metastases, high levels of IL13RA2 were associated with significantly worse overall survival19. In our study, CREB5 was increased in BLBCs and its overexpression in cell models promoted notable increases in IL13RA2 transcripts and cell surface protein expression (~100-fold). Further, in patients with BCBMs we observed increases in CREB5 along with dramatic increases in IL13RA2 mRNA expression. In patients with BLBCs, an astonishing 43.8% of the BCBM had increased IL13RA2 by over 100%. Altogether, our overexpression experiments and analysis of clinical samples support that IL13RA2 is a promising new target in BCs that are highly aggressive, such as BLBCs.
IL13RA2 CAR-T cell therapies, while still not broadly used against solid tumors, have already undergone several generations of development18,23. We found that CREB5 promoted IL13RA2 surface expression in 2 BLBC models, in which Lehmann et al. had classified as mesenchymal stem-like (MDA-MB-231) or was basal-like (HCC1806), indicating CREB5 has similar downstream effects across BLBCs with distinct transcriptional subtypes. This is appealing as BLBC are known to represent a collection of patients with highly heterogenous transcriptomes. Given there is a lack of common targets in these subsets of BLBCs, our study offers a promising solution in targeting IL13RA2.
Outside of BCs, studies have indicated that IL13RA2 has tumor promoting properties in several metastatic or advanced cancers24,25. Further, IL13RA2 is associated with therapy resistance in renal cell carcinomas26, and appears to be a key regulator of the mesenchymal-like glioma subtype21. As CREB5 is also a therapy resistant factor that is increased in metastatic prostate cancer, and a SCL gene in gliomas6,20,27, it is possible that regulation of the CREB5-IL13RA2 axis may be cancer type agnostic, a mechanism that should be interrogated in future studies. Given CREB5 overexpressed cell lines in 3D culture also are associated with KRAS signatures, future studies of CREB5 in BLBC may investigate EMT associated genes that are also regulated by KRAS, such as YAP128.
In summary, our study has demonstrated that CREB5 is a key driver of BLBC. On a clinical level, our findings depict that CREB5 high BLBC patients have worse survival outcomes, showing the continued need for novel targeted therapeutics. Further studies may inform novel therapies for advanced cancer types that rely on CREB5.
Methods
Expression analysis
The Cancer Genome Atlas (TCGA) raw read counts were downloaded using the TCGAbiolinks (version 2.30.4, project = “TCGA-BRCA”) R package. The raw reads were then filtered to include only protein-coding genes with expression in at least one sample. The data were converted to VST normalized counts using DESeq2 (1.42.0), which performs cross-sample normalization based on library size. METABRIC11 mRNA log2 intensity level expression was downloaded directly from cBioPortal, and genes with zero expression across all samples were filtered out. For determination of statistical significance, a t-test was used when data were normally distributed and a Wilcoxon’s test was used when data were not normally distributed, with corrections for multiple comparisons done using the Benjamini-Hochberg method to control the FDR at a significance level of 0.05. In the BCBM expression data from Cosgrove et al.12 (n = 44), the values are normalized and log transformed using edgeR, and then batch correction using a multiple linear regression model. We ran a Wilcoxon rank-sum test between the groups of percent change for each of the subtypes with FDR adjusted p-values.
Survival analysis
To compute the association with CREB5 expression and overall survival, we utilized the KMPlot.com Breast RNA-seq dataset29. We filtered the patient dataset based on PAM50 classification to Basal breast cancers. We applied no other filters to the patient cohort. To split patients into high and low CREB5 expression groups we utilized KMplot’s auto select feature. The overall survival Kaplan–Meier plot was made using the web portal with no modifications.
Gene set enrichment analysis (GSEA)
We conducted GSEA16 based on C6 oncogenic signatures from the MSigDB database17. From this we obtained normalized enrichment scores (NES) and compared GSEA oncogenic signatures across both BLBC cell lines in 3D culture conditions.
Algorithm for linking activity networks (ALAN) analysis
We utilized the Algorithm for Linking Activity Networks (ALAN)30 to examine the transcriptome of basal and luminal BC samples from both TCGA and METABRIC. Similarity scores from relevant genes were pulled from this output and visualized in a heatmap where maximum and minimum similarities of ALAN networks were numerically represented based on measurements between 1 and −1.
Generation of cell lines
The human BLBC cell lines MDA-MB-231 and HCC1806 were cultured in RPMI medium 1640 (Gibco), supplemented with 10% FBS (R&D Systems), 0.2% GlutaMax (Gibco), and 1% Penicillin Streptomycin (Gibco) grown at 37 °C and 5% CO2. All cell lines were purchased from the American Type Culture Collection. To produce the CREB5 and luciferase overexpressing cells, a mammalian expression lentiviral vector (VectorBuilder, vector IDs: VB240613-1734jvz and VB240917-1425tbq, respectively) was used for infecting each cell line in the presence of 10 μg/ml polybrene (VectorBuilder) followed by drug selection with 10 μg/ml puromycin (Gibco).
Proliferation experiments
In six-well plates, 15,000 cells were seeded with 1.5 mL of the RPMI media. 5, 7, and 10 days after being seeded, cells were harvested and counted with a Corning Cell Counter and CytoSMART software. For the 7- and 10-day plates, 0.5 mL media was added to each well on day 5. A Student’s t-test was utilized to determine significance.
Tumorsphere experiments
Adherent cells were washed with PBS and dissociated enzymatically in 0.25% trypsin for 2–5 min or until cells become unadhered to the plate. The cells were sieved through a 40-μm sieve (BD Falcon) and 18 G needle to obtain single cell suspensions. 1000 single cells were plated in 24-well ultra-low attachment plates (Corning) in a phenol red-free mammary epithelial basal medium (DMEM: F12; Thermo Scientific) containing 1% B27 Supplement (Invitrogen), 1% penicillin-streptomycin (Invitrogen), 5 μg/ml insulin (Invitrogen), 20 ng/ml EGF (Sigma Aldrich), 1 ng/ml hydrocortisone (Sigma Aldrich), and 100 μM β-mercaptoethanol. Tumorspheres were allowed to grow for 8 days and then were imaged with ZEN microscopy software. Sobel edge detection was used to find the edges of cells in the images, and areas were calculated by counting the number of pixels in each tumorsphere and a scaling by a factor to calculate the real area in micrometers2 of each tumorsphere. The number of tumorspheres in each image was calculated by counting the number of cell groups with a diameter greater than 50 micrometers.
RNA-sequencing experiments
2D and 3D cultures of CREB5 or luciferase overexpressed cell lines were cultured as stated above. RNA was extracted from the cell lines using a RNeasy Mini Kit (Qiagen). For RNA-seq experiments, library preparations, quality control and sequencing were performed by Novogene. All groups had three replicated expect the CREB5 overexpressed 2D cultured HCC1806 cell line had two.
Surfaceome experiments
Both cell lines were seeded in a 15 cm plate, then a Cell Surface Protein Biotinylation and Isolation Kit (Pierce) was utilized to isolate proteins. Biotinylated proteins were then subjected to mass spectrometry through the Center for Metabolomics and Proteomics at the University of Minnesota. Proteom Discoverer (Thermo Scientific) was used for peptide identification and label-free quantification (LFQ). Peptide sequences, and thus protein identities, were determined by matching them against the UniProt database. Significance was assessed using an ANOVA test with Benjamini Hochberg adjustment.
Data availability
Clinical datasets are derived from public resources and are available within the article. The data generated from this study are provided as supplemental data. The RNA-sequencing data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE283312 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE283312).
Code availability
The code generated from this study is available at https://github.com/alixx830/cell_quantification.
Change history
20 November 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41698-025-01168-1
References
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 73, 17–48 (2023).
Alluri, P. & Newman, L. Basal-like and triple negative breast cancers: searching for positives among many negatives. Surg. Oncol. Clin. N. Am. 23, 567–577 (2014).
Gastaldi, S., Comoglio, P. M. & Trusolino, L. The mMet oncogene and basal-like breast cancer: another culprit to watch out for?. Breast Cancer Res.12, 208 (2010).
Jovanović, B. et al. Heterogeneity and transcriptional drivers of triple-negative breast cancer. Cell Rep. 42, 113564 (2023).
Wu, Z. et al. Identification of CREB5 as a prognostic and immunotherapeutic biomarker in glioma through multi-omics pan-cancer analysis. Comput. Biol. Med. 173, (2024).
Hwang, J. H. et al. CREB5 promotes resistance to androgen-receptor antagonists and androgen deprivation in prostate cancer. Cell Rep. 29, 2355–2370.e6 (2019).
Wang, S. et al. CREB5 promotes invasiveness and metastasis in colorectal cancer by directly activating MET. J. Exp. Clin. Cancer Res. 39, 168 (2020).
He, S. et al. CREB5 promotes tumor cell invasion and correlates with poor prognosis in epithelial ovarian cancer. Oncol. Lett. 14, 8156–8161 (2017).
Contreras-Sanzón, E. et al. MicroRNA-204/CREB5 axis regulates vasculogenic mimicry in breast cancer cells. Cancer Biomark. Sect. Dis. Markers 35, 47–56 (2022).
Bhardwaj, A. et al. Regulation of miRNA-29c and its downstream pathways in preneoplastic progression of triple-negative breast cancer. Oncotarget 8, 19645–19660 (2017).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Cosgrove, N. et al. Mapping molecular subtype specific alterations in breast cancer brain metastases identifies clinically relevant vulnerabilities. Nat. Commun. 13, 514 (2022).
Lehmann, B. D. et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 121, 2750–2767 (2011).
He, L., Wick, N., Germans, S. K. & Peng, Y. The Role of breast cancer stem cells in chemoresistance and metastasis in triple-negative breast cancer. Cancers 13, 6209 (2021).
Dontu, G. et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17, 1253–1270 (2003).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Brown, C. E. et al. Optimization of IL13Rα2-Targeted chimeric antigen receptor T cells for improved anti-tumor efficacy against glioblastoma. Mol. Ther. J. Am. Soc. Gene Ther. 26, 31–44 (2018).
Márquez-Ortiz, R. A. et al. IL13Rα2 promotes proliferation and outgrowth of breast cancer brain metastases. Clin. Cancer Res. 27, 6209–6221 (2021).
Hwang, J. H. et al. CREB5 reprograms FOXA1 nuclear interactions to promote resistance to androgen receptor-targeting therapies. eLife. 11, e73223 (2022).
Brown, C. E. et al. Glioma IL13Rα2 is associated with mesenchymal signature gene expression and poor patient prognosis. PloS ONE 8, e77769 (2013).
Papageorgis, P. et al. Targeting IL13Ralpha2 activates STAT6-TP63 pathway to suppress breast cancer lung metastasis. Breast Cancer Res. 17, 98 (2015).
Gu, A. et al. IL13Rα2-targeted third-generation CAR-T cells with CD28 transmembrane domain mediate the best anti-glioblastoma efficacy. Cancer Immunol. Immunother. 72, 2393–2403 (2023).
Bartolomé, R. A., Jaén, M. & Casal, J. I. An IL13Rα2 peptide exhibits therapeutic activity against metastatic colorectal cancer. Br. J. Cancer 119, 940 (2018).
Okamoto, H. et al. Interleukin-13 receptor α2 is a novel marker and potential therapeutic target for human melanoma. Sci. Rep. 9, 1281 (2019).
Shibasaki, N. et al. Role of IL13RA2 in Sunitinib resistance in clear cell renal cell carcinoma. PLoS ONE 10, e0130980 (2015).
Kim, H. J. et al. CREB5 promotes the proliferation and self-renewal ability of glioma stem cells. Cell Death Discov. 10, 1–8 (2024).
Shao, D. D. et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158, 171–184 (2014).
Győrffy, B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innov. Camb. Mass. 5, 100625 (2024).
Bergom, H. E. et al. ALAN is a computational approach that interprets genomic findings in the context of tumor ecosystems. Commun. Biol. 6, 1–12 (2023).
Acknowledgements
A.M. is supported by the Undergraduate Research Opportunities Program and the Research and Travel Fund Award by the University of Minnesota. J.M.D. is supported by the NIH NCI R01CA269801. D.A.L. is supported by an American Cancer Society Research Professorship. J.H.O. is supported by the NCI/ NIH (5R01CA236948) and Hope for Change Funds by the University of Minnesota. J.H. is supported by the Randy Shaver Foundation Community Funds, Hope for Change Funds by the University of Minnesota, and NCI/NIH (R37 1R37CA288972-01). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The results shown here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Author information
Authors and Affiliations
Contributions
A.M. and J.H.H. conceptualized and designed the study, acquired data, analyzed data, drafted/revised manuscript, and approved final manuscript. E.B., L.W., A.A., K.I., C.L., M.L., and J.P.R. acquired data, analyzed data, drafted/revised manuscript, and approved final manuscript. A.G., H.E.B., E.J., S.T., S.K., A.D., F.I.D., C.R., L.C., D.M., J.M.D., E.S.A., D.A.L., and J.H.O. interpreted results, drafted/revised manuscript, and approved of final manuscript.
Corresponding author
Ethics declarations
Competing interests
A.M. consults for EMRGNSE LLC, but the business of this company is unrelated to the contents of this manuscript. E.B. consults for Tempus and EMRGNSE LLC, but the business of these companies is unrelated to the contents of this manuscript. H.E.B. is a co-founder of EMRGNSE LLC, but the business of this company is unrelated to the contents of this manuscript. D.M. consults for Tempus, but the business of these companies is unrelated to the contents of this manuscript. J.H. consults for Tempus and is a co-founder of EMRGNSE LLC, but the business of these companies is unrelated to the contents of this manuscript. J.M.D. has no conflicts relevant to this work. However, he serves as a consultant and Chief Scientific Officer of Astrin Biosciences. The interest related to J.M.D. has been reviewed and managed by the University of Minnesota in accordance with its Conflict-of-Interest policies. D.A.L. is the co-founder and co-owner of NeoClone Biotechnologies, Inc., Discovery Genomics, Inc. (recently acquired by Immusoft, Inc.), B-MoGen Biotechnologies, Inc. (recently acquired by Bio-Techne Corporation), and Luminary Therapeutics, Inc. D.A.L. holds equity in, serves as a Senior Scientific Advisor for and Board of Director member for Recombinetics, a genome editing company. The business of all these companies is unrelated to the contents of this manuscript. D.A.L. consults for Styx Therapeutics, Inc. and Genentech, Inc., which is funding some of his research. E.S.A. reports grants and personal fees from Janssen, Sanofi, Bayer, Bristol Myers Squibb, Curium, Merck, Pfizer, AstraZeneca, Clovis, Constellation; personal fees from Astellas, Amgen, Blue Earth, Exact Sciences, Invitae, Eli Lilly, and Foundation Medicine; grants from Novartis, Celgene, and grants from Orion outside the submitted work; and has a patent for an AR-V7 biomarker technology that has been licensed to Qiagen.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Makovec, A., Boytim, E., Gustafson, A. et al. CREB5 promotes tumorigenicity and upregulates druggable cell surface modalities in basal-like breast cancer. npj Precis. Onc. 9, 276 (2025). https://doi.org/10.1038/s41698-025-01051-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41698-025-01051-z