Abstract
We introduce a framework to adapt large language models for medicine: (1) Modeling: breaking down medical workflows into manageable steps; (2) Optimization: optimizing model performance via advanced adaptations; and (3) System engineering: developing agent or chain systems. Furthermore, we describe varied use cases, such as clinical trial design, clinical decision support, and medical imaging analysis. Finally, we discuss challenges and considerations for building medical AI with LLMs.
Similar content being viewed by others
Introduction
Artificial intelligence (AI) is increasingly being integrated into various medical tasks, including clinical risk prediction1, medical image understanding2, and synthetic patient records generation3. Typically, these models are designed for specific tasks and will struggle with unfamiliar tasks or out-of-distribution data4. Large language models (LLMs) are foundation models characterized by their extensive training data and enormous model scale. Unlike traditional AI models, LLMs demonstrate an emergent capability in language understanding and the ability to tackle new tasks through in-context learning5. For example, we can teach LLMs to conduct a new task by providing the text explanation of the task (or “prompts”), the input and output protocols, and several examples. This adaptability has sparked interest in employing generalist LLMs to medical AI applications such as chatbots for outpatient reception6. Contrary to the common belief that generalist LLMs will excel in many fields7, we advocate that domain-specific AI adaptations for medicine are more effective and safe. This paper will overview how various adaptation strategies can be developed for medical AI applications and the associated trade-offs.
Generalist LLMs such as ChatGPT can support broad tasks but may underperform in specialized domains8. One notable drawback is the occurrence of “hallucinations,” which are fabricated facts that look plausible yet incorrect9. High-stakes medical applications such as patient-facing diagnosis tools are especially vulnerable to such inaccurate information10. In response, adaptation methods in LLM for medicine have thrived, focusing on enhancing LLMs’ domain-specific capabilities (Fig. 1b). They include finetuning LLMs on medical data11, adding relevant medical information to the prompts for LLMs via retrieval-augmented generation (RAG)12,13, and equipping LLMs with external tools to building AI agents achieving autonomous planning and task execution14,15. With the increasing practice in developing LLM-based AI applications, it is becoming evident that cutting-edge performance is increasingly driven by the mixture of these adaptations16 and systematic engineering of multiple AI components17.
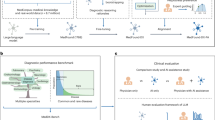
a Generalist AI models, such as proprietary systems (e.g., Open AI's GPT-4) and open-source models (e.g., LLaMA), serve as foundational technologies for developing specialized medical AI models. b Adapting generalist AI to medical tasks involves several techniques, including model finetuning, prompt optimization, and the development of AI agents or AI chains. These methods use diverse medical datasets, such as medical images, electronic health records (EHRs), clinical notes, publications, and omics data, to enhance AI model training and performance. c Effective system engineering for medical AI entails integrating AI modules into comprehensive chains to support tasks like cohort extraction, eligibility assessment, and result verification. This process emphasizes human interaction and AI, resulting in tailored AI modules for specific applications. d Generalist AI applications in medicine span various domains, including conversational diagnosis, radiology report generation, clinical note summarization, automated medical coding, drug design, patient-trial matching, and systematic literature reviews. All require advanced system integration for optimal performance.
In this Perspective, we present an overview of adaptation techniques for developing LLM-based medical AI and the workflow to solidify the development (Fig. 1). The adaptations can be concluded as: Model development focuses on designing model architectures and employing learning algorithms to adapt model parameters for medical tasks. Techniques include injecting medical knowledge into general-purpose models through continual pretraining on medical datasets18 and finetuning, which aligns the model’s outputs with domain-specific knowledge and human preferences19. Model optimization enhances model performance by optimizing its associated components, such as optimizing the input prompts16 and implementing retrievers accessing external data to enable RAG12. System engineering enhances medical AI performance by breaking down tasks into well-defined, narrow-scope components. LLMs can serve as the computational core for each specialized component, which can be linked together to support complex workflows20, or interact autonomously with other components to form agent-based systems21.
Next, we provide actionable guidelines on when and how to adopt these adaptation methods based on the specific task parameters, e.g., time and cost constraints. Additionally, we present concrete use cases that demonstrate the practical value of this framework. Finally, we discuss the associated challenges and opportunities for advancing LLM-based medical AI applications.
Adapting large language models for medical AI
Model development: building medical-specific LLMs
Generic LLMs such as ChatGPT benefit from vast parameters and are trained on vast, diverse datasets to develop a broad understanding of language5. A continual pretraining of generic LLMs on medical data, such as medical publications, clinical notes, and electronic health records (EHRs), can enhance LLMs’ alignment in medical languages. For example, MEDITRON is based on the open-source LLaMA model22, further trained on medical data, including PubMed articles and medical guidelines. It performs comparably to larger generalist models such as GPT-423 in medical question answering. Another model, PANACEA, shows the benefit of LLMs in clinical trial search, design, and patient recruitment via a continual pretraining of generic LLMs on clinical trial documents, including trial protocols and trial-related publications18. In addition, scaling model size using a mixture-of-experts strategy, where multiple LLMs (i.e., experts) are combined with a router network to select the appropriate expert during inference, has been found to yield superior performance at significantly lower computational cost compared to using a single, larger LLM24.
Instead of pretraining with medical data without specific task supervision, an LLM can also learn from paired custom queries and their expected responses to perform multiple target tasks. A widely used technique is instruction tuning25. For example, when PaLM, a 540-billion parameter LLM, underwent instruction tuning, it resulted in MedPaLM, which achieved 67.6% accuracy on US Medical Licensing Exam-style questions11. When the computational resources are limited, we can also update a subset of the model’s parameters or attach an additional small but learnable component to the model (e.g., prefix-tuning26 and LoRA27).
Another important finetuning objective is alignment, which ensures LLM outputs are consistent with human preferences, with high quality and safety. One prominent method for alignment is Reinforcement Learning from Human Feedback (RLHF), which was instrumental in developing ChatGPT28. RLHF starts with training a reward model based on human feedback, which then steers the LLM toward responses that align with human values and expectations using reinforcement learning. An example of a medical application of alignment is LLaMA-Clinic, where multiple clinicians provide feedback to guide the models for generating high-quality clinical notes19.
Model optimization: strategies for improving LLM performance
The prompt indicates the inputs to LLMs, which can include the task description, the expected input and output formats, and some input/output examples. For example, this input “Your task is to summarize the input clinical note. Please adhere to these summarization standards: [...list of criteria]. Refer to these examples: [...list of examples]. Keep in mind: [...list of important hints],” can guide LLMs towards generating a summary of patient notes. This practice leverages the principle of in-context learning, where LLMs adapt to new tasks based on the input prompts without additional training5. Research shows that the structure and content of the prompt significantly affect the model’s performance. For instance, chain-of-thought prompting29 encourages LLMs to engage in multiple steps of reasoning and self-reflection, thereby enhancing the output quality. Another strategy involves ensembling30,31, where outputs derived from multiple prompts are synthesized to produce a final, more robust response. In the medical domain, MedPrompt16 has demonstrated its ability to outperform domain-tuned LLMs by combining multiple prompting techniques. Additionally, TrialGPT32 effectively adapts LLMs to match patient notes to the eligibility criteria of a clinical trial through prompting.
Handcrafted prompts are highly dependent on domain knowledge and trial-and-error, but they can still perform suboptimally and cause reliability issues in applications. Techniques such as automatic prompt generation33 and optimization34 were proposed. These approaches can transform the adaptation of LLMs for medical tasks into a more structured machine-learning task. For example, requesting an LLM to summarize clinical notes could start with a simple prompt like “Summarize the input clinical note.” Using a collection of example notes and expert-written summaries, automatic summarization evaluation metrics can serve as the target for iterative prompt refinement. The prompt can ultimately evolve into a more advanced prompt consisting of a professional task description, representative examples, and a clear output format requirement description.
RAG is an extension of prompting. It dynamically incorporates information retrieved from external databases into the model’s inputs, supplementing LLM’s internal knowledge12. In medicine, it is extensively employed to improve the factual accuracy of LLM responses to medical questions by fetching the evidence from medical literature, clinical guidelines, or medical ontology13,35. In practice, the efficacy of RAG relies on two aspects: (1) the quality of the external database, which should be high-quality and up-to-date36,37, and (2) the performance of the retrieval systems, which are responsible for identifying and extracting the most relevant content to supplement the LLM’s output38,39.
System engineering: architecture development
Figure 2 illustrates the top-down system engineering process for adapting LLMs to medical AI applications. The process involves the following steps: (1) selecting the architecture to be developed (Fig. 2a); (2) designing the agent-based system (Fig. 2b) or the AI chain system (Fig. 2c); and (3) developing and finetuning LLMs to integrate with these systems (Fig. 2d).
a Selecting the overall architecture of the system should be based on the properties and requirements of the task at hand. In some scenarios, the best performance can be achieved with a fusion of the AI chain and AI agents. b A non-exhaustive list of example agent modules to be built in an agent-based system. LLMs act in different roles when equipped with different modules and interact with human experts to dynamically conduct the target task. c When building AI chain systems, we can first define the pipeline that decomposes the task into small steps following an expert workflow or professional guidelines, and then develop the module responsible for each step. d Adaptation techniques can be applied to enhance LLM's performance for the AI agent or for the AI module. Adaptation methods need to be selected according to data availability and task requirements.
The first step is to choose the system architecture: AI Chain40 and AI Agent41, depending on the nature of the tasks and requirements, as illustrated in Fig. 2a. AI Chain follows a structured, fixed workflow, making it ideal for repetitive tasks, professional practices, and scenarios requiring strict adherence to guidelines or compliance. Example applications include systematic literature reviews42 and patient-to-trial matching32. In contrast, AI Agents are more suitable for flexible workflows and exploratory tasks that tolerate uncertainty, cost, and time variations. This approach excels in dynamic and creative tasks such as data science code generation43 and brainstorming research hypotheses44. However, these two architectures are not mutually exclusive. A hybrid approach can be effective, where agents are embedded within structured workflows to handle steps requiring extensive reasoning or exploration. For instance, DeepResearch45 follows a systematic workflow of literature search, information processing, and summarization. However, within each step, a reasoning agent iteratively refines searches and processes information until sufficient material is gathered for summarization.
Developing AI agent systems boils down to designing and integrating key modules to facilitate the interactions between LLMs and human experts, as demonstrated in Fig. 2b. AI agents embody a vision where AI systems actively solve problems through autonomous planning, knowledge acquisition using diverse tools, and iterative self-reflection41. This concept is supported by various agent frameworks that facilitate these capabilities, such as AutoGPT and AutoGen46,47. Beyond external data sources, these agents can interact with various external tools, enhancing their operational scope. This includes interfaces for programming48, autonomous laboratory equipment21, and integration with other software programs49,50, broadening the potential applications of AI agents.
An AI agent typically consists of a base generalist AI, such as an LLM, along with one or more specialized modules that enable it to collaborate effectively with human experts. A non-exhaustive list of such modules includes Reasoning, which enhances decision-making through direct inference and iterative feedback from humans, external tools, or other agents; Perception, which enables the agent to process and interpret diverse data modalities such as text, images, and code; Memory, comprising internal memory for short-term context retention (e.g., within prompts) and external memory for long-term storage (e.g., in vector databases); and Interaction, which facilitates communication with external tools, coordination among multiple agents, and seamless human-AI collaboration. By integrating these modules, advanced multi-agent systems can efficiently decompose complex tasks into more manageable subtasks, such as brainstorming agents, expert consultation agents, research debate agents, self-driving lab agents, etc.51,52.
In medicine, AI agents are making remarkable progress in transforming healthcare delivery. For example, Polaris is an agent system designed to facilitate patient-facing, real-time healthcare conversations53. It integrates an LLM with automatic speech recognition for speech-to-text transcription and text-to-speech technology to convert LLM-generated text into audio, enabling seamless real-time interactions with patients. By leveraging agent-specific prompts and coordinating with specialized sub-AI models, such as those focused on medication management, laboratory and vital sign analysis, and clinical nutrition, Polaris functions as a multidisciplinary virtual specialist. Agents are also increasingly used in biomedical research, assisting in complex data analysis and hypothesis validation43,54. In addition, multi-agent systems, such as Virtual Lab55, embody the vision of automatic hypothesis generation, experimentation, and hypothesis validation in biomedical research.
For tasks that are usually fixed, repetitive, or strictly governed by professional guidelines, the AI chain is recommended to be developed (Fig. 2c). Rather than allowing LLMs to divide tasks and conduct them with several AI agents, experts can break down tasks into a chain of multiple steps. This approach allows multiple LLM calls to create an AI chain, which enhances both the transparency and controllability of the final output40. An illustrative example is WikiChat56, which addresses hallucinations by sequentially employing LLMs to query Wikipedia, summarize, filter, and extract the retrieved facts, perform additional fact-checking, and finally draft responses with a round of self-reflection for refinement. As a result, WikiChat surpassed GPT-4 by 55% in factual accuracy for question-answer tasks. In addition to optimizing performance, FrugalGPT57 optimizes query routing to reduce inference costs by using a cascade of LLMs with varying capabilities. This system utilizes weaker LLMs for initial processing and escalates to stronger LLMs only as necessary.
AI chain development, as shown in Fig. 2c, consists of three primary stages: pipeline development, module definition, and module development. The process begins with designing the pipeline based on expert workflows or professional guidelines, breaking it down into subtasks such as search, reasoning, and data extraction. Next, specialized modules are developed for each subtask. Each module is defined by its objectives (e.g., progress note generation) and constraints (e.g., response time and token usage) and is optimized using adjustable parameters such as the underlying models, prompts, and external tools or data sources. During module development, the focus is on the underlying model adaptations, considering the specifics of module inputs and outputs, including their format (e.g., text, image, code), length, and frequency (e.g., real-time or offline batch processing). For instance, the input length is an important factor for a module for summarizing multiple long documents, which requires an underlying model with a long context length optimized for improving fidelity to its inputs58. For even longer inputs, advanced adaptations such as map-reduce, i.e., segmenting inputs, summarizing each segment, and consolidating those summaries into a global summary59. This process also requires orchestrating data flows, integrating inputs from upstream modules, external components (e.g., knowledge bases, vector databases, or machine-learning models), and human requests. The outputs need to undergo automatic or human verification to ensure accuracy and reliability.
In medicine, AI chains have been developed to facilitate cohort extraction from EHR databases using natural language query60. As generating correct SQL queries aligned with complex EHR database schema is challenging, the authors break down the task into three steps: concept extraction, SQL query generation, and reasoning, where the prompt for each step is designed and optimized separately. Chaining AI calls was also proved effective in parsing eligibility criteria into a structured format61. This method makes the first LLM call to extract the basic target entity information, then routes to specialized extraction modules based on the entity type, where each prompt can be optimized, encoding expert knowledge. A post-processing module is attached to verify the outputs and aggregate the extraction results into a unified format.
System engineering: adaptations
In system engineering, the next step after deciding on the architecture development is the adaptation of LLMs, which includes two stages: model selection and model development and optimization, illustrated in Fig. 2d. In the model selection stage, the choice between proprietary and open-source models is guided by specific application requirements. Proprietary models, such as ChatGPT, are accessed via cloud services, enabling lightweight development and deployment without the need for local GPU resources but often at a higher cost, making them suitable for well-funded organizations. In contrast, open-source models such as finetuned LLaMa models offer greater flexibility for customization and are more cost-effective for high-volume tasks. They are particularly advantageous for local deployment scenarios due to privacy or security compliance requirements.
The second stage emphasizes optimizing the selected model based on data availability and task requirements. Prompt optimization is the recommended initial approach for cases with limited or no labeled data, particularly when working with proprietary models62. However, if sufficient domain-specific data is available, continual pretraining can be applied to inject domain knowledge into generalist LLMs, enhancing their adaptability to specialized tasks. Nonetheless, sometimes the gain may be marginal if the domain-specific data are publicly available because training data for generic LLMs may already include the domain-specific data63. When prompt optimization fails to meet performance expectations, we recommend collecting more task demonstrations (i.e., input-output pairs) and finetuning the generalist models64. Finetuning offers several advantages: (1) it enables the model to handle tasks that are rarely encountered in the public corpus, and (2) it reduces the reliance on lengthy and complex prompts during inference, leading to faster processing times and lower costs65.
Certain specialized tasks may require structured inputs and outputs. For instance, information extraction tasks or communication with physical devices require adherence to a specific input protocol and structured outputs, such as JSON objects, to ensure compatibility with machine parsing and processing21. Proprietary models often support features like JSON mode or function call capabilities66, enabling such tasks to be handled efficiently through prompt optimization alone. Alternatively, open-source models can be finetuned on structured input-output pairs to achieve similar capabilities and can be further enhanced by employing constraint decoding frameworks67. Additionally, for tasks requiring up-to-date knowledge, such as summarizing findings from the latest clinical trials, RAG is essential. It dynamically incorporates external knowledge into the LLM inference process, ensuring that outputs remain current and relevant. For instance, GeneGPT68 generates search queries to biomedical databases from the National Center for Biotechnology Information69 to enhance the factual accuracy of biomedical question answering, achieving an accuracy of 0.83 compared to ChatGPT’s 0.12.
Evaluation is critical when adopting LLMs for medical applications, with the choice of evaluation metrics playing a central role. In many cases, general-domain metrics may not suffice. For example, in clinical note summarization tasks, general natural language processing metrics such as ROUGE70 or BERTScore71 may fail to adequately capture the quality of summaries in the medical domain. Instead, metrics tailored to the professional standards of medicine, such as completeness, correctness, and conciseness, should be prioritized72. Designing these domain-specific metrics is often a collaborative effort between AI researchers and medical practitioners, and it is advisable to consult relevant medical guidelines or regulatory policies to ensure the chosen metrics are appropriate and comprehensive. Furthermore, real-world user studies serve as the gold standard for evaluation. Such studies typically involve two arms, one leveraging AI-assisted workflows and the other relying on manual efforts, to assess the practical value and effectiveness of medical AI systems6.
Use cases of LLMs for medical AI applications
Table 1 lists several studies that have explored adapting LLMs to medical AI tasks. In this section, we review a few example use cases, reinterpreting them through the lens of the proposed framework. Additionally, we discuss some potential improvements of those use cases.
Clinical note generation
Creating clinical notes is a routine task for physicians. These notes record the patient’s medical history, present conditions, and treatment plans. Recently, ambient documentation was introduced to record and transcribe provider-patient conversations and then ask LLMs to summarize them into a clinical note73,74. This method is gaining popularity with the recent adoption of automated scribe software75. However, the vanilla way of using LLMs is flawed in (1) Heterogeneity of note formats across specialties: The vanilla approach may not effectively adapt the note generation process to meet varying format preferences specific to different specialties (e.g., internal medicine versus general surgery), potentially leading to suboptimal documentation19. (2) Lack of quality control: Ensuring the accuracy and completeness of generated notes is challenging due to the absence of accuracy measures and hallucination control for reference-free note generation. (3) Limited electronic medical record (EMR) integration: LLMs primarily accept text inputs of limited length. They may not fully integrate with EMRs, often containing extensive and multimodal historical data. (4) Inadequate actionable insights: Unlike summarizing the inputs, LLMs usually perform unreliably in suggesting specific diagnostic tests or treatment actions following the generated summaries.
Next, we provide a deeper technology analysis for developing such a specialized medical AI system fusing AI chain and AI agents for clinical note generation (Fig. 3a). The primary objective is to produce accurate and concise notes that adhere to specific formatting requirements. This involves referencing “best practice” note formats across different medical specialties, clearly outlining the objectives and constraints for the output notes. Providing example inputs and outputs alongside LLM instructions can enhance clarity and improve the accuracy of the generated notes. To achieve this, integrating a knowledge base into the pipeline is crucial. This integration helps identify relevant sections of clinical guidelines and dynamically retrieves closely related examples to improve the instructions. Such adaptability ensures the application remains versatile across diverse clinical scenarios. For instance, LLMs can surface the latest clinical guidelines using an external knowledge base. If the notes describe symptoms of hypertension, LLMs can identify these symptoms, link them to relevant guidelines, and suggest appropriate diagnostic tests and treatments.
a AI chain crafted for clinical note generation, highlighting the expert involvement in selecting relevant patient data and adherence to external formatting guidelines. Agents can be integrated into the data retrieval and data selection steps to work with humans on iteratively retrieving and selecting the relevant pieces from medical records. Agents can also iterate the clinical notes by referencing formatting guidelines. b Automate medical coding can potentially benefit from an AI chain that employs two extraction modules designed for conditions and complications, respectively. Agents can be integrated so as to reference coding rules while filtering the input clinical notes. c A patient-trial matching pipeline adds a prescreening stage to reduce the candidate trial set. It also provides criterion-level assessment for users to select patients referring to various dimensions. Agents can dynamically retrieve medical knowledge when assessing patient eligibility. d Medical systematic review pipeline is built based on the established systematic review practice, e.g., PRISMA statement. An agent can optimize the generated search query by checking the identified studies before passing them to the screening stage.
Human oversight plays an essential role in validating the correctness of the generated notes. LLMs can be instructed to link each sentence in their summaries to specific indexed parts of the input, making it easier to trace the source. Incorporating an LLM self-reflection layer to compare the source and the generated text further streamlines this validation process. This feature provides clinicians with suggestions for addressing omissions or inaccuracies in the summaries.
Agents can be integrated into the workflow to improve performance. For instance, equipping LLMs with programming tools, such as an SQL interface to the underlying EMR database, can significantly enhance note generation. This capability enables LLMs to dynamically access and review previous notes, extracting only the necessary information to support the process. Additionally, it facilitates the development of provider copilots that assist clinicians in retrieving patient information using natural language queries. These copilots empower providers to focus more on patient care rather than navigating complex systems. AI agents can further reduce administrative workloads by assisting providers with specific tasks. For example, follow-up emails play a crucial role in reinforcing care plans, clarifying medical instructions, and providing patient education. LLM agents can create follow-up emails summarizing the visit, test results, and next steps. This ensures patients understand their health information, adhere to treatment plans, and feel supported throughout their care journey.
To structure these capabilities effectively, a pipeline can be developed to facilitate collaboration between physicians and AI. Upon receiving patient data, the application will initially present the identified formatting guidelines (from the RAG module) and relevant segments of historical EMR data (from the EMR integration module) for user verification and selection. Refined instructions will then prompt LLMs to generate the notes, followed by validation and action suggestion modules. A user-friendly interface will guide users through this process, ensuring an efficient and intuitive workflow.
Automated medical coding
Medical billing codes, such as ICD-10, CPT, and Diagnosis-Related Group (DRG) codes, are assigned by coding specialists through a time-consuming and manual process for healthcare payment systems. We can ask LLMs to translate EMR data into the most likely billing code76. Finetuning LLMs on pairs of EMR data and billing codes can further enhance the performance77. However, these methods are not yet satisfactory for production use because of (1) Inability to learn coding rules: Clinical coding goes beyond mere entity extraction. LLMs must be familiar with the potential list of billable codes relevant to specific input data. Moreover, the subtleties in the criteria for each billing code are crucial to avoid over-billing or under-billing. Compounding the complexity, each billing code system possesses its own set of nuanced rules and nomenclatures78,79,80. (2) Difficulty handling excessive EMR inputs: In inpatient settings, when a patient has a complex, multi-week hospital stay, it becomes problematic for an LLM to identify the most relevant sections of extensive medical record data without a sophisticated filtering strategy. (3) Lack of explainability: LLM-generated billing codes do not gain the trust of specialists without the rationale provided. We can approach these challenges considering (Fig. 3b):
The primary objective is to ensure precise coding from designated billable code sets, with a secondary objective of providing a rationale for the assigned codes. Achieving both objectives requires leveraging external knowledge of medical coding guidelines to ensure accuracy and compliance. Medical coding guidelines are essential for translating clinical notes into accurate billing codes. To meet these standards, coding rules must be integrated into LLM instructions. This can be accomplished through an agent augmented by RAG, which allows the agent to reference relevant rules throughout the coding process. By embedding coding rules directly into the workflow, this approach enhances the accuracy and relevance of the generated codes while enabling traceability back to the source guidelines.
When dealing with prolonged hospital stays, it is crucial to identify which clinical notes should be processed for billing code assignment. To optimize prediction performance, priority should be given to the most clinically relevant notes, such as admission notes and discharge summaries77. Additionally, implementing a filtering strategy to remove duplicate content, common in clinical documentation due to copy-pasting practices, improves efficiency and reduces costs by eliminating redundant information.
Building an advanced coding-specific extraction pipeline requires a deep understanding of billing regulations. For example, the DRG coding system for hospital stays involves two critical decision points: identifying the primary medical issue as the principal diagnosis and detecting any complications or comorbidities80. To address this, two tailored extraction modules can be developed: one dedicated to identifying the principal medical condition and another to detecting associated complications or comorbidities. The outputs of these modules are then combined to provide comprehensive coding results.
Patient-trial matching
Matching patients with appropriate clinical trials is crucial for ensuring sufficient trial recruitment and significantly impacting trial outcomes. The manual recruitment process, which involves screening surveys, is labor-intensive, time-consuming, and prone to errors. Efforts have been made to optimize prompts for powerful LLMs like GPT-4 to assess patient eligibility by matching patient records and trial eligibility criteria81. To mitigate concerns about Protected Health Information (PHI) leakage when using proprietary models, another approach has been proposed: leveraging the knowledge of these proprietary models to generate synthetic patient-trial pairs, which are then used to fine-tune smaller open-source LLMs for secure local deployment82. However, challenges remain, including (1) scalability: Existing methods typically assume a limited set of candidate trials or patients. However, with over 23,000 active trials on ClinicalTrials.gov and patient databases containing millions of records, screening on a patient-by-patient or trial-by-trial basis using LLMs becomes intractable. (2) Lack of medical expertise: Generalist LLMs may still struggle with medical reasoning when assessing eligibility. For example, a patient record noting a “prescription history of blood thinners due to recurrent chest pain” was incorrectly labeled as “not enough information” by GPT-4 in response to the criterion “patient must have a history of cardiovascular disease.”32. A system engineering can be made to approach this task (Fig. 3c).
The primary objective is to achieve accurate eligibility predictions, with secondary objectives aimed at optimizing speed and cost, particularly when managing large datasets. Enhancing the medical capabilities of LLMs can be achieved through finetuning on high-quality patient-trial pairs82. This process requires a streamlined method for annotating patient eligibility against trial criteria or access to historical records of participants in previous trials. A collaborative reasoning of AI agents and clinical guidelines, and medical ontologies can further enhance eligibility assessments83. Specifically, correlations between medical terms, including therapies and conditions, can be drawn from clinical guidelines, while medical ontologies provide hierarchical mappings from umbrella terms to specific subsets. These resources can be integrated into the LLM workflow to improve reasoning and ensure more accurate predictions.
Scalability challenges in eligibility prediction can be addressed through the addition of a prescreening module. For example, TrialGPT32 introduces a trial retrieval step by generating search queries based on patient summaries, reducing the candidate set by over 90% without compromising the recall of target trials. Similarly, prescreening can involve LLM-driven information extraction from both patient records and eligibility criteria, followed by entity matching to streamline the process84.
Building on these methodologies, a structured pipeline can be developed to optimize eligibility predictions. This pipeline may include the following steps: (1) finetuning underlying LLMs using patient-trial data; (2) initiating a prescreening process to narrow down the pool of clinical trials relevant to the target patient; (3) extracting key terms from patient records and retrieving supplementary knowledge from clinical guidelines and medical ontologies; and (4) performing the final eligibility assessment using LLM calls. This comprehensive approach ensures accuracy, scalability, and cost-effectiveness in processing large datasets.
Medical systematic review
Clinical evidence can be synthesized through the review of medical literature, but this process is increasingly challenged by the rapid growth of published research. LLMs have been employed to synthesize evidence from multiple articles using text summarization techniques58,85. However, this text summarization approach raises significant concerns about the quality of the outputs, including issues like a lack of specificity, fabricated references, and potential for misleading information86. Moreover, conducting a systematic review involves more than just summarization; it encompasses multiple steps, as outlined in PRISMA87. AI should be integrated into the established systematic review workflow with careful consideration of user experience optimization, such as offering verification, referencing, and human-in-the-loop42,88. From a systems engineering perspective, the following elements should be incorporated (Fig. 3d).
The primary objective is to generate trustworthy clinical evidence from medical literature, with sub-objectives focused on ensuring the comprehensiveness of collected studies and the accuracy of extracted results. To meet these objectives, strict formatting constraints should be established for outputs at each step. For example, synthesized evidence must be directly supported by the results of individual studies and should ideally be presented as a meta-analysis.
Generating effective search queries requires expertise in both medicine and library sciencess89. The search query building process can be performed by an AI agent in an iterative search and refinement process. Once citations are identified, a refined screening process is necessary to ensure relevance to the research question. This involves evaluating the population, intervention, comparison, and outcome elements. Additionally, a risk-of-bias assessment should be conducted to filter out low-quality citations. Developing specialized prompts incorporating itemized scoring guidelines can further improve the accuracy and consistency of this screening process.
A robust data extraction process is critical for handling the original study content. A PDF extraction module, leveraging Optical Character Recognition techniques, can extract essential details, including tables and figures. To maintain transparency and enable verification, LLM-generated outputs should include references to the original content, allowing users to identify and correct any potential misinformation. Synthesizing evidence from clinical studies requires accurate extraction of key numerical results, such as evaluated endpoints, sample sizes, and treatment effects. This task is particularly challenging but can be supported by equipping LLMs with an external program interface that facilitates numerical reasoning during evidence extraction and synthesis42.
Mapping data privacy legislations
The challenge of aligning diverse and evolving privacy, data protection, and AI legislation with standardized frameworks such as the National Institute of Standards and Technology (NIST) has become increasingly complex, especially when dealing with multiple jurisdictions90,91. For instance, in a recent U.S.-based project for a health application, an LLM pipeline is implemented by IQVIA to analyze privacy legislation across several states, identifying over 3000 overlaps between legislative requirements and NIST privacy actions92. This level of analysis, achieved in a short timeframe, would have been near impossible to accomplish manually at the necessary scale and depth. The same approach is being used to identify controls and governance for a data platform in the Middle East.
The LLM pipeline we devised automates identifying and mapping privacy, cybersecurity, and AI risk management actions, offering a scalable solution that can navigate a complex and rapidly changing regulatory landscape. The pipeline enables more efficient compliance management by processing extensive legal texts and implementation guidelines and aligning them with structured frameworks. Integrating a human-in-the-loop, expert verification and tuning process, while leveraging established NIST crosswalks to validate accuracy, provides a robust method for tackling large-scale data and AI compliance challenges93. The primary objective is to automate the mapping of regional and global legislation to the NIST Frameworks, providing a scalable and efficient approach to compliance management. Secondary objectives include ensuring the comprehensiveness of the legislative documents analyzed, the accuracy of identified compliance actions, and the risk-based prioritization of implementation activities.
The process begins with data collection and preprocessing, where relevant legislative and regulatory texts, including regional and sectoral guidelines, are gathered. This data is ingested into the LLM pipeline, which is optimized to process large volumes of documents efficiently. Preprocessing steps include document parsing, natural language normalization, and segmentation to ensure the model focuses on actionable items relevant to compliance management. After generating initial matches against the frameworks, a risk-based thresholding system is applied. Each compliance action identified by the LLM is scored based on its relevance and alignment with NIST compliance actions. The scoring incorporates a risk-based assessment, factoring in the overlap with sourced documents and weighting the importance of the business context, such as usability, implementation guardrails, and effort.
To enhance accuracy and relevance, the process integrates human-in-the-loop expert guidance. Experts validate the LLM’s outputs, resolving ambiguities and accounting for jurisdiction- and sector-specific variations that the model may misinterpret. They refine the mappings and scoring while interpreting and summarizing the results to align with client needs. This approach ensures evidence-based decision-making and tailored recommendations. The pipeline incorporates benchmarking and confidence validation to further ensure accuracy. Pre-established NIST crosswalks, developed by legal and other experts, serve as benchmarks for comparison, enabling the system to validate its outputs against known mappings. Advanced confidence elicitation strategies are used to address LLM overconfidence in ambiguous or novel situations.
Finally, the LLM pipeline synthesizes its outputs into a comprehensive report. This report includes detailed references to the relevant legislative texts and framework categories, ensuring traceability for every mapping decision. The synthesis process enables users to verify the source of each decision and provides evidence of how features are prioritized, ensuring a transparent and reliable compliance management solution.
Opportunities and challenges in LLMs for medical AI
Here, we describe the challenges that need to be addressed to embody the benefits of adapting LLMs to medical AI, with a focus on three critical areas: enhancing multimodal capabilities, ensuring trustworthiness and compliance, and managing system lifecycle through evaluation and continuous optimization.
Multimodality
Multimodal capabilities represent a key growth direction for LLMs in medical applications. A patient journey naturally comprises many information-rich modalities such as lab tests, imaging, genomics, etc. While generalist AI has demonstrated amazing capabilities in understanding and reasoning with biomedical text (e.g., MedPrompt94), competence gaps abound in the multimodal space, with vision-language models being a prominent example. E.g., GPT-4V performs poorly on identifying key findings from chest X-rays, even compared to much smaller domain-specific models (e.g., LLaVA-Rad95). Efficiently bridging such multimodal competency gaps thus represents a key growth frontier for medical AI. Progress is particularly fast in biomedical imaging, from harnessing public image-text data (e.g., BiomedCLIP96, PLIP97, CONCH98) to efficiently training vision-language models (e.g., LLaVA-Med99) to learn text-guided image generation and segmentation (e.g., BiomedJourney100, RoentGen101, BiomedParse). While promising, challenges abound in multimodal medical AI, from pretraining in challenging modalities (e.g., GigaPath102) to multimodal reasoning for precision health.
Trustworthiness and compliance
While adapting powerful AI models like LLMs to medicine holds great promise, significant challenges persist in building trustworthiness and ensuring compliance with their use. Here, we identified several challenges: (1) hallucinations, (2) privacy risks, (3) explainability, and (4) regulations.
The risk of “hallucinations” in LLM outputs, where the model generates plausible but incorrect or fabricated information, is widely mentioned103. In the medical domain, such errors can have severe consequences, including misdiagnoses, inappropriate treatments, and flawed research conclusions104. To address hallucinations, RAG is often employed to guide LLM outputs and ground them in verifiable citations51. However, failure modes persist, such as LLMs citing incorrect sources or hallucinating citations altogether105. Future efforts should focus on finetuning models with high-quality, domain-specific datasets, implementing rigorous validation mechanisms, and incorporating human-in-the-loop workflows106, where medical professionals review and correct AI-generated content to ensure accuracy and reliability.
The development and deployment of medical AI applications involve multiple stakeholders, including medical data providers (such as health systems, pharmaceutical companies, and real-world data companies), model providers (like healthcare AI startups), and users (comprising clinicians, patients, and these data-providing companies). AI developers play a crucial role in constructing pipelines that process data from these data providers into actionable knowledge, which is then made accessible to users by leveraging intelligence from model providers. It has been noted that LLMs are vulnerable to attacks using malicious prompts that aim to extract their training data107. This risk necessitates the de-identification of the PHI information from training data. When real data can now be acquired, synthetic training data can be incorporated108,109, which can be further protected with differential privacy110. Moreover, when AI developers handle data from multiple providers and present it to various users, it is essential to implement access controls within the AI applications. These controls ensure that users can only access specific datasets relevant to their queries. In addition, protecting user inputs is crucial, especially if they are utilized to optimize the pipeline and stored in prompts or databases for RAG. LLMs must prevent inadvertently disclosing one user’s data to another, maintaining strict confidentiality in user interactions.
LLM’s interpretability remains a critical challenge. It appears to show “emergent” intelligence, but that property may also disappear when we take different evaluation metrics111. As deep learning models, LLMs often function as black boxes, making it difficult to trace the reasoning behind specific outputs. This lack of transparency can hinder clinical decision-making and raise concerns about accountability. To address these issues, developing explainable AI methodologies is essential for gaining a deeper understanding of LLMs112. At present, techniques such as chain-of-thought and program-of-thought can be employed to reveal the step-by-step reasoning and operational processes behind LLM outputs, improving their interpretability in medical applications29,113. Researchers have also tried to dive into the inner workings of AI systems by introducing mechanistic interpretability, which is promising to provide a more interpretable and controlled LLM behavior114.
The development, deployment, and production of LLMs in medical practice must adhere to strict regulatory standards and compliance requirements to ensure patient safety and data privacy115. Regulatory bodies such as the FDA, EMA, and others have established guidelines for medical devices, which increasingly include AI-driven tools116. Developers of medical AI applications must navigate these regulations, establish robust practices, and process procedures to ensure that their models are validated, transparent, and compliant with relevant laws, such as HIPAA in the U.S. Additionally, as AI applications are continuously updated and retrained, maintaining compliance over time requires ongoing monitoring, documentation, and potentially re-certification.
Evaluation and continuous improvement
A medical AI system may comprise various modules and pipelines, each of which has the potential to malfunction, posing challenges in output assessment and debugging. Human oversight can be integrated to enhance validation, allowing users to confirm the accuracy of outputs through the provided references117. Furthermore, validation processes can be automated by leveraging LLM capabilities. For example, assertions can be embedded within the pipeline, enabling LLMs to self-correct their outputs during inference118. The increasing complexity of LLM-based systems, consisting of multiple interconnected components, can overwhelm any individual’s capacity to manage the entire architecture. LangSmith119 is a DevOps tool for AI applications that aims to support deploying and monitoring LLM-powered applications. It allows developers to visualize traces and debug problematic components. Additionally, it facilitates the collection and extraction of erroneous inputs and outputs, which can then be utilized to build and augment validation datasets to enhance the applications.
Maintaining medical AI applications poses significant challenges in ensuring system stability and reliability. One major concern is the variability of the underlying LLMs, as version updates can alter model behavior and capabilities, potentially disrupting the functionality of integrated systems120. Additionally, the interdependence of system components means that changes to one element can impact the overall performance, necessitating rigorous testing and calibration whenever modifications are made. Furthermore, managing distribution shifts, particularly in edge cases, remains a critical issue. An effective strategy to address these challenges is the construction of robust development datasets for evaluating and finetuning AI pipelines. These datasets can be sourced from real-world tasks or simulated scenarios generated by LLMs121. However, current approaches often rely on heuristic methods, underscoring the need for further research to enhance dataset creation. Another significant challenge lies in selecting appropriate evaluation metrics, as many standards are embedded in specialized medical guidelines or regulatory documents. Bridging the knowledge gap between AI scientists and medical practitioners through increased collaboration and interdisciplinary research is essential for establishing reliable evaluation frameworks.
User feedback is a crucial source of supervision for the continual optimization of AI pipelines. This feedback can take various forms, ranging from explicit expressions of preference, such as likes or dislikes, to more subtle indicators found in user interaction logs or direct textual comments on specific aspects of the system. Such feedback is valuable not only for refining the pipeline’s final outputs but also for improving any intermediate stages where user engagement occurs, whether through active participation or the generation of usage logs. Techniques like RLHF28 have been used to enhance LLM models based on ranked human preferences. Recently, a framework called TextGrad122 has shown promising results by enabling the backpropagation of textual feedback within LLM pipelines to optimize prompts across different components.
Discussion
Generalist AI models have the potential to revolutionize the medical field. A range of adaptation methods has been proposed to tailor these models for specialized applications. In this Perspective, we documented existing adaptation strategies and organized them within a framework designed to optimize the performance of medical AI applications from a systems engineering perspective. Our analysis and discussion of published use cases demonstrate the benefits of this framework as a systematic approach to developing and optimizing LLM-based medical AI. However, we also recognize the potential challenges that arise as the complexity of medical AI applications increases, particularly in monitoring, validation, and maintenance. Future research and development are essential to solidify the utility of LLM-driven medical AI applications, enhance patient outcomes, democratize access to quality healthcare, and reduce the workload on medical professionals.
Data availability
No datasets were generated or analyzed during the current study.
References
Choi, E. et al. Retain: an interpretable predictive model for healthcare using reverse time attention mechanism. In Proc. Advances in Neural Information Processing Systems. Vol. 29 (Curran Associates, Inc., 2016).
Wang, Z., Wu, Z., Agarwal, D. & Sun, J. MedCLIP: Contrastive learning from unpaired medical images and text. In Proc. 2022 Conference on Empirical Methods in Natural Language Processing. 3876–3887 (Association for Computational Linguistics, 2022).
Wang, Z. & Sun, J. PromptEHR: conditional electronic healthcare records generation with prompt learning. In Proc. Conference on Empirical Methods in Natural Language Processing. 2873–2885. (Association for Computational Linguistics, 2022).
Moor, M. et al. Foundation models for generalist medical artificial intelligence. Nature 616, 259–265 (2023).
Brown, T. et al. Language models are few-shot learners. In Proc. Advances in Neural Information Processing Systems. Vol. 33, 1877–1901 (Curran Associates, Inc., 2020).
Wan, P. et al. Outpatient reception via collaboration between nurses and a large language model: a randomized controlled trial. Nat. Med. 30, 2878–2885 (2024).
Yang, J. et al. Harnessing the power of LLMs in practice: a survey on ChatGPT and beyond. ACM Trans. Knowl. Discov. Data 18, 1–32 (2024).
Tian, S. et al. Opportunities and challenges for ChatGPT and large language models in biomedicine and health. Brief. Bioinform. 25, bbad493 (2024).
Au Yeung, J. et al. AI chatbots not yet ready for clinical use. Front. Digit. Health 5, 1161098 (2023).
Sarraju, A. et al. Appropriateness of cardiovascular disease prevention recommendations obtained from a popular online chat-based artificial intelligence model. JAMA 329, 842–844 (2023).
Singhal, K. et al. Large language models encode clinical knowledge. Nature 620, 172–180 (2023).
Lewis, P. et al. Retrieval-augmented generation for knowledge-intensive NLP tasks. In Proc. Advances in Neural Information Processing Systems. Vol. 33, 9459–9474 (Curran Associates, Inc., 2020).
Xiong, G., Jin, Q., Lu, Z. & Zhang, A. Benchmarking retrieval-augmented generation for medicine. In Proc. Findings of the Association for Computational Linguistics: ACL 2024 (eds Ku, L.-W., Martins, A. & Srikumar, V.) 6233–6251. https://aclanthology.org/2024.findings-acl.372 (Association for Computational Linguistics, Bangkok, Thailand, 2024).
Nakano, R. et al. WebGPT: Browser-assisted question-answering with human feedback. Preprint at arXiv https://arxiv.org/abs/2112.09332 (2021).
Jin, Q. et al. AgentMD: empowering language agents for risk prediction with large-scale clinical tool learning. Preprint at arXiv https://arxiv.org/abs/2402.13225 (2024).
Nori, H. et al. Can generalist foundation models outcompete special-purpose tuning? case study in medicine. Medicine 84, 77–3 (2023).
Zaharia, M. et al. The shift from models to compound AI systems. https://bair.berkeley.edu/blog/2024/02/18/compound-ai-systems/ (2024).
Lin, J., Xu, H., Wang, Z., Wang, S. & Sun, J. Panacea: a foundation model for clinical trial search, summarization, design, and recruitment. Preprint at https://arxiv.org/abs/2407.11007 (2024).
Wang, H. et al. Towards adapting open-source large language models for expert-level clinical note generation. Preprint at arXiv https://arxiv.org/abs/2405.00715 (2024).
Khattab, O. et al. DSPy: compiling declarative language model calls into self-improving pipelines. In Proc. R0-FoMo: Robustness of Few-shot and Zero-shot Learning in Large Foundation Models (2023).
Boiko, D. A., MacKnight, R., Kline, B. & Gomes, G. Autonomous chemical research with large language models. Nature 624, 570–578 (2023).
Touvron, H. et al. Llama 2: Open foundation and fine-tuned chat models. Preprint at arXiv https://arxiv.org/abs/2307.09288 (2023).
Chen, Z. et al. MEDITRON-70b: scaling medical pretraining for large language models. Preprint at arXiv https://arxiv.org/abs/2311.16079 (2023).
Jiang, A. Q. et al. Mixtral of experts. Preprint at arXiv https://arxiv.org/abs/2401.04088 (2024).
Wei, J. et al. Finetuned language models are zero-shot learners. In Proc. International Conference on Learning Representations. OpenReview.net (2021).
Li, X. L. & Liang, P. Prefix-Tuning: optimizing continuous prompts for generation. In Proc. 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing. Vol. Long Papers, 4582–4597 (Association for Computational Linguistics (ACL), 2021).
Hu, E. J. et al. LoRA: low-rank adaptation of large language models. In Proc. International Conference on Learning Representations. OpenReview.net (2023).
Ouyang, L. et al. Training language models to follow instructions with human feedback. In Proc. Advances in Neural Information Processing Systems. Vol. 35, 27730–27744 (Curran Associates, Inc., 2022).
Wei, J. et al. Chain-of-thought prompting elicits reasoning in large language models. In Proc. Advances in Neural Information Processing Systems. Vol. 35, 24824–24837 (Curran Associates, Inc., 2022).
Wang, X. et al. Self-consistency improves chain of thought reasoning in language models. In Proc. Eleventh International Conference on Learning Representations. OpenReview.net (2022).
Chen, L. et al. Are more LLM calls all you need? towards scaling laws of compound inference systems. Preprint at arXiv https://arxiv.org/abs/2403.02419 (2024).
Jin, Q. et al. Matching patients to clinical trials with large language models. Nat. Commun. 15, 9074 (2024).
Shin, T., Razeghi, Y., Logan IV, R. L., Wallace, E. & Singh, S. AutoPrompt: eliciting knowledge from language models with automatically generated prompts. In Proc. 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP). 4222–4235 (Association for Computational Linguistics (ACL), 2020).
Cheng, J. et al. Black-box prompt optimization: aligning large language models without model training. Preprint at arXiv https://arxiv.org/abs/2311.04155 (2023).
Wen, Y., Wang, Z. & Sun, J. MindMap: knowledge graph prompting sparks graph of thoughts in large language models. Preprint at arXiv https://arxiv.org/abs/2308.09729 (2023).
Zakka, C. et al. Almanac-retrieval-augmented language models for clinical medicine. NEJM AI 1, AIoa2300068 (2024).
Arasteh, S. T. et al. RadioRAG: factual large language models for enhanced diagnostics in radiology using dynamic retrieval augmented generation. Preprint at arXiv https://arxiv.org/abs/2407.15621 (2024).
Wang, Z. & Sun, J. Trial2vec: Zero-shot clinical trial document similarity search using self-supervision. In Proc. Findings of the Association for Computational Linguistics: EMNLP 2022 6377–6390 (2022).
Jin, Q. et al. MedCPT: contrastive pre-trained transformers with large-scale PubMed search logs for zero-shot biomedical information retrieval. Bioinformatics 39, btad651 (2023).
Wu, T., Terry, M. & Cai, C. J. AI chains: Transparent and controllable human-ai interaction by chaining large language model prompts. In Proc. 2022 CHI Conference on Human Factors in Computing Systems 1–22 (2022).
Yao, S. et al. ReAct: synergizing reasoning and acting in language models. In Proc. Eleventh International Conference on Learning Representations. OpenReview.net (2022).
Wang, Z. et al. Accelerating clinical evidence synthesis with large language models. https://arxiv.org/abs/2406.17755 (2024).
Wang, Z., Danek, B., Yang, Z., Chen, Z. & Sun, J. Can large language models replace data scientists in clinical research? Preprint at arXiv https://arxiv.org/abs/2410.21591 (2024).
Asai, A. et al. OpenScholar: synthesizing scientific literature with retrieval-augmented language models. Preprint at Arxiv https://arxiv.org/abs/2411.14199 (2024).
Introducing deep research. https://openai.com/index/introducing-deep-research/ (2025).
Gravitas, S. Autogpt. https://agpt.co (2023).
Wu, Q. et al. AutoGen: enabling next-gen LLM applications via multi-agent conversation framework. Preprint at arXiv https://arxiv.org/abs/2308.08155 (2023).
Tayebi Arasteh, S. et al. Large language models streamline automated machine learning for clinical studies. Nat. Commun. 15, 1603 (2024).
Trinh, T. H., Wu, Y., Le, Q. V., He, H. & Luong, T. Solving olympiad geometry without human demonstrations. Nature 625, 476–482 (2024).
Gu, Y. et al. Middleware for LLMs: tools are instrumental for language agents in complex environments. Preprint at arXiv https://arxiv.org/abs/2402.14672 (2024).
Gao, T., Yen, H., Yu, J. & Chen, D. Enabling large language models to generate text with citations. In Proc. 2023 Conference on Empirical Methods in Natural Language Processing 6465–6488 (2023).
Kim, Y. et al. MDAgents: an adaptive collaboration of LLMs for medical decision-making. In Proc. Advances in Neural Information Processing Systems. Vol. 37, 79410–79452 (2025).
Mukherjee, S. et al. Polaris: a safety-focused llm constellation architecture for healthcare. Preprint at arXiv https://arxiv.org/abs/2403.13313 (2024).
Huang, K. et al. Automated hypothesis validation with agentic sequential falsifications. Preprint at arXiv https://arxiv.org/abs/2502.09858 (2025).
Swanson, K., Wu, W., Bulaong, N. L., Pak, J. E. & Zou, J. The virtual lab: AI agents design new SARS-CoV-2 nanobodies with experimental validation. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/2024.11.11.623004v1 (2024).
Semnani, S., Yao, V., Zhang, H. C. & Lam, M. WikiChat: Stopping the hallucination of large language model chatbots by few-shot grounding on Wikipedia. In Proc. 2023 Conference on Empirical Methods in Natural Language Processing (2023).
Chen, L., Zaharia, M. & Zou, J. FrugalGPT: How to use large language models while reducing cost and improving performance. Preprint at arXiv https://arxiv.org/abs/2305.05176 (2023).
Tang, L. et al. Evaluating large language models on medical evidence summarization. NPJ Digit. Med. 6, 158 (2023).
Bhaskar, A., Fabbri, A. & Durrett, G. Prompted opinion summarization with GPT-3.5. In Proc. Findings of the Association for Computational Linguistics: ACL 2023 9282–9300 (2023).
Park, J. et al. Criteria2query 3.0: leveraging generative large language models for clinical trial eligibility query generation. J. Biomed. Inform. 154, 104649 (2024).
Datta, S. et al. AutoCriteria: a generalizable clinical trial eligibility criteria extraction system powered by large language models. J. Am. Med. Inform. Assoc. 31, 375–385 (2024).
Agrawal, M., Hegselmann, S., Lang, H., Kim, Y. & Sontag, D. Large language models are few-shot clinical information extractors. In Proc. Conference on Empirical Methods in Natural Language Processing 1998–2022 (2022).
Jeong, D., Garg, S., Lipton, Z. C. & Oberst, M. Medical adaptation of large language and vision-language models: Are we making progress? In Proc. 2024 Conference on Empirical Methods in Natural Language Processing, 12143–12170 (2024).
Zhang, G. et al. Closing the gap between open source and commercial large language models for medical evidence summarization. npj Digit. Med. 7, 239 (2024).
Klang, E. et al. A strategy for cost-effective large language model use at health system-scale. npj Digit. Med. 7, 320 (2024).
OpenAI. Function calling and other api updates. https://openai.com/index/function-calling-and-other-api-updates/ (2023).
Chase, H. LangChain. https://github.com/langchain-ai/langchain (2022).
Jin, Q., Yang, Y., Chen, Q. & Lu, Z. GeneGPT: augmenting large language models with domain tools for improved access to biomedical information. Bioinformatics 40, btae075 (2024).
Sayers, E. W. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 47, D23 (2019).
Lin, C-Y. ROUGE: a package for automatic evaluation of summaries. In Proc. Text Summarization Branches Out. 74–81 (Association for Computational Linguistics, 2004).
Zhang, T., Kishore, V., Wu, F., Weinberger, K. Q. & Artzi, Y. BERTscore: evaluating text generation with BERT. In Proc. International Conference on Learning Representations. OpenReview.net (2019).
Van Veen, D. et al. Adapted large language models can outperform medical experts in clinical text summarization. Nat. Med. 30, 1134-1142 (2024).
Abacha, A. B., Yim, W.-w., Adams, G., Snider, N. & Yetisgen-Yildiz, M. Overview of the MEDIQA-Chat 2023 shared tasks on the summarization & generation of doctor-patient conversations. In Proc. 5th Clinical Natural Language Processing Workshop. 503–513 (Association for Computational Linguistics, 2023).
Nelson, H. Epic announces ambient clinical documentation EHR integration. Accessed 5 September 2024, https://www.techtarget.com/searchhealthit/news/366564355/Epic-Announces-Ambient-Clinical-Documentation-EHR-Integration. Accessed on Jun 2025 (2023).
Yim, W.-w. et al. Aci-bench: a novel ambient clinical intelligence dataset for benchmarking automatic visit note generation. Sci. Data 10, 586 (2023).
Soroush, A. et al. Large language models are poor medical coders-benchmarking of medical code querying. NEJM AI 1, AIdbp2300040 (2024).
Wang, H., Gao, C., Dantona, C., Hull, B. & Sun, J. Drg-llama: tuning llama model to predict diagnosis-related group for hospitalized patients. npj Digit. Med. 7, 16 (2024).
Topaz, M., Shafran-Topaz, L. & Bowles, K. H. ICD-9 to ICD-10: evolution, revolution, and current debates in the united states. Perspect. Health Inf. Manag. 10, 1d (2013).
Peggy, D. CPT® Codes: What Are They, Why Are They Necessary, and How Are They Developed? Advances in Wound Care 2, 583–587 (2013).
ICD-10-CM/PCS MS-DRG V40.1 Definitions Manual. Accessed June 23, 2025. https://www.cms.gov/icd10m/fy2023-version40.1-fullcode-cms/fullcode_cms/P0006.html.
Wornow, M. et al. Zero-shot clinical trial patient matching with LLMs. Preprint at arXiv https://arxiv.org/abs/2402.05125 (2024).
Nievas, M., Basu, A., Wang, Y. & Singh, H. Distilling large language models for matching patients to clinical trials. J. Am. Med. Inform. Assoc. 31, 1953–1963 (2024).
Unlu, O. et al. Retrieval-augmented generation–enabled GPT-4 for clinical trial screening. NEJM AI 1, AIoa2400181 (2024).
Wong, C. et al. Scaling clinical trial matching using large language models: a case study in oncology. In Proc. Machine Learning for Healthcare Conference. 846–862 (Proceedings of Machine Learning Research (PMLR), 2023).
Shaib, C. et al. Summarizing, simplifying, and synthesizing medical evidence using GPT-3 (with varying success). In Proc. Annual Meeting Of The Association For Computational Linguistics. (Association for Computational Linguistics, 2023).
Yun, H., Marshall, I., Trikalinos, T. & Wallace, B. Appraising the potential uses and harms of LLMs for medical systematic reviews. In Proc. Conference on Empirical Methods in Natural Language Processing (Association for Computational Linguistics, 2023).
Page, M. J. et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. Syst. Rev. 10, 89 (2021).
Wang, Z. et al. A foundation model for human-ai collaboration in medical literature mining. Preprint at arXiv https://arxiv.org/abs/2501.16255 (2025).
Wang, S., Harrisen, S., Bevan, K. & Guido Z. Can ChatGPT Write a Good Boolean Query for Systematic Review Literature Search? In Proceedings of the 46th International ACM SIGIR Conference on Research and Development in Information Retrieval. 1426–36 (NewYork, NY, USA: ACM, 2023).
National Institute of Standards and Technology. NIST Privacy Framework: A Tool for Improving Privacy through Enterprise Risk Management, Version 1.0. Technical Report (National Institute of Standards and Technology, 2020).
National Institute of Standards and Technology. The NIST Cybersecurity Framework (CSF) 2.0. Technical Report NIST CSWP 29 (U.S. Department of Commerce, 2024).
Quentin, C., Steinhagen, D., Francis, M. & Streff, K. Towards a Triad for Data Privacy. In Proceedings of the Annual Hawaii International Conference on System Sciences. (Hawaii International Conference on System Sciences (2020) https://doi.org/10.24251/hicss.2020.535.
Tabassi, E. Artificial Intelligence Risk Management Framework (AI RMF 1.0): AI RMF (1.0). Technical Report (2023).
Nori, H. et al. Can generalist foundation models outcompete special-purpose tuning? Case study in medicine. Preprint at arXiv [cs.CL] https://arxiv.org/abs/2311.16452 (2023).
Chaves, J. M. Z. et al. Towards a clinically accessible radiology foundation model: open-access and lightweight, with automated evaluation. Preprint at arXiv [cs.CL] https://arxiv.org/abs/2403.08002 (2024).
Zhang, S. et al. Large-scale domain-specific pretraining for biomedical vision-language processing. Preprint at arXiv https://www.microsoft.com/en-us/research/publication/large-scale-domain-specific-pretraining-for-biomedical-vision-language-processing/ (2023).
Huang, Z., Bianchi, F., Yuksekgonul, M., Montine, T. J. & Zou, J. A visual-language foundation model for pathology image analysis using medical Twitter. Nat. Med. 29, 2307–2316 (2023).
Lu, M. Y. et al. A visual-language foundation model for computational pathology. Nat. Med. 30, 863–874 (2024).
Li, C. et al. LLaVA‑Med: Training a Large Language‑and‑Vision Assistant for Biomedicine in One Day. In Advances in Neural Information Processing Systems 36 (NeurIPS 2023) (Datasets & Benchmarks Spotlight) (2023).
Gu, Y. et al. BiomedJourney: Counterfactual biomedical image generation by instruction-learning from multimodal patient journeys. Preprint at arXiv [cs.CV] https://arxiv.org/abs/2310.10765 (2023).
Bluethgen, C. et al. A vision-language foundation model for the generation of realistic chest X-ray images. Nat. Biomed. Eng. 9, 494–506 (2025) .
Xu, H. et al. A whole-slide foundation model for digital pathology from real-world data. Nature 630, 181–188 (2024).
Lei, H. et al. A Survey on Hallucination in Large Language Models: Principles, Taxonomy, Challenges, and Open Questions. ACM Transactions on Information Systems. 43, 1–55 (2025).
Thirunavukarasu, A. J. et al. Large language models in medicine. Nat. Med. 29, 1930–1940 (2023).
Kevin, W. et al. An Automated Framework for Assessing How Well LLMs Cite Relevant Medical References. Nat. Commun. 16, 3615 (2025).
Sams, C. M., Fanous, A. H. & Daneshjou, R. Human-artificial intelligence interaction research is crucial for medical artificial intelligence implementation. J Investig. Dermatol. https://www.sciencedirect.com/science/article/pii/S0022202X24019766 (2024).
Carlini, N. et al. Extracting training data from large language models. In Proc. 30th USENIX Security Symposium (USENIX Security 21). 2633–2650 (USENIX Association, 2021).
Theodorou, B., Xiao, C. & Sun, J. Synthesize high-dimensional longitudinal electronic health records via hierarchical autoregressive language model. Nat. Commun. 14, 5305 (2023).
Das, T., Wang, Z. & Sun, J. TWIN: Personalized clinical trial digital twin generation. In Proceedings of the 29th ACM SIGKDD Conference on Knowledge Discovery and Data Mining. 402–413 (Association for Computing Machinery (ACM), 2023). https://doi.org/10.1145/3580305.3599370.
Torfi, A., Fox, E. A. & Reddy, C. K. Differentially private synthetic medical data generation using convolutional GANs. Inf. Sci. 586, 485–500 (2022).
Schaeffer, R., Miranda, B. & Koyejo, S. Are emergent abilities of large language models a mirage? In Proc. Advances in Neural Information Processing Systems. Vol. 36 (Curran Associates, Inc., 2024).
Zini, J. E. & Awad, M. On the explainability of natural language processing deep models. ACM Comput. Surv. 55, 1–31 (2022).
Chen, W., Ma, X., Wang, X. & Cohen, W. W. Program of thoughts prompting: Disentangling computation from reasoning for numerical reasoning tasks. Trans. Mach. Learn. Res. https://openreview.net/forum?id=YfZ4ZPt8zd (2023).
Bereska, L. & Gavves, E. Mechanistic interpretability for AI safety—a review. Preprint at arXiv https://arxiv.org/abs/2404.14082 (2024).
Li, X. & Zhang, T. An exploration on artificial intelligence application: From security, privacy and ethic perspective. In Proc. IEEE International Conference on Cloud Computing and Big Data Analysis 416–420 (IEEE, 2017).
Wu, K. et al. Characterizing the clinical adoption of medical AI devices through us insurance claims. NEJM AI 1, AIoa2300030 (2023).
Suzanne, B. AI in Health: Keeping the Human in the Loop. Journal of the American Medical Informatics Association: JAMIA 30, 1225–26 (2023).
Singhvi, A. et al. DSPy assertions: Computational constraints for self-refining language model pipelines. Preprint at arXiv https://arxiv.org/abs/2312.13382 (2023).
Chase, H. Langsmith. https://www.langchain.com/langsmith (2024).
Lingjiao, L., Zaharia, M. & Zou, J. How Is ChatGPT’s Behavior Changing Over Time? Harvard Data Science Review. 6 (2024) https://doi.org/10.1162/99608f92.5317da47.
AI, C. DeepEval. https://github.com/confident-ai/deepeval (2023).
Mert, Y. Optimizing Generative AI by Backpropagating Language Model Feedback. Nature 639, 609–16 (2025).
Habicht, J. et al. Closing the accessibility gap to mental health treatment with a personalized self-referral chatbot. Nat. Med. 30, 595–602 (2024).
Pais, C. et al. Large language models for preventing medication direction errors in online pharmacies. Nat. Med. 30, 1574–1582 (2024).
Jin, Q. et al. Hidden flaws behind expert-level accuracy of multimodal GPT-4 vision in medicine. npj Digit. Med. 7, 190 (2024).
Zhou, J. et al. Pre-trained multimodal large language model enhances dermatological diagnosis using SKINGPT-4. Nat. Commun. 15, 5649 (2024).
Zhang, K. et al. A generalist vision–language foundation model for diverse biomedical tasks. Nat. Med. 1–13 (2024).
Kresevic, S. et al. Optimization of hepatological clinical guidelines interpretation by large language models: a retrieval augmented generation-based framework. NPJ Digit. Med. 7, 102 (2024).
Jiang, L. Y. et al. Health system-scale language models are all-purpose prediction engines. Nature 619, 357–362 (2023).
Sandmann, S., Riepenhausen, S., Plagwitz, L. & Varghese, J. Systematic analysis of ChatGPT, Google Search and Llama 2 for clinical decision support tasks. Nat. Commun. 15, 2050 (2024).
Keloth, V. K. et al. Advancing entity recognition in biomedicine via instruction tuning of large language models. Bioinformatics 40, btae163 (2024).
Huang, J. et al. A critical assessment of using ChatGPT for extracting structured data from clinical notes. npj Digit. Med. 7, 106 (2024).
He, H. et al. De novo generation of SARS-CoV-2 antibody CDRH3 with a pre-trained generative large language model. Nat. Commun. 15, 6867 (2024).
Acknowledgements
Thanks for the feedback from the IQVIA team, especially Jay Nanavati, for providing the privacy use case. This work was supported by NSF award SCH-2205289.
Author information
Authors and Affiliations
Contributions
All authors contributed to this work. Z.W. and J.S. drafted the initial manuscript. H.W., B.D., Y.L., C.M., D.B., L.A., H.P., and Y.W. contributed domain-specific use cases and provided editorial feedback. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Z., Wang, H., Danek, B. et al. A perspective for adapting generalist AI to specialized medical AI applications and their challenges. npj Digit. Med. 8, 429 (2025). https://doi.org/10.1038/s41746-025-01789-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-01789-7
This article is cited by
-
Holistic AI in medicine; improved performance and explainability
npj Digital Medicine (2026)
-
A foundation model for human-AI collaboration in medical literature mining
Nature Communications (2025)