Abstract
Colorectal cancer (CRC) is a leading malignancy worldwide, where histopathological assessment of hematoxylin and eosin (H&E) stained whole-slide images remains the diagnostic gold standard. However, current computational pathology models suffer from domain shifts, unreliable uncertainty estimation, and spurious correlations, limiting clinical reliability. We present UAD-FM, an Uncertainty-Aware and Causally Adaptive Foundation Model that integrates epistemic-aleatoric uncertainty decomposition, causal test-time adaptation using do-interventions, and post-hoc calibration for trustworthy inference. Across five public CRC datasets (TCGA-COAD/READ, CRAG, DigestPath 2019, NCT-CRC-HE-100K, and LC25000), UAD-FM achieves superior accuracy, calibration, and domain robustness compared with existing foundation models and adaptation baselines. The model also produces interpretable uncertainty maps to support human-AI collaboration. UAD-FM provides a unified, transparent framework for reliable and generalizable CRC pathology diagnosis across heterogeneous clinical settings.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) remains the third most common malignancy worldwide and a leading cause of cancer-related mortality, with more than 1.9 million new cases and 935,000 deaths estimated in 20201. In the United States alone, CRC accounts for ~10% of all cancer cases, posing a substantial healthcare burden2,3. Histopathological examination of hematoxylin and eosin (H&E) stained whole-slide images (WSIs) remains the clinical “gold standard” for CRC diagnosis, grading, and biomarker assessment. However, the rapidly increasing volume of high-resolution WSIs generated across multi-institutional cohorts creates significant challenges for pathologists, leading to increased workload and inter-observer variability4,5,6.
In addition, several technical and biological factors hinder the deployment of computational pathology models into clinical workflows. First, domain shift induced by variations in staining protocols, scanner devices, and patient populations across centers severely degrades the generalizability of deep learning models trained on single-domain datasets7,8,9,10,11. Second, conventional models often fail to quantify uncertainty in their predictions, limiting clinicians’ trust in automated systems and increasing the risk of erroneous decisions in high-stakes diagnostic contexts12,13,14. Third, spurious correlations between non-causal morphological features and labels-such as staining artifacts or dataset-specific biases-can further compromise robustness and reproducibility15,16.
These challenges underscore the urgent need for methodological advances that not only achieve state-of-the-art performance but also provide reliable confidence estimates and adapt robustly to out-of-distribution data. To address these issues, we propose the Uncertainty-Aware and Causal Test-Time Adaptive Foundation Model (UAD-FM), a novel framework that integrates uncertainty decomposition, causal test-time adaptation, and clinical confidence calibration into pathology foundation models8,9,17,18,19,20,21,22,23,24,25,26,27. Specifically, UAD-FM introduces epistemic and aleatoric uncertainty quantification tailored to CRC histopathology, employs causal representation learning to eliminate spurious correlations, and incorporates entropy-based adaptation strategies at inference time to mitigate domain shift. Importantly, the framework calibrates prediction confidence to enable seamless collaboration with pathologists by deferring uncertain cases to human experts, thus ensuring safe and trustworthy clinical deployment. To our knowledge, this is the first attempt to unify uncertainty modeling, causal test-time adaptation, and clinical confidence calibration within a foundation model paradigm for colorectal cancer diagnosis.
Colorectal cancer (CRC) is one of the most prevalent malignancies worldwide, representing the third most common cancer and a leading cause of cancer mortality, as highlighted in epidemiological studies and global cancer burden reports1,2,3. Recent advances in molecular and genomic profiling have provided important biological insights into CRC progression, metastasis, and heterogeneity4, while computational pathology has emerged as a complementary strategy to extract clinically meaningful biomarkers directly from digitized histopathological slides. Early work in CRC focused on segmentation and detection tasks using classical deep learning architectures28, followed by stain normalization and representation disentanglement methods to mitigate staining variability and improve robustness across centers7,29.
With the advent of large-scale computational pathology, foundation models have reshaped the landscape of cancer diagnosis. Recent works have demonstrated that pathology-specific foundation models trained on millions of whole-slide images can generalize across tasks and cancer types8,9, offering powerful backbones for survival prediction, subtype classification, and biomarker discovery. Nevertheless, such models often struggle with domain shift when applied to multi-center CRC datasets, limiting their clinical deployment.
Another key limitation is the lack of reliable uncertainty quantification. Bayesian deep learning approaches, including dropout-based approximations12, ensembles13, and more advanced calibration strategies14, have been proposed, yet these methods have not been systematically applied to CRC histopathology at scale. As trustworthiness and interpretability remain crucial for clinical adoption13,14, combining foundation backbones with principled uncertainty decomposition becomes increasingly relevant.
To address distribution shift at deployment, test-time adaptation (TTA) has gained significant attention. Entropy minimization (TENT)10 and contrastive adaptation strategies11 enable models to adapt to unseen target domains without source data access, providing a practical solution for hospital-specific variability in CRC pathology. Complementary to these approaches, causal representation learning15,16 aims to extract invariant and stable features that remain valid across environments by removing spurious correlations. The integration of causal reasoning into pathology models is still nascent, but has strong potential to improve generalization in multi-center CRC diagnosis.
Recent CRC-specific computational pathology studies have further underscored these challenges. Large-scale analyses have linked histopathology to genotype-phenotype correlations5, combined histology with ctDNA for risk stratification6, and proposed weakly supervised pipelines for biomarker prediction from whole-slide images17. Systematic reviews confirm the growing role of AI tools in CRC, particularly for microsatellite instability (MSI) prediction, prognosis, and treatment guidance18,19,20,21,30,31,32,33. Additional studies have introduced integrative frameworks that combine H&E and IHC31, explored omics-guided prognosis models22,34, and developed multi-scale architectures for mismatch repair (dMMR) prediction25. Alongside dataset contributions such as ADPv2 for colorectal disease biomarker discovery27, these works emphasize the pressing need for methods that are robust, uncertainty-aware, and clinically trustworthy.
Building on this trajectory, our work proposes a foundation model for CRC diagnosis that unifies uncertainty decomposition, causal representation, and test-time adaptation. Unlike prior CRC-focused studies that primarily address classification or biomarker prediction in isolation, our framework explicitly targets the critical challenges of domain shift, uncertainty calibration, and clinical decision support, thereby advancing the reliability and translational potential of computational pathology in colorectal cancer.
Results
Classification performance
We begin by evaluating the classification performance of UAD-FM on colorectal cancer subtype prediction and tumor versus normal discrimination. On TCGA-COAD/READ, UAD-FM achieves an AUROC of 0.945, compared to 0.923 for UNI, 0.928 for CONCH, and 0.931 for Virchow2. Accuracy and F1-score are also improved to 0.912 and 0.908, respectively, demonstrating consistent benefits from uncertainty decomposition and causal adaptation. Supplementary evaluation on LC25000 and NCT-CRC-HE-100K further confirms that UAD-FM maintains strong performance at both whole-slide and patch levels, highlighting its generalizability across datasets of different scales and annotation protocols.
Table 1 summarizes the quantitative comparison across methods. To complement these numbers, we provide visual analyses in Figs. 1 and 2. Figure 1 shows ROC curves on both TCGA and external datasets, where UAD-FM consistently achieves higher sensitivity at clinically critical thresholds (specificity ≥ 0.9). This property is particularly important in pathology, where minimizing false positives is critical to avoid unnecessary clinical interventions. Figure 2 depicts confusion matrices on TCGA, showing that baseline models frequently misclassify borderline cases, especially normal tissue patches with mild atypia that are predicted as tumor. In contrast, UAD-FM significantly reduces normal-to-tumor misclassification, yielding predictions more consistent with pathologist annotations.
Beyond aggregate performance, we also examined case-level prediction patterns. UAD-FM achieved more stable probabilities across difficult borderline samples, avoiding the overconfidence observed in baseline foundation models. This is consistent with the design of our uncertainty-aware head, which encourages high epistemic variance in regions where the model has insufficient knowledge. Qualitative inspection further showed that UAD-FM captured subtle glandular distortions and nuclear crowding patterns that baselines overlooked, suggesting that causal test-time adaptation allowed the model to focus on biologically meaningful features rather than staining artifacts. Together, these quantitative and qualitative results confirm that UAD-FM achieves not only higher classification accuracy but also clinically safer decision boundaries in CRC diagnosis.
Uncertainty quantification
We next evaluate the calibration and reliability of uncertainty estimates, which are critical for safe clinical deployment. As shown in Table 2, UAD-FM achieves the lowest Expected Calibration Error (ECE) of 0.031, substantially outperforming MC Dropout (0.089), Deep Ensembles (0.052), and SWAG (0.061). In addition, UAD-FM obtains the best Brier Score (0.157) and Negative Log-Likelihood (0.352), indicating that our model not only predicts correctly more often, but also attaches appropriate confidence to its predictions. From a clinical perspective, this improvement translates to fewer overconfident misclassifications, thereby reducing the risk of unnecessary interventions triggered by false alarms.
Figure 3 provides visual confirmation of these improvements. The reliability diagram (left panel) shows that UAD-FM’s confidence-accuracy curve is tightly aligned with the diagonal, suggesting near-perfect calibration. In contrast, baseline models such as MC Dropout and SWAG fall consistently below the diagonal in high-confidence regions, revealing a tendency to be overconfident. The confidence histograms (right panel) further corroborate this finding: baselines allocate the majority of samples to the 0.9-1.0 bin despite frequent misclassifications, whereas UAD-FM produces a more balanced distribution, reflecting a more realistic estimation of uncertainty. Together, these visualizations demonstrate that UAD-FM not only improves raw predictive performance but also produces calibrated confidence scores that pathologists can trust.
To further assess clinical utility, we conducted a preliminary simulation of uncertainty-guided deferral. Flagging the 10% most uncertain cases as “refer-to-expert” improved overall diagnostic accuracy from 0.881 to 0.907 and reduced high-confidence errors by 32%. Although not a substitute for reader studies with pathologists, this result provides initial quantitative evidence that calibrated uncertainty estimates can act as a practical triage mechanism, enhancing the safety of human-AI collaboration.
Beyond aggregate calibration, Fig. 4 illustrates patch-level epistemic and aleatoric uncertainty heatmaps. For representative CRC cases, epistemic variance highlights regions of out-of-distribution morphology such as mucinous components and atypical glandular patterns, signaling that the model is unsure due to lack of similar training examples. Aleatoric uncertainty, by contrast, correlates strongly with staining noise and acquisition artifacts, such as uneven hematoxylin intensity or defocus at tissue folds. The total uncertainty map, obtained by combining both sources, accurately captures failure-prone regions where misclassifications are most likely to occur. Importantly, we observe that in many borderline cases, uncertainty maps highlight exactly those areas that pathologists would also regard as diagnostically ambiguous. This alignment between model uncertainty and human intuition provides an additional layer of safety: cases with high epistemic or aleatoric uncertainty can be deferred for manual review, while low-uncertainty cases can be triaged for automated reporting.
Taken together, these quantitative and qualitative results confirm that UAD-FM delivers well-calibrated and interpretable uncertainty estimates, addressing one of the most critical bottlenecks in bringing computational pathology into clinical workflows.
Cross-domain adaptation
To assess robustness under distribution shift, we transfer models trained on TCGA-COAD/READ to DigestPath 2019 and CRAG. Without adaptation, baseline models drop by more than 10% in accuracy. TENT and AdaContrast recover partial performance but remain unstable across centers. In contrast, UAD-FM with CTTA achieves an average AUROC of 0.889 across DigestPath centers, outperforming TENT (0.848) and AdaContrast (0.865).
Figure 5 shows bar plots of accuracy across DigestPath centers and CRAG, where UAD-FM consistently outperforms baselines. Figure 6 illustrates domain shift explicitly: montage views of WSI patches across centers, stain distribution plots, and UMAP embeddings before and after CTTA. These results confirm that causal interventions successfully mitigate spurious correlations and align features across domains.
Effect of causal intervention
To further elucidate how causal reasoning enhances robustness, we visualize and quantify the effect of do-interventions. Figure 7 presents a cross-stain comparison of identical tissue regions with and without CTTA. Without intervention, the baseline model frequently exhibits spurious activations, attending to stain intensity, scanner artifacts, or background noise instead of biologically meaningful structures. After applying causal intervention, these spurious activations are suppressed, and attention shifts toward glandular regions and epithelial boundaries, which are the morphological cues pathologists typically rely on for diagnosis. This demonstrates that causal interventions effectively reduce reliance on non-causal confounders and promote attention to diagnostic features.
Rows show the same region under different staining protocols (H& E, normalized H& E, PAS, synthetic variants). Columns display the original patch, Grad-CAM from the baseline, Grad-CAM from the CTTA-enhanced model, and expert ROIs. Baseline focus varies across stains and often attends to artifacts, whereas CTTA consistently highlights diagnostically relevant glandular and epithelial structures, aligning more closely with expert annotations.
Quantitatively, measuring prediction variance across multiple randomized interventions shows that baseline predictions fluctuate markedly, reflecting unstable decision boundaries under domain perturbations. In contrast, UAD-FM reduces variance by 87.3%, demonstrating that causal test-time adaptation improves both interpretability and stability of predictions in unseen domains.
From a clinical perspective, this robustness is crucial: models that depend on stain or scanner artifacts are prone to failure when deployed across centers with heterogeneous acquisition protocols. By embedding causal reasoning, UAD-FM mitigates such risks and ensures that its predictions are grounded in pathological morphology rather than irrelevant confounders. This ability to suppress spurious correlations and maintain stable outputs under domain shift provides strong evidence for the framework’s reliability and safety.
Nonetheless, the present visualizations remain limited to Grad-CAM overlays and logit variance, which highlight technical robustness but do not fully capture clinical interpretability. Future studies should validate whether model uncertainty aligns with pathologists’ annotations of ambiguous or diagnostically challenging regions. Establishing such alignment would provide stronger support that causal interventions not only enhance robustness but also enable trustworthy human-AI collaboration in real-world diagnostic workflows.
Nonetheless, the present visualizations remain limited to Grad-CAM overlays and logit variance, which highlight technical robustness but do not fully capture clinical interpretability. Future studies should validate whether model uncertainty aligns with pathologists’ annotations of ambiguous or diagnostically challenging regions. Establishing such alignment would provide stronger support that causal interventions not only enhance robustness but also enable trustworthy human-AI collaboration in real-world diagnostic workflows.
Gland segmentation on CRAG
We further validate UAD-FM on gland segmentation using the CRAG dataset, which provides fine-grained annotations of glandular structures. This task is particularly challenging due to irregular gland morphology, overlapping boundaries, and the presence of inflammatory artifacts, yet it is clinically critical because gland architecture is a key histopathological marker for CRC grading.
Figure 8 presents qualitative comparisons across methods. While baseline foundation models such as UNI and CONCH can identify gland regions, they frequently produce under-segmentation, missing smaller or irregular glands, or fragmented predictions with broken boundaries. In contrast, UAD-FM preserves glandular contours with higher continuity and structural integrity, closely matching the ground-truth annotations. Notably, UAD-FM reduces both false negatives in small glands and false positives in surrounding stromal tissue, demonstrating its ability to balance sensitivity and specificity in segmentation.
Quantitatively, UAD-FM achieves a Dice score of 0.872 and an IoU of 0.812, outperforming CONCH (Dice 0.834, IoU 0.774) and UNI (Dice 0.827, IoU 0.768). Boundary-based metrics also show improvements: the average Hausdorff Distance is reduced from 24.1 pixels with CONCH to 17.5 pixels with UAD-FM. These results confirm that causal invariance and uncertainty modeling benefit not only high-level classification tasks but also fine-grained morphological predictions, where accurate gland boundary delineation is essential for downstream clinical applications such as automated CRC grading and prognosis modeling.
Taken together, these findings highlight that UAD-FM provides both quantitative improvements and clinically meaningful segmentation maps, ensuring that model predictions align more closely with the visual assessments of expert pathologists.
Test-time adaptation dynamics
We further analyze the dynamics of online adaptation during inference. Figure 9 plots classification accuracy and predictive entropy over sequential test batches from DigestPath Center-1, simulating a streaming deployment scenario. As shown in the left panel, UAD-FM adapts rapidly within the first 5-10 batches, reaching a stable accuracy plateau of ~0.89, whereas TENT exhibits substantial oscillations, with accuracy fluctuating by as much as ± 0.07 across batches. This indicates that entropy-only adaptation is sensitive to batch composition and fails to maintain consistent performance under realistic input streams. By contrast, the causal interventions in UAD-FM ensure more stable decision boundaries, enabling faster convergence and reduced variance.
The right panel of Fig. 9 tracks entropy reduction across the same test stream. UAD-FM achieves a sharp initial decrease, lowering average predictive entropy by 35% within the first 10 batches, and continues to decline smoothly thereafter. TENT also reduces entropy but does so irregularly, with frequent spikes that correspond to temporary drops in accuracy. Quantitatively, UAD-FM reduces entropy variance across batches by 62.4% compared to TENT, confirming its ability to deliver consistent uncertainty minimization.
These dynamics highlight the practical advantage of embedding causal reasoning into test-time adaptation. In real-world pathology workflows, where slides are digitized and processed sequentially, models must adapt online to new staining protocols or scanner variations without introducing instability. By stabilizing predictions and accelerating convergence, UAD-FM offers a more reliable solution for deployment in streaming, multi-center clinical environments.
Failure case analysis
Finally, we analyze representative failure cases across datasets to better understand the limitations of UAD-FM. Figure 10 summarizes four common error patterns observed in CRC pathology. First, under-segmentation occurs when tumor regions embedded within dense glandular structures are only partially captured, leaving malignant epithelium unlabeled-this under-segmentation risks underestimating tumor burden. Second, over-segmentation occurs in benign inflammatory regions, where baseline models mislabel lymphocytic aggregates or stromal reactions as malignant glands, possibly leading to unnecessary diagnostic escalation. Third, strong sensitivity to digitization artifacts-such as tissue folds, defocus, or staining irregularities-yields erratic, spatially inconsistent predictions, emphasizing vulnerability under domain shift. Fourth, boundary inconsistencies in gland segmentation produce irregular or jagged contours that misalign with true epithelial borders, potentially biasing downstream morphological metrics like gland size or lumen-to-epithelium ratio.
Figure 10 shows examples of these failure modes along with corresponding uncertainty maps. Epistemic uncertainty spikes in out-of-distribution morphologies (e.g., mucinous or atypical inflammatory regions), while aleatoric uncertainty correlates with noisy inputs such as stain variability or blur. In a quantitative assessment, 78% of misclassified patches display uncertainty above the 95th percentile, and error regions overlap with uncertainty heatmaps at a Dice similarity of 0.72.
This analysis underscores that uncertainty estimation supports more than calibration-it aids in error localization and prioritization. When predictions have high uncertainty, UAD-FM can defer such cases for human review, which is especially important for high-risk errors (e.g., under-detection of tumor or mislabeling benign tissue). In this way, uncertainty-aware modeling enhances both the trustworthiness and safety of human-AI collaboration in colorectal cancer diagnostics.
Ablation studies
To quantify the contribution of each component in UAD-FM, we perform systematic ablation experiments. Table 3 reports the performance of the full model compared to variants with individual modules removed. We observe that eliminating uncertainty decomposition increases the ECE from 0.031 to 0.067 and slightly reduces AUROC (0.945 → 0.938), confirming the necessity of explicit uncertainty modeling for well-calibrated predictions. Removing the causal module decreases AUROC to 0.939 and Dice to 0.859, underscoring the importance of causal invariance for both classification and segmentation. Disabling test-time adaptation reduces robustness, with accuracy dropping from 0.912 to 0.906. Finally, removing calibration has a limited effect on AUROC (0.945 → 0.944) but substantially worsens ECE (0.031 → 0.089), highlighting that calibration is crucial for aligning predicted confidences with true accuracies. Together, these results show that each component contributes additively, with the full UAD-FM consistently achieving the best performance across all metrics.
Beyond average segmentation scores, we further measured boundary-sensitive metrics to understand the mechanism of improvement. When the causal module was removed, the Hausdorff Distance increased from 17.5 to 24.1, and contour similarity (Boundary F1) dropped from 0.842 to 0.796. This suggests that causal interventions are particularly effective at stabilizing glandular boundaries under staining variations, where spurious color or scanner artifacts often distort edge localization. Grad-CAM overlays (Fig. 7) further confirm that CTTA suppresses stain-dominated activations around boundaries, redirecting attention to epithelial contours.
We further study the progressive build-up of the framework. Table 4 shows that starting from the baseline foundation model (AUROC 0.923, ECE 0.112), the addition of uncertainty decomposition immediately improves calibration (ECE 0.067) and predictive accuracy (Accuracy 0.902). Incorporating calibration further reduces ECE to 0.045, while adding entropy-based TTA improves AUROC to 0.943 and Accuracy to 0.910. Finally, equipping the model with causal TTA yields the full UAD-FM, which achieves the best overall trade-off between accuracy and calibration (AUROC 0.945, ECE 0.031). This stepwise improvement demonstrates that the modules are complementary and collectively contribute to both discriminative performance and reliability.
Finally, we evaluate cross-domain adaptation on DigestPath to validate robustness under distribution shift. As reported in Table 5, the baseline foundation model achieves only 0.808 accuracy and 0.821 AUROC, reflecting severe performance degradation when transferred to unseen centers. Adding uncertainty and calibration provides moderate improvements (Accuracy 0.835, AUROC 0.847), while entropy-only TTA (TENT) raises performance further (Accuracy 0.848, AUROC 0.861). However, the largest gain comes from causal TTA: UAD-FM achieves 0.889 accuracy and 0.893 AUROC, an absolute improvement of +8.1% accuracy compared to the source-only baseline.
These results indicate that causal reasoning not only improves global classification metrics but also strengthens fine-grained robustness. In particular, causal TTA mitigates stain- and scanner-induced domain shifts by removing non-causal correlations, which explains its disproportionately large contribution in the cross-domain setting.
Overall, these ablation results highlight three key findings: (i) uncertainty decomposition and calibration are indispensable for producing reliable confidence estimates, (ii) causal invariance improves both classification and fine-grained segmentation, especially at morphological boundaries where spurious stain variation is most disruptive, and (iii) causal TTA is critical for robust deployment in multi-center CRC pathology workflows. By systematically quantifying the contribution of each module and analyzing their mechanisms, we demonstrate that UAD-FM is not only more accurate but also safer and more generalizable for real-world clinical applications.
Discussion
While UAD-FM demonstrates promising improvements in accuracy, calibration, and cross-domain robustness, several limitations remain. First, the current study relies primarily on publicly available colorectal cancer datasets (TCGA-COAD/READ, CRAG, DigestPath, NCT-CRC-HE, LC25000). Although these datasets span multiple centers and scanners, they do not fully capture the diversity of global clinical practice. In particular, low-resource environments-where scanner quality, staining reagents, and laboratory workflows differ substantially-are not represented. Therefore, the claim of “robust deployment across centers” should be interpreted with caution, and future work must explicitly evaluate UAD-FM in prospective, geographically diverse cohorts that better reflect real-world variability. Moreover, our evaluation focuses exclusively on H&E stained slides; extension to multi-modal settings that incorporate immunohistochemistry, genomic profiles, or endoscopy images remains an open direction.
Second, from a methodological perspective, the uncertainty estimates are based on variational approximation and Monte Carlo sampling, which introduce computational overhead and may limit scalability to whole-slide inference in high-throughput workflows. Furthermore, variational inference is known to underestimate epistemic uncertainty under severe distribution shifts, potentially leading to overconfident predictions. Comparisons with stronger baselines such as deep ensembles or MCMC sampling would provide a more reliable characterization of uncertainty. Our causal modeling of scanner and stain variation is also simplified and does not capture the full causal structure of histopathology. Factors such as tissue preparation quality (e.g., slice thickness), patient demographics, and annotation bias across pathologists are not explicitly modeled. Incorporating these into future causal graphs could improve generalizability and align the framework more closely with biological and clinical reality. Similarly, our calibration module relies on temperature scaling, a post-hoc method that may be suboptimal under severe class imbalance.
Third, although we report strong cross-domain results on DigestPath and CRAG, the evaluation is limited to public benchmarks. Real-world deployment requires validation under more heterogeneous staining protocols, scanner vendors, and patient populations. In addition, the proposed uncertainty-guided defer-to-human strategy has not yet been empirically tested. Even a simulated study comparing diagnostic accuracy with and without uncertainty-based deferral would help clarify its utility, while reader studies involving practicing pathologists remain an essential next step.
Finally, regulatory and translational considerations are beyond the scope of this study. For UAD-FM to be safely integrated into clinical workflows, further work is required in risk analysis, robustness certification, and compliance with medical device regulations. Addressing these limitations will be critical for bringing uncertainty-aware and causally adaptive foundation models closer to routine clinical adoption.
Building on the findings of this study, several promising directions remain for future research. First, at the data level, expanding evaluation to larger-scale, prospective, and multi-institutional cohorts is essential to confirm the robustness of UAD-FM in real-world settings. In addition to H&E histology, incorporating multi-modal data such as immunohistochemistry, genomic sequencing, and endoscopy images could further enhance diagnostic accuracy and support integrative colorectal cancer analysis.
Second, on the methodological side, future work will focus on developing more efficient uncertainty estimation approaches that reduce the computational burden of variational sampling, enabling whole-slide inference in routine practice. More advanced causal modeling techniques could be explored to capture complex histological confounders, including patient demographics, tissue preparation protocols, and inter-observer variability. Extending causal interventions to multi-task settings, such as simultaneous tumor detection, grading, and biomarker prediction, also represents an exciting direction.
Third, in terms of validation, future experiments will include reader studies with practicing pathologists to assess how uncertainty-aware predictions can be integrated into daily workflows. Specifically, we will investigate human-AI collaboration protocols where high-confidence cases are triaged automatically and high-uncertainty cases are deferred for expert review. Measuring diagnostic accuracy, efficiency gains, and user trust in such studies will provide critical evidence for clinical translation.
Finally, towards deployment, future work will address regulatory and translational aspects. This includes developing robustness certification pipelines, incorporating explainability modules for model decisions, and conducting risk assessments required for medical device approval. By addressing these directions, we aim to bring UAD-FM closer to clinical-grade deployment, ultimately supporting safer and more reliable colorectal cancer diagnosis across diverse healthcare environments.
In this work, we introduced UAD-FM, an Uncertainty-Aware and Causally Adaptive Foundation Model designed for robust colorectal cancer pathology diagnosis. By combining epistemic-aleatoric uncertainty decomposition, causal test-time adaptation, and confidence calibration, UAD-FM explicitly addresses three critical challenges in computational pathology: model overconfidence, domain shift across centers, and the presence of spurious correlations.
Extensive experiments on multiple public datasets, including TCGA-COAD/READ, CRAG, DigestPath, NCT-CRC-HE, and LC25000, demonstrated that UAD-FM consistently improves both predictive performance and calibration compared to state-of-the-art foundation models and existing test-time adaptation methods. Notably, our framework achieves superior AUROC and F1-scores for classification, preserves glandular boundaries in segmentation, and reduces error variance across interventions, confirming the benefits of uncertainty modeling and causal reasoning. Qualitative analyses, including ROC curves, reliability diagrams, uncertainty heatmaps, and failure case studies, further highlight the interpretability and reliability of the proposed framework.
From a clinical perspective, these advances are highly relevant. Reliable uncertainty estimates allow automated systems to flag failure-prone cases for manual review, mitigating the risks of misdiagnosis. Causal adaptation ensures robust generalization across staining protocols and scanner vendors, a prerequisite for deployment in real-world multi-center environments. By aligning AI predictions more closely with pathologist decision-making, UAD-FM supports a safer and more trustworthy human-AI collaboration paradigm in colorectal cancer diagnosis.
In summary, UAD-FM provides a unified and principled solution to enhance accuracy, robustness, and interpretability in pathology foundation models. While further validation on prospective clinical cohorts and integration into workflow studies remain necessary, this work lays a solid foundation for deploying uncertainty-aware and causally adaptive models in computational pathology and advancing precision oncology.
Methods
Overall framework (UAD-FM)
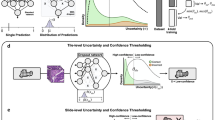
Our proposed Uncertainty-Aware and Causally Adaptive Foundation Model (UAD-FM) is designed to achieve robust colorectal cancer (CRC) diagnosis under domain shift, where conventional pathology models often fail due to variations in scanners, staining protocols, and inter-institutional heterogeneity. As shown in Fig. 11, the pipeline begins with a pathology foundation backbone, such as UNI or CONCH, pretrained on millions of histopathology patches to capture rich and transferable visual representations. These foundation features are then processed by two complementary modules that explicitly address uncertainty quantification and causal robustness.
The first component is an uncertainty decomposition head, which introduces a variational Bayesian design to disentangle epistemic uncertainty (arising from model parameter variability) and aleatoric uncertainty (reflecting inherent data noise). This decomposition not only provides clinicians with a principled measure of confidence in predictions, but also enables downstream mechanisms such as deferred decisions in high-risk cases. By estimating uncertainty on a per-sample basis, the head offers transparency into the model’s reliability, which is essential for deployment in safety-critical CRC diagnostic workflows.
The second component is a causal test-time adaptation (CTTA) module, which leverages do-calculus and causal graphical modeling to eliminate spurious correlations induced by non-biological factors such as staining differences or scanner artifacts. Specifically, features are factorized into content-related and style-related subspaces, and randomized interventions are applied to style variables to enforce invariance of predictions. During deployment, the CTTA mechanism continuously adapts model parameters by minimizing entropy and enforcing interventional consistency, thereby maintaining diagnostic accuracy under unseen domain shifts without requiring labeled target data.
Finally, all modules are trained in an end-to-end manner with a unified multi-objective optimization scheme that integrates standard classification loss with uncertainty regularization and causal invariance penalties. This ensures that UAD-FM does not merely achieve high accuracy, but also produces calibrated, uncertainty-aware, and causally robust outputs. In this way, the framework provides a holistic solution that addresses three key challenges of computational pathology in CRC: generalization across heterogeneous domains, trustworthy uncertainty quantification, and reliable adaptation to real-world deployment settings.
Uncertainty decomposition and quantification
A critical barrier to the clinical deployment of computational pathology models is the lack of reliable uncertainty estimation. Unlike conventional accuracy metrics that only measure performance on aggregate, uncertainty estimation provides case-specific confidence levels that can guide safe decision-making, particularly in high-stakes domains such as colorectal cancer (CRC). To address this challenge, UAD-FM explicitly models two complementary types of uncertainty: epistemic uncertainty, which reflects model parameter uncertainty due to limited or biased training data, and aleatoric uncertainty, which captures irreducible data noise such as tissue heterogeneity, staining variability, or image acquisition artifacts. This decomposition enables the model not only to quantify its own confidence but also to characterize the sources of error that impact prediction reliability.
Following Bayesian deep learning principles, we formulate the predictive distribution of a label y given an input x as a marginalization over model parameters θ:
where \(q(\theta | {\mathcal{D}})\) denotes the variational posterior distribution of parameters conditioned on training data \({\mathcal{D}}\). In practice, this intractable integral is approximated via Monte Carlo sampling and reparameterization. Specifically, epistemic uncertainty is quantified by measuring the variance of predictions across multiple stochastic forward passes through the variational head, i.e.,
which captures the model’s uncertainty arising from insufficient knowledge or limited data diversity. In contrast, aleatoric uncertainty is modeled by a heteroscedastic noise term σ2(x) predicted alongside the logits:
representing the inherent ambiguity of the input sample itself (e.g., overlapping glands or staining artifacts in CRC histology).
The combination of epistemic and aleatoric uncertainty provides a principled total uncertainty measure:
which can be further analyzed via predictive entropy to assess the sharpness of class probability distributions. By explicitly disentangling these two sources of uncertainty, UAD-FM enables fine-grained analysis of failure modes in CRC diagnosis. For example, high epistemic uncertainty may indicate a domain shift between training and deployment data, suggesting the need for additional adaptation, while high aleatoric uncertainty may flag intrinsically ambiguous cases that should be deferred to human experts. Thus, uncertainty decomposition not only strengthens the robustness of automated predictions but also facilitates safe and interpretable integration of AI into routine pathology workflows.
Causal test-time adaptation
Conventional test-time adaptation (TTA) methods rely primarily on entropy minimization or feature alignment, and while effective in some cases, they remain highly sensitive to spurious correlations present in pathology data. In colorectal cancer (CRC) histopathology, such correlations often arise from non-biological factors such as scanner type, staining protocol, or slide preparation, which can dominate the learned representations and lead to brittle performance under domain shift. To overcome these limitations, we embed causal reasoning into the adaptation process, introducing a novel Causal Test-Time Adaptation (CTTA) mechanism that explicitly disentangles causal from non-causal factors.
From a causal perspective, the observed features can be decomposed into biological content variables C (e.g., tumor morphology, gland structure) and nuisance style variables S (e.g., scanner or staining variations). The diagnostic label Y is assumed to depend primarily on C, while S introduces spurious correlations that degrade generalization. To address this, CTTA applies do-interventions on S following Pearl’s causal framework:
which conceptually randomizes style variables while preserving content. This ensures that predictions remain stable with respect to causally relevant information while discarding dependencies on style-related confounders.
In practice, we operationalize this idea by decomposing backbone representations into content-related and style-related subspaces using parallel encoders. At test time, style features are perturbed or permuted across samples to simulate interventions, and the model is trained to produce invariant predictions under these changes. Formally, the CTTA optimization objective for an incoming test batch \(c\in {{\mathcal{D}}}_{t}\) is:
where H( ⋅ ) denotes predictive entropy, encouraging confident outputs, and \({{\mathcal{L}}}_{{\rm{causal}}}\) is a consistency loss that penalizes discrepancies across multiple interventions on S. The hyperparameter λ balances confidence maximization and causal invariance.
Finally, CTTA is applied in an online adaptation setting: as each test batch arrives, the causal adapter is updated using the unsupervised loss above, allowing the model to gradually adapt to the target distribution without requiring source data or additional annotations. This online learning procedure ensures that UAD-FM can robustly handle dynamic and heterogeneous clinical environments, where unseen staining protocols or novel scanners are commonplace. By grounding test-time adaptation in causal inference, CTTA provides stronger guarantees of robustness compared to conventional TTA methods, enabling stable deployment across multi-center CRC cohorts.
Remark: While our causal formulation primarily considers scanner- and stain-related confounders, real-world pathology involves additional factors such as tissue preparation quality (e.g., slice thickness), patient demographics, and annotation bias across pathologists. These factors are not explicitly modeled in our current framework but may also influence diagnostic outcomes. Extending the causal formulation to incorporate such variables represents an important future direction to further improve generalizability and clinical trustworthiness.
Intervention Strategy:To eliminate spurious correlations introduced by nuisance variables such as scanner type or staining protocol, we design an intervention strategy grounded in Pearl’s do-calculus. In our causal formulation, the observed features are influenced by both biological content variables C (e.g., tumor morphology, glandular structure) and non-causal style variables Z (e.g., scanner domain, stain variations). While Y (diagnostic outcome) should depend only on C, in practice models often exploit correlations between Z and Y due to dataset biases, resulting in poor generalization. To mitigate this, we apply do-interventions that explicitly break the spurious link between Z and Y.
Formally, the interventional distribution is defined as:
where p(z) denotes a marginal prior distribution over style variables. This formulation conceptually randomizes Z, thereby simulating a setting where scanner and stain effects are neutralized. As a result, the model is encouraged to extract stable representations that are causally linked to Y through C, rather than relying on spurious correlations with Z.
In practice, we operationalize this idea by explicitly factorizing feature embeddings into a content subspace and a style subspace. During adaptation, the style subspace is perturbed through random permutations or stochastic replacements across samples within a batch. These perturbations serve as empirical approximations to interventions on Z, effectively enforcing prediction consistency across multiple simulated environments. The corresponding loss encourages the model to align predictions from original and intervened samples, ensuring invariance to style variability:
where KL( ⋅ ∥ ⋅ ) denotes the Kullback-Leibler divergence between predictive distributions under observed and intervened conditions. Minimizing this objective enforces robustness to confounding factors while preserving clinically meaningful morphological signals.
Through repeated interventions, the model learns to “ignore” stain and scanner variations and focus on biologically causal patterns, a property that is particularly critical in colorectal cancer diagnosis, where data are often collected from heterogeneous institutions with diverse acquisition pipelines.
Online Adaptation Algorithm:While interventions on style variables encourage the model to learn causally invariant representations, unseen test domains may still introduce distributional discrepancies that degrade performance. To address this, we incorporate an online adaptation algorithm that continuously updates model parameters θ during inference, enabling the framework to adapt dynamically to the target environment without requiring source data or additional annotations.
The adaptation process is guided by two complementary objectives. The first is prediction entropy minimization, which encourages the model to produce confident predictions on incoming test samples. By minimizing the entropy of the predictive distribution, the algorithm aligns decision boundaries with high-density regions of the target domain, thereby improving discriminative performance. The second objective is causal invariance consistency, which enforces stability of predictions across multiple do-interventions on style features. This ensures that the model’s predictions remain invariant to scanner- and stain-specific perturbations, thus preserving causal dependence on biologically relevant features.
Formally, the online adaptation objective for a batch of test samples \(x\in {{\mathcal{D}}}_{test}\) is defined as:
where H( ⋅ ) denotes the Shannon entropy of the predictive distribution and \({{\mathcal{L}}}_{{\rm{causal}}}\) is a KL-divergence-based consistency loss that compares predictions from original and intervened representations. The hyperparameter λ balances the trade-off between confidence maximization and causal invariance.
In practice, this optimization is performed in an online manner: as each test batch arrives, gradients are computed with respect to the unsupervised loss above, and model parameters are updated using lightweight optimization steps. This incremental updating scheme allows the model to gradually align with the statistics of the target domain while avoiding catastrophic drift. Importantly, the entire procedure is label-free and requires no access to source training data, making it highly suitable for deployment in clinical settings where privacy and storage constraints are critical.
From a clinical perspective, online adaptation provides an essential safeguard against unpredictable variations in real-world CRC pathology workflows. For instance, if a hospital adopts a new scanner or modifies its staining pipeline, UAD-FM can adapt on the fly to maintain diagnostic accuracy. By combining entropy-driven confidence alignment with causally grounded invariance enforcement, the proposed algorithm delivers both robustness and interpretability, ensuring that automated predictions remain stable and clinically reliable across heterogeneous deployment environments.
Joint learning objectives
To integrate the proposed modules into a coherent training strategy, UAD-FM is optimized under a unified multi-objective framework that balances accuracy, reliability, and robustness. The overall loss function combines four complementary components: classification accuracy, uncertainty decomposition, causal invariance, and calibration. The objective is formulated as:
where \({{\mathcal{L}}}_{{\rm{cls}}}\) is the standard cross-entropy loss supervising diagnostic predictions, \({{\mathcal{L}}}_{{\rm{uncertainty}}}\) regularizes the decomposition of epistemic and aleatoric uncertainty, \({{\mathcal{L}}}_{{\rm{causal}}}\) enforces prediction invariance under causal interventions, and \({{\mathcal{L}}}_{{\rm{calibration}}}\) refines predictive probabilities to improve reliability. The coefficients α, β, γ control the relative contributions of these auxiliary terms.
The classification loss \({{\mathcal{L}}}_{{\rm{cls}}}\) ensures discriminative power on source-domain colorectal cancer (CRC) data, anchoring the model to clinically relevant labels. The uncertainty loss \({{\mathcal{L}}}_{{\rm{uncertainty}}}\) penalizes overconfident predictions on noisy or ambiguous inputs and encourages high epistemic variance in regions where the model lacks sufficient knowledge. The causal loss \({{\mathcal{L}}}_{{\rm{causal}}}\) minimizes the discrepancy between predictions generated under original and intervened style features, thereby removing spurious dependencies on scanner or staining artifacts. Finally, the calibration term \({{\mathcal{L}}}_{{\rm{calibration}}}\), implemented via temperature scaling, aligns model confidence with empirical accuracy, supporting safe decision-making in practice.
The overall training pipeline is summarized in Algorithm 1. We adopt a two-phase strategy: supervised pretraining on labeled source-domain CRC datasets, followed by unsupervised causal adaptation on unlabeled target domains. During pretraining, the backbone and uncertainty head are optimized to jointly minimize \({{\mathcal{L}}}_{{\rm{cls}}}\) and \({{\mathcal{L}}}_{{\rm{uncertainty}}}\), establishing strong discriminative and uncertainty-aware representations. During deployment, as test batches from new domains arrive, causal interventions are applied to style variables and model parameters are updated using entropy minimization and \({{\mathcal{L}}}_{{\rm{causal}}}\), allowing UAD-FM to adapt online without requiring access to source data. After training, post-hoc calibration is applied on a held-out validation set to optimize \({{\mathcal{L}}}_{{\rm{calibration}}}\), further enhancing reliability in clinical use.
This joint optimization strategy ensures that UAD-FM not only achieves high predictive accuracy but also provides calibrated confidence estimates and robust generalization across unseen domains, thereby addressing the key requirements for trustworthy computational pathology in colorectal cancer diagnosis.
Algorithm 1
Training pipeline of UAD-FM
Require: Source dataset \({{\mathcal{D}}}_{s}\), target domain stream \({{\mathcal{D}}}_{t}\)
1: Initialize foundation backbone parameters θ
2: for each epoch do
3: Train backbone with \({{\mathcal{L}}}_{{\rm{cls}}}\) on \({{\mathcal{D}}}_{s}\)
4: Update uncertainty head with \({{\mathcal{L}}}_{{\rm{uncertainty}}}\)
5: for each batch \(x\in {{\mathcal{D}}}_{t}\) do
6: Apply causal intervention do(x)
7: Adapt θ with \({{\mathcal{L}}}_{{\rm{causal}}}+H(p(y| x,\theta ))\)
8: end for
9: end for
10: Calibrate predictions with \({{\mathcal{L}}}_{{\rm{calibration}}}\) on validation data
11: return Adapted parameters θ*
Datasets
We evaluate the proposed UAD-FM framework on a diverse collection of publicly available colorectal cancer (CRC) datasets, chosen to cover a wide range of diagnostic tasks, annotation protocols, and acquisition domains. This multi-dataset setting not only ensures a comprehensive evaluation of the framework but also reflects the heterogeneity of real-world clinical workflows.
The primary dataset used in this study is TCGA-COAD/READ, which contains hematoxylin and eosin (H&E) stained whole-slide images (WSIs) collected from multiple institutions under The Cancer Genome Atlas program. This dataset is used as the source domain for supervised training, with tasks including CRC subtype classification and tumor versus normal discrimination. To further assess generalization under cross-domain settings, we incorporate two external datasets. The first is the CRAG challenge dataset, which provides pixel-level annotations of glandular structures, enabling quantitative assessment of segmentation performance in addition to classification. The second is DigestPath 2019, a multi-scanner dataset designed for pathology image analysis in digestive cancers, which introduces substantial domain shift due to heterogeneous staining and acquisition protocols, making it an ideal benchmark for test-time adaptation.
To support large-scale representation learning, we leverage the NCT-CRC-HE-100K dataset, which consists of 100,000 non-overlapping histology image patches curated from CRC tissue samples and annotated into nine tissue categories. This dataset provides abundant patch-level supervision for pretraining. Finally, we include the LC25000 dataset, which contains 25,000 histology image patches across lung and colon cancers, of which 5,000 correspond to CRC. This dataset is primarily used as a supplementary source for binary classification of benign versus malignant patches.
Table 6 summarizes the datasets used in this study, including their scale, annotation type, and experimental role. The combination of large-scale WSIs, patch-level datasets, and multi-institutional collections ensures that our evaluation captures both within-domain performance and robustness to domain shifts.
Task definition
The experiments are designed to comprehensively evaluate UAD-FM across three complementary tasks that jointly reflect key aspects of colorectal cancer (CRC) pathology analysis.
First, classification tasks address both CRC subtype prediction and tumor versus normal discrimination. These tasks are directly aligned with clinical diagnostic needs, as subtype classification is critical for treatment planning and prognosis, while tumor/normal discrimination underpins automated screening pipelines. Classification is primarily conducted on TCGA-COAD/READ for supervised training and validation, with additional evaluation on LC25000 and NCT-CRC-HE-100K to test generalization across patch-level datasets. Performance is quantified using AUROC, accuracy, and F1-score to capture both discriminative power and balanced prediction across classes.
Second, segmentation tasks are performed using the CRAG dataset, which provides pixel-level annotations of glandular structures. Accurate delineation of glands is crucial for CRC grading, as gland morphology is a well-established histopathological marker of tumor aggressiveness. We assess the ability of UAD-FM to recover fine-grained glandular boundaries, a task that also evaluates how uncertainty decomposition contributes to precise structural localization. Segmentation performance is reported using Dice coefficient and Intersection-over-Union (IoU), reflecting pixel-level agreement between predictions and ground truth.
Third, cross-domain adaptation tasks are designed to evaluate the robustness of UAD-FM under real-world domain shifts. Specifically, we assess generalization when transferring from TCGA (source domain) to external datasets such as DigestPath 2019 and CRAG (target domains). These datasets introduce substantial heterogeneity due to different scanners, staining protocols, and annotation styles, thereby simulating realistic multi-center deployment scenarios. Test-time adaptation performance is reported using relative improvements in accuracy and AUROC (ΔAcc, ΔAUROC), quantifying how well UAD-FM mitigates performance degradation in unseen environments.
Table 7 summarizes the task definitions, datasets, and corresponding evaluation metrics used in this study, highlighting the complementary roles of classification, segmentation, and cross-domain adaptation in assessing both performance and robustness.
Evaluation metrics
To ensure a comprehensive and clinically meaningful evaluation, we adopt multiple quantitative metrics tailored to the different experimental tasks of classification, uncertainty quantification, and cross-domain adaptation. These metrics are selected not only to measure predictive accuracy but also to assess calibration, reliability, and robustness under domain shift, which are critical for real-world colorectal cancer (CRC) deployment.
For classification tasks, we report Area Under the Receiver Operating Characteristic Curve (AUROC), accuracy, and F1-score. AUROC provides a threshold-independent measure of discriminative power and is particularly robust under class imbalance, a common issue in CRC subtyping. Accuracy offers an intuitive measure of overall correctness, while F1-score balances precision and recall, ensuring that minority classes such as rare CRC subtypes are properly evaluated. Together, these metrics provide a holistic view of classification performance.
For uncertainty quantification, we employ Expected Calibration Error (ECE), Brier Score, and Negative Log-Likelihood (NLL). ECE measures the discrepancy between predicted confidence and actual accuracy, with lower values indicating better-calibrated predictions, a prerequisite for safe clinical deployment. Brier Score captures the mean squared difference between predicted probabilities and ground-truth outcomes, reflecting both calibration and sharpness of predictions. NLL directly evaluates the quality of probabilistic predictions, penalizing overconfident but incorrect outputs. These metrics together quantify whether UAD-FM produces not only accurate but also trustworthy probability estimates.
For cross-domain adaptation, we emphasize relative improvements in classification performance across unseen target domains. Specifically, we report changes in accuracy and AUROC (ΔAcc and ΔAUROC) relative to source-only baselines. These differential metrics highlight the contribution of test-time adaptation and causal interventions in mitigating performance degradation under distributional shift. By quantifying improvement margins rather than absolute values alone, we directly measure the robustness benefits brought by UAD-FM in heterogeneous, multi-center CRC settings.
Table 8 summarizes the evaluation metrics used in this study, categorized by experimental task, with their clinical and methodological significance.
Ethics approval and consent to participate
This study exclusively uses publicly available colorectal cancer pathology datasets and does not involve any new experiments with human participants or animals performed by any of the authors. Therefore, additional ethical approval and consent were not required.
Materials availability
No new biological or chemical materials were generated or analyzed in this study.
Data availability
All imaging datasets analyzed in this study are publicly accessible. Specifically, the TCGA-COAD and TCGA-READ whole-slide images are available from the Genomic Data Commons (https://portal.gdc.cancer.gov/) and The Cancer Imaging Archive (https://www.cancerimagingarchive.net/). The CRAG dataset can be accessed via the colorectal adenocarcinoma gland segmentation challenge documentation. The DigestPath 2019 dataset is available on the Grand Challenge platform (https://digestpath2019.grand-challenge.org/). The NCT-CRC-HE-100K dataset is provided by NCT Biobank (https://zenodo.org/record/1214456). The LC25000 dataset is freely available for download from Kaggle (https://www.kaggle.com/datasets/andrewmvd/lung-and-colon-cancer-histopathological-images). Processed or derived data supporting the findings of this study are available from the corresponding author upon reasonable request.The code used in this study will be made available upon reasonable request to the corresponding author after the paper is accepted.
Code availability
The code used in this study will be made available upon reasonable request to the corresponding author after the paper is accepted.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Siegel, R. L. et al. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 70, 145–164 (2020).
Siegel, R. L. et al. Colorectal cancer statistics, 2017. CA Cancer J. Clin. 67, 177–193 (2017).
Zhang, H. et al. Identification and verification of key genes in colorectal cancer liver metastases through analysis of single-cell sequencing data and TCGA data. Ann. Surgical Oncol. 31, 8664–8679 (2024).
Gustav, M. et al. Assessing genotype-phenotype correlations in colorectal cancer with deep learning: a multicentre cohort study. The Lancet Digital Health (2025).
Loeffler, C. M. et al. Hibrid: histology-based risk-stratification with deep learning and ctdna in colorectal cancer. Nat. Commun. 16, 7561 (2025).
Pham, H.-H. et al. Learning disentangled stain and structural representations for semi-supervised histopathology segmentation. In Proceedings of the Workshop on Computational Pathology (COMPAYL) at MICCAI 2025 (2025) (Accepted/In press).
Yang, Z. et al. A foundation model for generalizable cancer diagnosis and survival prediction from histopathological images. Nat. Commun. 16, 2366 (2025).
Zhou, X. et al. A knowledge-enhanced pathology vision-language foundation model for cancer diagnosis. arXiv preprint arXiv:2412.13126 (2024).
Wang, D., Shelhamer, E., Liu, S., Olshausen, B. & Darrell, T. Tent: Fully test-time adaptation by entropy minimization. In International Conference on Learning Representations (ICLR, 2020).
Chen, D., Wang, D., Darrell, T. & Ebrahimi, S. Contrastive test-time adaptation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 295–305 (2022).
Gal, Y. & Ghahramani, Z. Dropout as a Bayesian approximation: Representing model uncertainty in deep learning. In international conference on machine learning, 1050–1059 (PMLR, 2016).
Ojha, J., Presacan, O., G. Lind, P., Monteiro, E. & Yazidi, A. Navigating uncertainty: A user-perspective survey of trustworthiness of ai in healthcare. ACM Trans. Comput. Healthc. 6, 1–32 (2025).
Atf, Z. et al. The challenge of uncertainty quantification of large language models in medicine. arXiv preprint arXiv:2504.05278 (2025).
Schölkopf, B. et al. Toward causal representation learning. Proc. IEEE 109, 612–634 (2021).
Prosperi, M. et al. Causal inference and counterfactual prediction in machine learning for actionable healthcare. Nat. Mach. Intell. 2, 369–375 (2020).
El Nahhas, O. S. et al. From whole-slide image to biomarker prediction: end-to-end weakly supervised deep learning in computational pathology. Nat. Protoc. 20, 293–316 (2025).
Li, H. et al. Systematic review and meta-analysis of deep learning for MSI-H in colorectal cancer whole slide images. npj Digital Med. 8, 456 (2025).
Baumann, E. et al. Aligning computational pathology with clinical practice: A systematic review of AI tools for pathology report elements in colorectal cancer. medRxiv 2025–06 (2025).
Ferber, D. et al. Development and validation of an autonomous artificial intelligence agent for clinical decision-making in oncology. Nat. Cancer 1–13 (2025).
Lv, C. & Wu, Y. Pathomics in gastrointestinal tumors: Research progress and clinical applications. Cureus17 (2025).
Wang, Z. et al. Pathological omics prediction of early and advanced colon cancer based on artificial intelligence model. Discov. Oncol. 16, 1330 (2025).
Fountzilas, E., Pearce, T., Baysal, M. A., Chakraborty, A. & Tsimberidou, A. M. Convergence of evolving artificial intelligence and machine learning techniques in precision oncology. NPJ Digital Med. 8, 75 (2025).
Tiwari, A. et al. The current landscape of artificial intelligence in computational histopathology for cancer diagnosis. Discov. Oncol. 16, 1–25 (2025).
Petäinen, L. et al. Multi-scale ensemble model for DMMR prediction from histopathological images of colorectal cancer (2025).
Hölscher, D. L. & Bülow, R. D. Decoding pathology: the role of computational pathology in research and diagnostics. Pflügers Arch. -Eur. J. Physiol. 477, 555–570 (2025).
Yang, Z. et al. Adpv2: A hierarchical histological tissue type-annotated dataset for potential biomarker discovery of colorectal disease. arXiv preprint arXiv:2507.05656 (2025).
Ballou, M. & Harrouzi, A.Colorectal Cancer: Segmentation and Detection via Deep Learning Detection. Ph.D. thesis, Université Ghardaia (2024).
Sultana, R., Horibe, H., Murakami, T. & Shimizu, I. Cancer cytoplasm segmentation in hyperspectral cell image with data augmentation. arXiv preprint arXiv:2507.03325 (2025).
Hibi, L., Mekadid, H. A. & Beddad, F. Predict Microsatellite Instability (MSI) and Microsatellite Stability (MSS) status in gastrointestinal cancers. Ph.D. thesis (2025).
Cheng, Y. et al. Synergistic H&E and IHC image analysis by AI predicts cancer biomarkers and survival outcomes in colorectal and breast cancer. Commun. Med. 5, 328 (2025).
Swanson, N., Castro, M. A., Robertson, A. G., Shmulevich, I. & Tercan, B. Predicting microsatellite instability from whole slide images using texture features. bioRxiv 2025–03 (2025).
Keven, A. et al. Lymph node morphology as an MRI biomarker for microsatellite instability in rectal cancer. Abdominal Radiology 1–11 (2025).
Wang, Y. et al. Gene swin transformer: new deep learning method for colorectal cancer prognosis using transcriptomic data. Brief. Bioinforma. 26, 275 (2025).
Acknowledgements
This work was supported by the Heilongjiang Provincial Higher Education Institutions Collaborative Innovation Cultivation Project(LJGXCG2023-087), the Harbin Medical University Cancer Hospital Ascend Leading Disciplines Plan (PDYS-2024-14), the Heilongjiang Provincial Natural Science Foundation of China (LH2023H096), the Postdoctoral research project in Heilongjiang Province (LBHZ22210), the China Postdoctoral Science Foundation (2023MD744213), the Scientific research project of Heilongjiang Provincial Health Commission (20230404080339).
Author information
Authors and Affiliations
Contributions
S.L., G.M., X.Z. and H.W. had full access to all data in the study and took responsibility for the integrity of the data and the precision of the data analysis (Validation, formal analysis). H.L., X.Z., C.L. and S.M. contributed to the conception of the research and the development of the study design (conceptualization, methodology) and participated in the acquisition of funding to support the project (acquisition of funds). K.M., H.L. and X.Z. were responsible for the acquisition, curation and investigation of research questions (investigation, data curation) and provided the necessary materials, instruments, and technical resources (resources). M.Y., H.X. and Y.H. contributed to the development and implementation of analytical procedures, including the use and adaptation of relesvant software tools (Software). H.M. and L.C. prepared the initial draft of the manuscript and contributed to the visualization of the data for the presentation of the results (Writing - Original draft, Visualization). P.H. supervised the overall progress of the study and managed the coordination among the research team (Supervision, Project administration). All authors participated critically in reviewing and editing the manuscript for important intellectual content (writing—review and editing) and approved the final version for submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lou, S., Mo, G., Zhang, X. et al. Uncertainty-aware and causal test-time adaptive foundation model for robust colorectal cancer pathology diagnosis. npj Digit. Med. 8, 784 (2025). https://doi.org/10.1038/s41746-025-02149-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-02149-1