Abstract
As large language models (LLMs) become increasingly integrated into clinical decision-making, ensuring trustworthy reasoning is paramount. However, current evaluation strategies of LLMs’ medical reasoning capability either suffer from unsatisfactory assessment or poor scalability, and a rigorous benchmark remains absent. To address this, we present MedThink-Bench, a benchmark designed for rigorous and scalable assessment of LLMs’ medical reasoning. MedThink-Bench comprises 500 high-complexity questions spanning ten medical domains, accompanied by expert-authored, step-by-step rationales that elucidate intermediate reasoning processes. Further, we introduce LLM-w-Rationale, an evaluation framework that combines fine-grained rationale assessment with an LLM-as-a-Judge paradigm, enabling expert-level fidelity in evaluating reasoning quality while preserving scalability. Results show that LLM-w-Rationale correlates strongly with expert evaluation (Pearson coefficient up to 0.87) while requiring only 1.4% of the evaluation time. Overall, MedThink-Bench establishes a rigorous and scalable standard for evaluating medical reasoning in LLMs, advancing their safe and responsible deployment in clinical practice.
Similar content being viewed by others
Introduction
Large language models (LLMs) have made remarkable progress in clinical decision-making, demonstrating the ability to perform complex reasoning tasks such as disease diagnosis1,2, treatment planning3, and patient management4,5. Despite their impressive capabilities, the opaque and black-box nature of LLMs limits their reliability in high-stakes clinical scenarios6,7. For instance, an LLM may arrive at the correct diagnosis based on parametric knowledge without providing evidence grounded in clinical guidelines or considering a comprehensive differential diagnosis8,9,10. Moreover, LLMs are prone to hallucinations, generating plausible but factually incorrect information that can mislead clinical decision-making11,12,13. Such behavior poses potential risks to patient safety and undermines the reliability of clinical workflows14. Therefore, deploying LLMs in clinical practice requires not only high prediction accuracy but also transparent and explainable reasoning processes15.
Evaluating the medical reasoning capabilities of LLMs is crucial for establishing trust and ensuring safe integration into healthcare settings16,17,18. Recent efforts in this direction have followed two main approaches. The first approach involves assessing performance on complex medical exercises, such as multiple-choice questions (MCQs), by measuring prediction accuracy19,20,21. While this method aligns with the prevailing approaches to evaluating LLMs’ medical capabilities and offers a coarse measurement22,23, it fails to capture the depth and validity of the reasoning process that underpins clinical decisions and cannot identify flawed reasoning24,25. The second approach focuses on evaluating the rationales provided by LLMs, which can well address the above issues. Within this category, evaluation strategies can be further classified into three types: (1) text-similarity metrics, which compare LLM-generated rationales with reference rationales26,27; (2) human expert evaluation, which relies on domain experts’ manual efforts to assess the reasoning quality15,25,28,29; and (3) LLM-as-a-Judge, where a separate LLM is used to assess the quality of the reasoning process30,31.
Despite these advances, existing evaluation strategies either suffer from unsatisfactory assessment or poor scalability. Specifically, while conventional text similarity metrics are scalable and cost-efficient6, such as those based on lexical-level overlap (e.g., BLUE32, ROUGE33) or semantic similarity (e.g., BERTScore34), they fail to capture medical semantics or nuanced logics and lack robustness to the variance of the text’s expression styles6. In contrast, human evaluation remains the gold standard for assessing factuality and nuance but is labor-intensive and limited in scale25. Additionally, LLM-as-a-Judge offers a scalable alternative and can comprehend medical knowledge30,35, yet it is vulnerable to hallucinations and evaluative bias36. As such, the challenge of conducting automated, scalable evaluations of LLMs’ medical reasoning while maintaining expert-level factuality remains unresolved. A further barrier is the lack of a benchmark designed to evaluate LLM-generated medical rationales rigorously. Existing datasets often suffer from narrow clinical scenarios37 or rely on LLM-generated rationales as reference answers31,37,38, which may involve incorrect knowledge or flawed rationale. This is because the credibility of such artificial intelligence (AI)-generated rationales is uncertain9,39,40, and the alignment with human expert judgment remains unclear41.
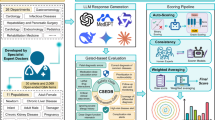
To address these gaps, we introduce MedThink-Bench, a benchmark tailored for rigorous, explainable, and scalable evaluation of LLMs’ medical reasoning (Fig. 1). MedThink-Bench comprises 500 challenging medical question-answer (QA) pairs across 10 representative domains, each annotated by medical professionals with fine-grained, step-by-step reasoning trajectories that mirror real-world clinical logic. Building on this resource, we propose LLM-w-Rationale, an evaluation framework that integrates the expert-curated fine-grained rationales with an LLM-as-a-Judge mechanism, thus combining their complementary strengths. Specifically, by calibrating the LLM-based evaluator with nuanced reasoning trajectories, our framework can accurately assess the intermediate reasoning to achieve expert-level factual consistency while maintaining scalability.
a Data collection. Medical questions were sourced from ten publicly available datasets, each accompanied by ground-truth answers. b Data preprocessing. Duplicate entries and questions involving medical images were removed. Medical experts then manually curated a subset of complex questions requiring multi-step reasoning. c Expert annotation. A team of ten medical experts annotated the questions into ten distinct medical domains and collaboratively generated fine-grained reasoning trajectories through consensus. d Medical reasoning evaluation. We rigorously evaluated the medical reasoning capabilities of twelve LLMs, comparing them against expert evaluations, text-similarity metrics, LLM-as-a-Judge, and reference-based LLM-as-a-Judge (LLM-w-Rationale). Additionally, we analyzed the correlation between these automated metrics and expert evaluations. Icons adapted from flaticon.com, used under royalty-free license.
In this study, we demonstrate that LLM-w-Rationale correlates strongly with expert evaluation (Pearson coefficient up to 0.87) and remains robust across different prompts and judge models. Moreover, our benchmark comparison of twelve state-of-the-art LLMs reveals two surprising findings: reasoning performance shows disparity with prediction accuracy, which could truly reflect the medical capability of LLMs; smaller models such as MedGemma-27B42 and HuatuoGPT-o1-70B43 can outperform larger proprietary models like OpenAI-o3 and DeepSeek-R144 in medical reasoning. Our contributions are threefold. First, we address the longstanding challenge of scalable and expert-level evaluation of LLM-generated medical rationales. Second, we construct a high-quality dataset featuring 500 expert-annotated questions with nuanced reasoning trajectories across 10 medical domains. Third, we provide a comprehensive comparison of twelve LLMs in terms of their medical reasoning capabilities. Overall, MedThink-Bench offers a foundational resource for assessing the trustworthiness of LLMs in medical decision-making, thereby advancing their safe and responsible integration into clinical practice.
Results
We developed LLM-w-Rationale, an evaluation framework that integrates expert-curated, fine-grained rationales with the LLM-as-a-Judge paradigm to assess medical reasoning. This section summarizes the key findings, including an overview of MedThink-Bench, comparisons with conventional metrics across twelve LLMs, and analyses of the framework’s fidelity, robustness, and efficiency.
Dataset
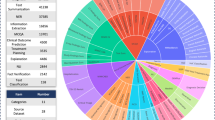
We created MedThink-Bench, a medical QA dataset with expert-derived reasoning annotations, comprising 500 complex questions across 10 medical domains: Pathology, Discharge, Disease Diagnosis, Anatomy & Physiology, Treatment, Public Health, Policy & Ethics, Prognosis, Diagnostic Workup, and Pharmacology. All questions were sourced from publicly available medical QA datasets (Supplementary Note 1). Our expert annotation team manually selected questions requiring multi-step reasoning and provided fine-grained annotations of the underlying reasoning processes (Supplementary Notes 2 and 3). The data statistics are presented in Fig. 2.
Comparison of evaluation metrics on LLM reasoning
We evaluated the reasoning capabilities of twelve LLMs on MedThink-Bench using zero-shot Chain-of-Thought (CoT) prompting45 to assess their generated rationales. Performance was assessed through expert evaluations and eight automated metrics (Fig. 3a). Prediction accuracy results are provided in Supplementary Note 7. As shown in Fig. 3a, the expert evaluation scores ranged from 0.453 (95% confidence interval (CI): 0.419–0.485) for Med42-70B to 0.759 (95% CI: 0.730–0.789) for MedGemma-27B. For the reference-based LLM-as-a-Judge (LLM-w-Rationale), scores ranged from 0.482 (95% CI: 0.450–0.514) for Med42-70B to 0.769 (95% CI: 0.742–0.798) for MedGemma-27B. Meanwhile, the performance of reference-free LLM-as-a-Judge (LLM-w/o-Rationale) ranged from 0.823 (95% CI: 0.812–0.834) for Med42-70B to 0.907 (95% CI: 0.896–0.918) for Qwen3-32B. Among the text-similarity metrics, BLUERT and BERTScore generally outperformed the other metrics. Specifically, BLUERT scores ranged from 0.395 (95% CI: 0.388–0.403) for OpenAI-o3 to 0.599 (95% CI: 0.589–0.608) for MedGemma-27B, while BERTScore ranged from 0.554 (95% CI: 0.551–0.557) for HuatuoGPT-o1-70B to 0.630 (95% CI: 0.625–0.635) for Med42-70B.
a Comparison of overall medical reasoning performance, including expert evaluation, five text-similarity metrics, and the proposed LLM-w-Rationale framework under zero-shot prompting. The automated reasoning assessments were obtained by comparing ground-truth reasoning annotations with the predicted annotations. Error bars represent the 95% CI of the mean, calculated via bootstrapping. b Breakdown of medical reasoning performance across the ten medical domains in the MedThink-Bench dataset.
Domain-specific reasoning performance
LLM performance varied considerably across the 10 medical domains (Fig. 3b). DeepSeek-R1 achieved top performance in Anatomy & Physiology, Public Health, and Treatment, outperforming OpenAI-o3 by 0.047, 0.159, and 0.063, respectively. MedGemma-27B led in four clinically complex domains, Pathology, Diagnostic Workup, Disease Diagnosis, and Discharge, with gains over OpenAI-o3 of 0.075, 0.091, 0.110, and 0.144. HuatuoGPT-o1-70B excelled in Policy & Ethics, Pharmacology, and Prognosis, with an average margin of 0.140 over OpenAI-o3. These findings highlight that model strengths are often domain-specific, and MedThink-Bench enables fine-grained evaluation of domain-specific capabilities.
Correlation analysis between expert and automated evaluation
We computed Pearson correlation coefficients between the expert evaluation scores and all the automated metrics across all LLMs, as shown in Fig. 4a. The results reveal weak correlations for metrics such as BLEU, ROUGE-L, METEOR, BLEURT, and BERTScore, with Pearson coefficients ranging from −0.17 to 0.45. Similarly, LLM-w/o-Rationale showed weak correlation, with coefficients ranging from 0.01 to 0.27. In contrast, LLM-w-Rationale demonstrated a strong correlation with expert evaluations, with Pearson coefficients ranging from 0.68 to 0.87. We further employed Kendall’s tau correlation to assess the concordance between model rankings derived from expert evaluations and those obtained from automated metrics (Fig. 4b). Both LLM-w/o-Rationale (τ = 0.06) and text-similarity metrics (τ ranging from −0.39 to 0) exhibited weak or negative associations with expert-derived rankings. In contrast, LLM-w-Rationale achieved a markedly strong positive correlation with expert assessments (τ = 0.88), indicating its close alignment with human judgment. Additionally, we visualized the individual evaluation scores of the LLMs in Fig. 5 and Supplementary Note 8. The results demonstrated that data points for LLM-w-Rationale generally closely align with the dashed line, indicating agreement with expert scores. In contrast, BLEURT, BERTScore, and LLM-w/o-Rationale show greater divergence from expert evaluations.
a Pearson correlation analysis of predicted rationales against expert assessments and various automated metrics. These metrics include text-similarity measures (BLEU, ROUGE-L, METEOR, BLEURT, BERTScore), LLM-w/o-Rationale (which does not use ground-truth rationales as a reference), and LLM-w-Rationale (which uses our annotated fine-grained rationales as a reference). We take GPT-4o-mini as the judge model. Warmer colors (red tones) denote stronger positive correlations, while cooler colors (blue tones) indicate weaker or negative correlations. The results indicate a strong correlation between LLM-w-Rationale and expert evaluations, while LLM-w/o-Rationale and text-based metrics show weaker correlations with expert assessments. b Kendall’s tau correlation analysis on the ranking of models based on expert evaluations and the automated metrics. Using GPT-4o-mini as the judge model, LLM-w-Rationale demonstrates a very strong positive correlation (τ = 0.88) with expert rankings, whereas LLM-w/o-Rationale (τ = 0.06) and text-similarity metrics (with τ values ranging from −0.39 to 0, indicating negative or weak correlations) show much weaker associations with expert-derived model performance rankings.
The plot includes results from GPT-4o, Llama-3.3-70B, and MedGemma-27B; the judge model in the two LLM-based evaluation metrics is GPT-4o-mini. Each point represents an individual sample, with dashed lines indicating equal performance between expert and automated scores. For LLM-w-Rationale, data points tend to align closely with the dashed line, suggesting strong agreement with expert evaluations. In contrast, BLEURT, BERTScore, and LLM-w/o-Rationale exhibit greater divergence from the dashed line, indicating a weaker alignment with expert assessments.
Stratified discrimination analysis of evaluation metrics
Discriminative power is a key property of an effective evaluation metric. To assess this, we conducted a stratified discrimination analysis, categorizing data samples into three quality levels, i.e., low, medium, and high, based on human evaluation scores. Figure 6 presents a heatmap of p-values from Kruskal–Wallis H tests applied to each evaluation metric. Traditional text similarity metrics such as BLEU and ROUGE-L, as well as LLM-w/o-Rationale, demonstrated limited discriminative capability, with several p-values exceeding the 0.05 threshold, indicating low sensitivity to quality differences. In contrast, LLM-w-Rationale, which incorporates fine-grained rationales, consistently yielded the lowest p-values (p < 0.001) across multiple judge models, highlighting its superior ability to distinguish among the three quality levels.
Human-evaluated samples are grouped into three quality levels: “low score,” “medium score,” and “high score.” For each metric, the test evaluates whether the score distributions significantly differ across these quality strata. Lower p-values (e.g., p < 0.001) indicate stronger discriminative power, reflecting the metric’s ability to distinguish between different levels of medical reasoning quality.
Impact of the judge model in LLM-w-Rationale
We evaluated the robustness of LLM-w-Rationale to the judge model. Specifically, predicted rationales generated by four LLMs (Llama-3.3-70B, OpenAI-o3, Qwen3-32B, and MedGemma-27B) were assessed using 10 different LLMs serving as judge models (Fig. 7a and Supplementary Note 10). As shown in Fig. 7a, when rationales predicted by Llama-3.3-70B were evaluated, the reasoning scores ranged from 0.522 (95% CI: 0.492–0.550) for Gemini-2.5-flash to 0.553 (95% CI: 0.515–0.587) for HuatuoGPT-o1-70B. However, when using smaller models as the judge models, such as Llama-3-8B-Instruct, Llama-3.2-3B-Instruct, and Llama-3.2-1B-Instruct, the performance is higher than that of the large models, which range from 0.704 to 0.891.
a Robustness of LLM-w-Rationale across different judge models. Predicted rationales from Llama-3.3-70B were evaluated using 10 LLMs of varying scales as judge models. b Sensitivity of LLM-w-Rationale to prompt variations. Five semantically similar prompt variations were tested to assess the framework’s robustness. Error bars represent the 95% CI of the mean, calculated via bootstrapping.
Prompt sensitivity in LLM-w-Rationale
We also examined the sensitivity of the LLM-w-Rationale framework to different prompt formulations by composing five prompt variations with similar semantics (Supplementary Note 9). The reasoning performance of each variation was compared using the predicted rationales from Llama-3.3-70B, with GPT-4o-mini fixed as the judge model. As shown in Fig. 7b, the performance varied from 0.538 (95% CI: 0.500–0.571) for the second prompt variant to 0.567 (95% CI: 0.533–0.599) for the fifth prompt.
Efficiency comparison
To evaluate scalability, we compared the time efficiency of different evaluation strategies. Specifically, we recorded the running times for human evaluation, text-similarity metrics, and LLM-w-Rationale evaluations on MedThink-Bench. As shown in Table 1, the average assessment time for text-similarity metrics was 9.0 min, while LLM-w-Rationale took 310.7 min when using HuatuoGPT-o1-70B as the judge model. In contrast, human evaluation required an average of 3708.3 min, significantly longer than the automated metrics.
Case study
Case studies were conducted to demonstrate the effectiveness of LLM-w-Rationale in measuring the medical reasoning capability of LLMs. As shown in Fig. 8, Llama-3.3-70B produced an incorrect answer, yet followed a partially correct medical reasoning trajectory. In another case (Supplementary Note 11), flawed reasoning led to a correct answer. By measuring medical reasoning with expert-curated fine-grained rationales, LLM-w-Rationale captured flawed reasoning patterns and offered a more nuanced evaluation of an LLM’s medical capabilities. These findings underscore the importance of assessing not only the final prediction but also the underlying reasoning process.
This case demonstrates that while the prediction model Llama-3.3-70B produced an incorrect answer, it followed partially correct medical reasoning trajectories (highlighted in red). This underscores the advantage of the LLM-w-Rationale framework, which, in conjunction with the fine-grained rationale annotations in MedThink-Bench, provides a more nuanced evaluation of the medical reasoning abilities of LLMs compared to merely assessing prediction accuracy.
Correlation between reasoning performance and prediction accuracy
We computed Pearson correlation coefficients between the medical QA prediction accuracy and reasoning evaluation metrics (Fig. 9). The results revealed weak or moderate correlations for text-similarity metrics and LLM-w/o-Rationale, with coefficients ranging from −0.04 to 0.40. Similarly, both LLM-w-Rationale and expert evaluation exhibited moderate correlations with expert assessments, with average Pearson correlation coefficients of 0.462 and 0.436, respectively.
Warmer colors (red tones) denote stronger positive correlations, while cooler colors (blue tones) indicate weaker or negative correlations. Most automated metrics (e.g., BLEU, ROUGE-L, METEOR) show weak to moderate positive correlations with prediction accuracy. Overall, both LLM-w-Rationale and expert evaluation exhibit higher positive correlations with prediction accuracy, though not strong. This finding aligns with observations that prediction accuracy alone inadequately captures reasoning quality, as correct multiple-choice answers may stem from flawed reasoning and incorrect answers can contain partially valid rationale.
Analysis of data leakage
Given that MedThink-Bench was constructed from publicly available datasets, we examined potential data contamination that might have occurred during LLM pre-training and could influence evaluation outcomes. Since the rationale annotations were independently curated by medical experts, their leakage was not assessed. As shown in Table 2, MedGemma-27B and Llama-3.3-70B exhibited relatively high data contamination ratios of 0.252 and 0.118, respectively, substantially higher than those observed for Qwen3-32B, Baichuan-M1-14B, and HuatuoGPT-o1-70B. These results indicated that the training data of certain LLMs, particularly MedGemma-27B and Llama-3.3-70B, likely included portions of public medical QA datasets. To evaluate the impact of contamination on reasoning performance and QA performance, we further analyzed a subset of 200 uncontaminated samples. As shown in Fig. 10 and Supplementary Fig. 1, reasoning performance remained largely consistent across all models, indicating minimal influence from data leakage. In contrast, QA prediction accuracy declined notably in a few models (e.g., MedGemma-27B and Llama-3.3-70B). Overall, these findings suggest that while data contamination may affect QA prediction to some extent, it does not substantially influence the reasoning evaluation or alter the main conclusions of this study.
The bar plot illustrates reasoning scores obtained via LLM-w-Rationale on the full dataset (blue bars) and an uncontaminated subset (red bars). Error bars represent the 95% CI of the mean, calculated via bootstrapping. Reasoning performance shows minimal variation between the full dataset and the uncontaminated subset across models, indicating limited influence of data leakage on reasoning evaluation.
Error analysis of reasoning evaluation
We conducted an error detection analysis to quantify the degree of divergence between the LLM-w-Rationale framework and expert evaluations. Expert assessments of rationale correctness were used as the ground truth. As summarized in Table 3, the proposed LLM-w-Rationale framework demonstrated strong concordance with expert evaluation. Across 12 tested models, Precision, Recall, and F1-scores were all ≥0.755, with overall averages of 0.849, 0.839, and 0.843, respectively. These results indicate that the framework makes only minor classification errors and maintains high consistency with human judgment in medical reasoning assessment. To further understand sources of disagreement, we analyzed representative error cases where LLM-w-Rationale’s evaluations diverged from expert assessments. As shown in Fig. 11, a false-positive occurred when LLM-w-Rationale assigned a higher score than experts because it treated a tentative reasoning path as correct, overlooking its later rejection. Conversely, a false-negative case (Supplementary Note 12) arose when the judge model lowered the score, assuming key information was missing, while experts agreed that the reasoning contained essential elements and should be marked correct.
Discussion
In this study, we presented MedThink-Bench, a curated dataset comprising complex medical questions spanning 10 clinical scenarios, each accompanied by fine-grained rationale annotations from domain experts. MedThink-Bench was designed to address a critical challenge in evaluating LLMs for medical reasoning: enabling automated assessment while maintaining expert-level factual accuracy. The key findings and insights are summarized below.
First, MedThink-Bench effectively differentiated the reasoning capabilities of the LLMs (Fig. 3a). Our benchmarking results showed that MedGemma-27B, HuatuoGPT-o1-70B, and DeepSeek-R1 achieved the highest overall reasoning performance. Surprisingly, smaller open-source models like MedGemma-27B outperformed larger commercial counterparts, including OpenAI-o3 and DeepSeek-R1. Likewise, Qwen3-32B significantly outperformed Gemini-2.5-flash and DeepSeek-R1 (p < 0.001). Among commercial LLMs, DeepSeek-R1, OpenAI-o3, and Gemini-2.5-flash emerged as the top performers, substantially surpassing GPT-4o and Claude-sonnet-3.5. The domain-wise breakdown (Fig. 3b) further highlighted nuanced differences in model capabilities across clinical areas, offering practical guidance for model selection in clinical applications.
Second, the proposed evaluation strategy, LLM-w-Rationale, exhibited strong alignment with expert evaluations (Fig. 4a, b). Specifically, the Pearson correlation coefficients between LLM-w-Rationale and expert scores ranged from 0.68 to 0.87, while Kendall’s tau correlation for model ranking reached τ = 0.88, both indicating a strong concordance with expert judgment. Visualization of the score distributions (Fig. 5) further confirmed this consistency. Importantly, LLM-w-Rationale demonstrated significantly higher efficiency compared to human evaluation (Table 1) and was cost-effective, making it well-suited for scalable, automated evaluation. Together, these results demonstrated that integrating fine-grained rationale annotations as references within the LLM-as-a-Judge paradigm enabled efficient, reliable, and expert-aligned evaluation of medical reasoning.
Third, our analysis revealed that conventional reasoning evaluation methods, including text similarity metrics and LLM-w/o-Rationale, showed weak correlation with expert judgments (Fig. 4a). This was primarily because metrics such as BLEU and ROUGE relied on surface-level word overlap and failed to capture semantic or logical equivalence. Although BERTScore incorporated word embeddings, it remained limited in two key aspects. First, it operates at the word level rather than reasoning-chain level. Second, it cannot comprehend the underlying logical structure of complex medical justifications. As a result, these metrics often produced narrow score ranges (Fig. 5), since LLMs tended to repeat contextual information, generating superficially plausible rationales that were misinterpreted as accurate (Supplementary Figs. 5 and 6). In contrast, LLM-w/o-Rationale lacked access to ground-truth rationales and highly depended on the judge model’s capacity and biases, undermining its reliability. Overall, these findings underscore the limitations of existing strategies for assessing medical reasoning performance.
Additionally, LLM-w-Rationale exhibited superior discriminative ability in evaluating reasoning quality (Fig. 6). It assigned significantly different scores to outputs of the three human-rated quality levels (low, medium, and high scores), demonstrating its ability to distinguish reasoning levels. In contrast, text-similarity metrics, such as METEOR and BERTScore, and LLM-w/o-Rationale failed to make such distinctions. These findings empirically support the discriminative validity of the LLM-w-Rationale as a more reliable, human-aligned evaluation method for medical reasoning.
We further demonstrated that LLM-w-Rationale was robust to variations in both the judge model and prompt phrasing. As shown in Fig. 7a and Supplementary Fig. 3, evaluation outcomes remained stable when using LLMs with strong instruction-following capabilities, such as GPT-4o-mini or MedGemma-27B, as judge models. This robustness likely stems from these models’ ability to faithfully execute instructions (Supplementary Note 6), such as rejecting rationales that deviate from or contradict the reference standard (Supplementary Figs. 5 and 6). To secure evaluation reliability, we recommend selecting judge models with demonstrated instruction-following proficiency. Likewise, results in Fig. 7b indicated that prompt variants with similar semantics yielded consistent evaluation outcomes, suggesting robustness to prompt engineering. This stability enhanced the practical applicability of LLM-w-Rationale for widespread use in reasoning evaluation.
Finally, we observed a discrepancy between reasoning performance and prediction accuracy (Figs. 3a, b and 9, and Supplementary Fig. 1). For instance, while OpenAI-o3 underperformed MedGemma-27B and HuatuoGPT-o1-70B in reasoning (Fig. 3a, b), it achieved the highest prediction accuracy at 0.692 (95% CI: 0.652–0.732), compared to 0.384 (95% CI: 0.342–0.426) and 0.49 (95% CI: 0.446–0.532), respectively. This divergence could be attributed to two factors. First, incorrect multiple-choice answers sometimes contained partially valid reasoning, which was captured by rationale-based evaluation but not prediction accuracy (Fig. 8). Second, some LLMs produced correct multiple-choice answers through flawed reasoning, leading to inflated accuracy despite weak justifications (Supplementary Figs. 5 and 6). These findings underscore the limitations of the prediction accuracy in evaluating medical reasoning. In contrast, rationale-based evaluation provides a more nuanced and faithful representation of an LLM’s reasoning quality, offering deeper insights into its alignment with expert-level thinking.
Despite these advances, our study has several limitations. First, the LLM-w-Rationale framework currently assesses only two critical dimensions of reasoning: correctness and comprehensiveness, while other dimensions, such as fairness, potential harm, and readability, remain unexplored14,46. Future research should investigate methods to systematically evaluate these additional dimensions to better align with expert-level expectations. Second, although LLM-w-Rationale correlates highly with expert evaluation (Figs. 4a, b and 5), discrepancies remain, manifesting as false-negative or false-positive cases (Fig. 11 and Supplementary Fig. 7). These could potentially be mitigated by employing a more capable judge model with stronger instruction-following abilities47 or by integrating advanced prompt-engineering strategies23,26. Third, our evaluation framework relies on expert-curated annotations as reference standards, which are labor-intensive and consequently limit the size of the MedThink-Bench dataset. Future work may explore reference-free approaches to achieve high-fidelity and scalable assessments.
In summary, this study addressed the critical challenge of accurately and efficiently evaluating the medical reasoning capabilities of LLMs. To this end, we curated MedThink-Bench, a medical QA dataset covering 10 clinical scenarios with fine-grained rationale annotations from domain experts. We introduced an evaluation framework, LLM-w-Rationale, that combines the nuanced annotations with the LLM-as-a-Judge paradigm to enable automated yet expert-aligned reasoning assessment. Using this framework, we benchmarked the reasoning performance of twelve LLMs and systematically compared multiple evaluation strategies. Our findings demonstrated that LLM-w-Rationale achieved strong concordance with expert assessments while offering substantially greater efficiency. Overall, this work provided a robust and scalable solution for evaluating LLMs’ medical reasoning, reducing the burden of manual evaluation and paving the way for their integration into clinical practice.
Methods
Data curation
We collected comprehensive medical questions from 10 existing datasets, filtered out hard questions that required multi-step reasoning (Supplementary Note 2), and annotated the reasoning rationales by experts. Specifically, the medical QA datasets from which we collected questions included MedBullets48, MMLU-Pro49, MedExQA50, MedXpertQA21, Humanity’s Last Exam51, MedQA-USMLE52, PubMedQA53, MedMCQA54, MMLU-Medicine55, and HEAD-QA56. The statistics of these datasets are shown in Fig. 2. Then, we pre-processed the collected data by removing duplicates and filtering out the questions involving images for prediction.
Data annotation
We built a well-annotated dataset to facilitate automated expert-level reasoning evaluation. The medical questions were divided into 10 medical domains: Pathology, Discharge, Disease Diagnosis, Anatomy & Physiology, Treatment, Public Health, Policy & Ethics, Prognosis, Diagnostic Workup, and Pharmacology. We employed 10 medical experts to curate the dataset manually. Two independent physicians annotated each medical question. When disagreement existed in the annotation, a third physician examined the case and made the final annotation. We checked the inter-annotator agreement on the reasoning and question types (Supplementary Note 3).
Evaluation framework
To evaluate medical rationales, we applied three evaluation strategies: human evaluation, LLM-as-a-Judge, and text-similarity metrics. Notably, we proposed an evaluation framework, LLM-w-Rationale, that enabled scalable, step-level assessment of clinical reasoning abilities of LLMs. Built upon MedThink-Bench’s nuanced rationales, LLM-w-Rationale compared model-generated rationales against expert-annotated reasoning trajectories to evaluate their logical correctness and stepwise completeness. Additionally, the prediction accuracy was computed to compare LLMs’ medical capability. The following section describes the implementation and evaluation protocol in detail.
To validate the reliability of our evaluation framework, we conducted a human evaluation and compared its results with those of automated metrics. Specifically, for each question, domain experts were provided with the medical question, answer, model-generated rationale, and a grading scheme (Supplementary Note 5), which functioned as a protocol to ensure consistency and uniformity in assessment. Experts examined each rationale step-by-step, recording (1) the number of reasoning steps they considered necessary to reach the correct answer for that question, and (2) how many of those necessary steps were present in the model’s rationale. The instance-level reasoning score was defined as:
Here \({r}_{{model}}^{(i)}\) is the model-generated rationale for the question \({q}^{\left(i\right)}\), and \({ExpertRequired}({q}^{(i)})\) is the number of reasoning steps the experts deem necessary for that question in real-time assessment, while \({ExpertCovered}{(r}_{{model}}^{(i)},{q}^{(i)})\) denotes the number of those required steps that experts judge to appear in the model’s rationale.
To achieve fine-grained and scalable evaluation aligned with expert reasoning, we adopted a reference-based LLM-as-a-Judge approach (LLM-w-Rationale) that operates at the reasoning-step level. For each question, expert-annotated rationales consist of discrete reasoning steps provided by medical specialists during dataset construction. To assess step-level correctness, the judge model receives the question, the model-generated rationale (as a whole), and each expert reasoning step individually. It is then prompted to determine whether the generated rationale adequately supports each expert step. This design enables robust evaluation even when the model and expert rationales differ in length or granularity. Instead of enforcing step-by-step alignment, we adopt a one-to-many comparison, where each expert step is independently matched against the complete model rationale. This grading scheme differs from prior studies and is customized for complex medical reasoning evaluation. The instance-level reasoning score is calculated as the proportion of expert steps that are successfully supported by the model rationale:
Here \({{S}^{(i)}}_{{expert}}\) is the set of expert-annotated reasoning steps for the question \({q}^{\left(i\right)}\), and \({{r}^{(i)}}_{{model}}\) is the model-generated rationale, as previously defined.
To examine the necessity of expert-annotated rationales for accurate evaluation, we implemented a commonly adopted baseline: LLM-as-a-Judge without reference. This setting allowed us to compare grounded and ungrounded evaluations and to better understand the role of reference rationales in ensuring the reliable assessment of clinical reasoning. Specifically, in this setting, the judge model was provided only with the medical question, answer, and the model-generated rationale. The judge model was first asked to estimate the number of reasoning steps required to answer the question, and then to determine how many of those steps were sufficiently supported by the model-generated rationale (Supplementary Note 6). The instance-level score was calculated as:
\({LLMRequired}\left({q}^{\left(i\right)}\right)\) denotes the number of reasoning steps the LLM estimates are necessary to answer the question \({q}^{\left(i\right)}\), \({LLMCovered}\left({r}_{{model}}^{(i)},{q}^{(i)}\right)\) and indicates how many of those steps the LLM judges to be sufficiently reflected in the rationale.
To contextualize our evaluation framework, we reported baseline performance using widely adopted metrics, including BLEU32, ROUGE-L33, METEOR57, BLEURT58, and BERTScore34. Metrics such as BLEU and ROUGE-L rely on surface-level token overlap, using n-gram precision or longest common subsequence to quantify similarity. While computationally efficient, they are insensitive to paraphrasing and semantic equivalence. More recent metrics like METEOR, BLEURT, and BERTScore incorporate semantic information through synonym matching, pretrained language models, or human-annotated supervision. Although these approaches better capture general linguistic similarity, they remain limited in evaluating factual accuracy, logical soundness, and clinical relevance. While these metrics offer convenient and scalable evaluation, they do not explicitly assess the logical structure, clinical validity, or step-by-step correctness of medical reasoning. Therefore, they are used here primarily for baseline comparison.
For all step-level evaluation methods described above, we reported both instance-level and dataset-level scores. Given instance-level reasoning scores \(R(i)\) for each question, \(i=1,...,N,\) the final dataset-level score was computed as the average:
Notably, an effective reasoning evaluation should faithfully reflect the model’s genuine performance rather than pursue higher absolute “scores.” Therefore, we prioritized the evaluation methods that best aligned with human assessments, rather than those yielding the highest numerical results.
Apart from reasoning evaluation, we followed related studies and evaluated the performance of the predicted answer with accuracy using an exact match. Formally, the instance-level accuracy is:
where \({{a}_{{pred}}}^{(i)}\) and \({{a}_{{gold}}}^{(i)}\) denotes the predicted and golden answer for the ith question, the indicator function 1(⋅) returns 1 if the condition is satisfied, and 0 otherwise. The final accuracy is the average over the dataset:
For each sample in MedThink-Bench, we extracted the model-generated rationale and final answer using rule-based regular expressions. In the human evaluation, 10 medical professionals assessed whether each annotated reasoning step was present in the generated rationale. For automatic evaluation, we employed GPT-4o-mini as the judge model (LLM-w-Rationale), with prompts detailed in Supplementary Note 6. The model received the medical question, the generated rationale, and the fine-grained reasoning trajectories. Decoding was performed with a temperature of 0.1, a commonly adopted setting in LLM-as-a-Judge evaluations59, using a maximum decoding length of 4096 tokens and a fixed random seed (42) for reproducibility. In the reference-free setting, all parameters remained unchanged. For text similarity evaluation, we computed BLEU, ROUGE-L, METEOR, BERTScore, and BLEURT using standard toolkits with default configurations.
LLM baselines
We benchmarked representative LLMs from both commercial closed-source LLMs and open-source LLMs. Since this work aimed to assess the medical reasoning capabilities of LLMs, we included both reasoning and non-reasoning LLMs from each category. Specifically, the commercial closed-source LLMs included GPT-4o60, OpenAI-o3, Claude-3.5-sonnet61, Gemini-2.5-flash62, DeepSeek-R144, and open-source LLMs include Baichuan-M1-14B63, HuatuoGPT-o1-70B64, MedGemma-27B42, Med42-70B65, Llama-3.3-70B66, Qwen3-32B67, and QwQ-32B68. In the experiment, we obtained responses from closed-source LLMs through APIs. For open-source LLMs, we used the package vLLM to load them for faster inference. For inference, we adopted zero-shot CoT to prompt LLMs to generate medical reasoning (Supplementary Note 4).
The LLM inference configuration employs carefully selected hyperparameters to ensure optimal performance and reproducibility. The temperature parameter is set to 0, implementing deterministic sampling to eliminate randomness in token selection and ensure consistent outputs across multiple runs. A fixed random seed of 42 is specified to guarantee reproducible results, which is essential for scientific rigor and experimental validation. The maximum token limit is configured to 4096, providing sufficient generation capacity for comprehensive reasoning. These parameter settings collectively establish a controlled inference environment that prioritizes consistency and reliability over creative variation, aligning with the systematic evaluation requirements of our benchmarking framework.
Stratified discrimination analysis
To assess the discriminative power of each evaluation metric, we conducted a stratified discrimination analysis, which evaluates how well a metric distinguishes between system outputs of varying quality. Specifically, the data samples were stratified into three quality levels, low (0.1–0.4), medium (0.4–0.6), and high (0.6–0.9), based on the human evaluation scores. Then, for each evaluation metric, we computed the average metric scores within each human quality stratum. To statistically assess whether the metric scores significantly differ across the three quality levels, we applied the Kruskal–Wallis H test69. Lower p-values (especially those <0.001) indicate stronger stratified discrimination capability, that is, the metric assigns significantly different scores to outputs of different human-rated quality levels. Metrics with better stratified discrimination can more reliably reflect human judgment and are therefore more suitable for practical evaluation scenarios.
Data leakage analysis
To ensure MedThink-Bench evaluated clinical reasoning rather than memorized content, we adopted the CDD framework70 to detect potential data leakage. Although our benchmark has no direct overlap with public datasets, some medical knowledge (e.g., common procedures) may appear in LLM training corpora. For each prompt, we generated 50 outputs via nucleus sampling with temperature t = 0.8 and one greedy decoding, then computed token-level edit distances between each sample and the greedy output. We defined peakedness as the fraction of samples within an edit distance threshold α = 0.05 × length. A prompt was flagged as contaminated if its peakedness exceeded a threshold of 0.01. The overall contamination rate was calculated as the proportion of such prompts across the test set. Using this procedure, we identified clean and contaminated samples for the LLMs and further selected 200 uncontaminated samples to assess the impact of contamination on both reasoning and QA prediction performance.
Efficiency analysis
For each evaluation approach, we recorded the evaluation time once per LLM and calculated the average runtime across all evaluated LLMs.
Error detection of reasoning evaluation
We performed an error detection analysis to evaluate the alignment between the errors identified by LLM-w-Rationale and those annotated by experts. Expert assessments of rationale correctness served as the ground truth, while the model’s assessments were treated as predictions. Based on these, we calculated the numbers of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN). Subsequently, precision, recall, and F1-score were computed to quantify the accuracy of LLM-w-Rationale in detecting reasoning errors. The evaluation metrics are defined as:
Statistical analysis
A non-parametric bootstrap procedure with 1000 iterations was employed to estimate the mean and 95% confidence intervals of the evaluation metrics. In each iteration, a resampled dataset equal in size to the test set was generated via random sampling with replacement.
Data availability
The medical questions are sourced from public datasets, which may be available for research purposes upon reasonable request. 1. Medbullets: https://huggingface.co/datasets/LangAGI-Lab/medbullets, 2. MMLU-Pro: https://huggingface.co/datasets/TIGER-Lab/MMLU-Pro, 3. MedExQA: https://huggingface.co/datasets/bluesky333/MedExQA, 4. MedXpertQA: https://huggingface.co/datasets/TsinghuaC3I/MedXpertQA, 5. Humanity’s Last Exam: https://github.com/centerforaisafety/hle, 6. MedQA-USMLE: https://huggingface.co/datasets/bigbio/med_qa, 7. PubMedQA: https://huggingface.co/datasets/bigbio/pubmed_qa, 8. MedMCQA: https://huggingface.co/datasets/lighteval/med_mcqa, 9. MMLU-Medicine: https://huggingface.co/datasets/cais/mmlu, 10. HEAD-QA: https://huggingface.co/datasets/dvilares/head_qa. The expert-curated data with fine-grained rationale annotation is released at https://github.com/plusnli/MedThink-Bench.
Code availability
The code is publicly available at https://github.com/plusnli/MedThink-Bench.
References
Zhou, S. et al. Large language models for disease diagnosis: a scoping review. npj Artif. Intell. 1, 9 (2025).
Chen, X. et al. Enhancing diagnostic capability with multi-agents conversational large language models. npj Digit. Med. 8, 159 (2025).
Truhn, D. et al. A pilot study on the efficacy of GPT-4 in providing orthopedic treatment recommendations from MRI reports. Sci. Rep. 13, 20159 (2023).
Goh, E. et al. GPT-4 assistance for improvement of physician performance on patient care tasks: a randomized controlled trial. Nat. Med. 31, 1233–1238 (2025).
Chen, C. et al. ClinicalBench: Can LLMs beat traditional ML models in clinical prediction? Preprint at arXiv https://doi.org/10.48550/arXiv.2411.06469 (2024).
Abbasian, M. et al. Foundation metrics for evaluating effectiveness of healthcare conversations powered by generative AI. npj Digit. Med. 7, 82 (2024).
Li, J. et al. Fact or guesswork? Evaluating large language model’s medical knowledge with structured one-hop judgment. Preprint at arXiv https://doi.org/10.48550/arXiv.2502.14275 (2025).
Zhou, S. et al. Uncertainty-aware large language models for explainable disease diagnosis. npj Digit. Med. 8, 690 (2025).
Hager, P. et al. Evaluation and mitigation of the limitations of large language models in clinical decision-making. Nat. Med. 30, 2613–2622 (2024).
McDuff, D. et al. Towards accurate differential diagnosis with large language models. Nature 642, 451–457 (2025).
Zhang, Y. et al. Siren’s Song in the AI Ocean: A Survey on Hallucination in Large Language Models. Comput. Linguist. 1–46 (2025).
Liu, W. et al. Mitigating hallucination through theory-consistent Symmetric Multimodal Preference Optimization. Preprint at arXiv https://doi.org/10.48550/arXiv.2506.11712 (2025).
Mündler, N., He, J., Jenko, S. & Vechev, M. T. Self-contradictory hallucinations of large language models: evaluation, detection and mitigation. In Proc. International Conference on Learning Representations (ICLR, 2024).
Haltaufderheide, J. & Ranisch, R. The ethics of ChatGPT in medicine and healthcare: a systematic review on Large Language Models (LLMs). npj Digit. Med. 7, 183 (2024).
Savage, T., Nayak, A., Gallo, R., Rangan, E. & Chen, J. H. Diagnostic reasoning prompts reveal the potential for large language model interpretability in medicine. npj Digit. Med. 7, 20 (2024).
Kim, H. et al. Small language models learn enhanced reasoning skills from medical textbooks. npj Digit. Med. 8, 240 (2025).
Wang, W. et al. Medical reasoning in the era of LLMs: a systematic review of enhancement techniques and applications. Preprint at arXiv https://doi.org/10.48550/arXiv.2508.00669 (2025).
Peng, Q. et al. Aligning clinical needs and AI capabilities: a survey on LLMs for medical reasoning. Preprint at techrxiv https://doi.org/10.36227/techrxiv.175790907.73315176/v1 (2025).
Zhu, Y. et al. DiagnosisArena: benchmarking diagnostic reasoning for large language models. Preprint at arXiv https://doi.org/10.48550/arXiv.2505.14107 (2025).
Tang, X. et al. MedAgentsBench: benchmarking thinking models and agent frameworks for complex medical reasoning. Preprint at arXiv https://doi.org/10.48550/arXiv.2503.07459 (2025).
Zuo, Y. et al. MedXpertQA: Benchmarking Expert-Level Medical Reasoning and Understanding. In Proc. Forty-Second International Conference on Machine Learning, International Conference on Machine Learning (ICML, 2025).
Singhal, K. et al. Large language models encode clinical knowledge. Nature 620, 172–180 (2023).
Yang, H. et al. Large language model Synergy for ensemble learning in Medical Question Answering: design and evaluation study. J. Med. Internet Res. 27, e70080 (2025).
Jin, Q. et al. Hidden flaws behind expert-level accuracy of multimodal GPT-4 vision in medicine. npj Digit. Med. 7, 190 (2024).
Liévin, V., Hother, C. E., Motzfeldt, A. G. & Winther, O. Can large language models reason about medical questions? Patterns 5, 100943 (2024).
Zhou, S. et al. Explainable differential diagnosis with dual-inference large language models. npj Health Syst. 2, 12 (2025).
Kim, Y. et al. MedExQA: Medical Question Answering Benchmark with Multiple Explanations. In Proc. 23rd Workshop on Biomedical Natural Language Processing, 167–181 (Association for Computational Linguistics, 2024).
Brodeur, P. G. et al. Superhuman performance of a large language model on the reasoning tasks of a physician. Preprint at arXiv https://doi.org/10.48550/arXiv.2412.10849 (2024).
Singhal, K. et al. Toward expert-level medical question answering with large language models. Nat. Med. 31, 943–950 (2025).
Li, D. et al. ExplainCPE: A Free-text Explanation Benchmark of Chinese Pharmacist Examination. Findings of the Association for Computational Linguistics: EMNLP 2023, 1922–40 (Association for Computational Linguistics, 2023).
Qiu, P. et al. Quantifying the reasoning abilities of LLMs on clinical cases. Nat. Commun. 16, 9799 (2025).
Papineni, K., Roukos, S., Ward, T. & Zhu, W.-J. BLEU. In Proc. 40th Annual Meeting on Association for Computational Linguistics - ACL ’02. https://doi.org/10.3115/1073083.1073135 (Association for Computational Linguistics, 2001).
Lin, C.-Y. ROUGE: A Package for Automatic Evaluation of Summaries. In Text Summarization Branches Out. 74–81 (Association for Computational Linguistics, Barcelona, Spain, 2004).
Zhang, T. et al. BERTScore: Evaluating Text Generation with BERT. International Conference on Learning Representations (2020).
Liu, L. et al. Towards automatic evaluation for LLMs’ clinical capabilities: Metric, data, and algorithm. In Proc. 30th ACM SIGKDD Conference on Knowledge Discovery and Data Mining Vol. 614, 5466–5475 (ACM, 2024).
Croxford, E. et al. Current and future state of evaluation of large language models for medical summarization tasks. npj Health Syst. 2, 6 (2025).
Wu, K. et al. MedCaseReasoning: evaluating and learning diagnostic reasoning from clinical case reports. Preprint at arXiv https://doi.org/10.48550/arXiv.2505.11733 (2025).
Ding, C. et al. Building a human-verified clinical reasoning dataset via a human LLM hybrid pipeline for trustworthy medical AI. Preprint at arXiv https://doi.org/10.48550/arXiv.2505.06912 (2025).
Chen, X. et al. FFA-GPT: an automated pipeline for fundus fluorescein angiography interpretation and question-answer. npj Digit. Med. 7, 111 (2024).
Griot, M., Hemptinne, C., Vanderdonckt, J. & Yuksel, D. Large Language Models lack essential metacognition for reliable medical reasoning. Nat. Commun. 16, 642 (2025).
Chen, H. et al. Benchmarking Large Language Models on Answering and Explaining Challenging Medical Questions. In Proc. 2025 Conference of the Nations of the Americas Chapter of the Association for Computational Linguistics: Human Language Technologies (Volume 1: Long Papers), 3563–3599 (Association for Computational Linguistics, 2025).
Sellergren, A. et al. MedGemma Technical Report. Preprint at arXiv https://doi.org/10.48550/arXiv.2507.05201 (2025).
Zhang, H. et al. HuatuoGPT, Towards Taming Language Model to Be a Doctor. Findings of the Association for Computational Linguistics: EMNLP 2023, 10859–10885 (Association for Computational Linguistics, 2023).
Guo, D. et al. DeepSeek-R1 incentivizes reasoning in LLMs through reinforcement learning. Nature 645, 633–638 (2025).
Wei, J. et al. Chain-of-Thought Prompting Elicits Reasoning in Large Language Models. In Proc. 36th International Conference on Neural Information Processing Systems (Curran Associates Inc., 2022).
Zhou, S. et al. Mitigating ethical issues for large language models in oncology: a systematic review. JCO Clin. Cancer Inform. 9, e2500076 (2025).
Gu, J. et al. A survey on LLM-as-a-Judge. arXiv https://doi.org/10.48550/arXiv.2411.15594 (2024).
Chen, H., Fang, Z., Singla, Y. & Dredze, M. Benchmarking large language models on answering and explaining challenging medical questions. In Proc. 2025 Conference of the Nations of the Americas Chapter of the Association for Computational Linguistics: Human Language Technologies (Volume 1: Long Papers) (eds Chiruzzo, L., Ritter, A. & Wang, L.) 3563–3599 (Association for Computational Linguistics, 2025).
Wang, Y. et al. MMLU-Pro: A More Robust and Challenging Multi-Task Language Understanding Benchmark. In Proc. 38th International Conference on Neural Information Processing Systems, vol. 37, 95266–95290 (Curran Associates Inc., 2024).
Kim, Y., Wu, J., Abdulle, Y. & Wu, H. MedExQA: medical question answering benchmark with multiple explanations. In Proc. 23rd Workshop on Biomedical Natural Language Processing (eds Demner-Fushman, D., Ananiadou, S., Miwa, M., Roberts, K. & Tsujii, J.) 167–181 (Association for Computational Linguistics, 2024).
Phan, L. et al. Humanity’s last exam. Preprint at arXiv https://doi.org/10.48550/arXiv.2501.14249 (2025).
Jin, D. et al. What Disease Does This Patient Have? A Large-Scale Open Domain Question Answering Dataset from Medical Exams. Appl. Sci. 11, 6421 (2021).
Jin, Q., Dhingra, B., Liu, Z., Cohen, W. & Lu, X. PubMedQA: A Dataset for Biomedical Research Question Answering. In Proc. 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP) (eds Inui, K., Jiang, J., Ng, V. & Wan, X.) 2567–2577 (Association for Computational Linguistics, 2019).
Pal, A., Umapathi, L. K. & Sankarasubbu, M. MedMCQA : a large-scale multi-subject multi-choice dataset for medical domain question answering. CHIL 174, 248–260 (2022).
Hendrycks, Dan, Collin Burns, Steven Basart, Andy Zou, Mantas Mazeika, Dawn Song, and Jacob Steinhardt. Measuring Massive Multitask Language Understanding. In Proc. International Conference on Learning Representations (ICLR, 2021).
Vilares, D. & Gómez-Rodríguez, C. HEAD-QA: a healthcare dataset for complex reasoning. In Proc. 57th Annual Meeting of the Association for Computational Linguistics (eds Korhonen, A., Traum, D. & Màrquez, L.) 960–966 (Association for Computational Linguistics, 2019).
Banerjee, S. & Alon, L. METEOR: An Automatic Metric for MT Evaluation with Improved Correlation with Human Judgments. In Proc. ACL Workshop on Intrinsic and Extrinsic Evaluation Measures for Machine Translation and/or Summarization, 65–72 (Association for Computational Linguistics, 2005).
Sellam, T. et al. BLEURT: Learning Robust Metrics for Text Generation. In Proc. 58th Annual Meeting of the Association for Computational Linguistics, 7881-7892 (Association for Computational Linguistics, 2020) .
Wei, H. et al. Systematic evaluation of LLM-as-a-judge in LLM alignment tasks: explainable metrics and diverse prompt templates. Preprint at arXiv https://doi.org/10.48550/arXiv.2408.13006 (2024).
Open AI et al. GPT-4o System Card. Preprint at arXiv https://doi.org/10.48550/arXiv.2408.13006 (2024).
Anthropic. Claude 3.5 Sonnet model card addendum. Available online at: https://www-cdn.anthropic.com/fed9cc193a14b84131812372d8d5857f8f304c52/Model_Card_Claude_3_Addendum.pdf
Comanici, G. et al. Gemini 2.5: Pushing the frontier with advanced reasoning, multimodality, long context, and next generation agentic capabilities. 207, 06261 (2025).
Wang, B. et al. Baichuan-M1: pushing the medical capability of large language models. Preprint at arXiv https://doi.org/10.48550/arXiv.2502.12671 (2025).
Chen, J. et al. Towards Medical Complex Reasoning with LLMs through Medical Verifiable Problems. Findings of the Association for Computational Linguistics: ACL 2025, 14552–14573 (Association for Computational Linguistics, 2025).
Christophe, C., Kanithi, P. K., Raha, T., Khan, S. & Pimentel, M. A. F. Med42-v2: A suite of clinical LLMs. Preprint at arXiv https://doi.org/10.48550/arXiv.2408.06142 (2024).
Grattafiori, A. et al. The Llama 3 herd of models. Preprint at arXiv https://doi.org/10.48550/arXiv.2407.21783 (2024).
Yang, A. et al. Qwen3 Technical Report. Preprint at arXiv https://doi.org/10.48550/arXiv.2505.09388 (2025).
Team, Q. Qwq-32b: Embracing the Power of Reinforcement Learning. https://qwenlm.github.io/blog/qwq-32b/ (2025).
Kruskal, W. H. & Wallis, W. A. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 47, 583 (1952).
Dong, Y. et al. Generalization or Memorization: Data Contamination and Trustworthy Evaluation for Large Language Models. Findings of the Association for Computational Linguistics: ACL 2024, 12039–12050 (Association for Computational Linguistics, 2024).
Acknowledgements
This work was supported by the National Institutes of Health’s National Center for Complementary and Integrative Health under grant number R01AT009457 and U01AT012871, National Institute on Aging under grant number R01AG078154, and National Cancer Institute under grant number R01CA287413. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health. The authors also acknowledge the support from the Center for Learning Health System Sciences.
Author information
Authors and Affiliations
Contributions
S.Z. and R.Z. conceptualized and led the study design. S.Z., W.X. and Y.S. conducted the literature search. S.Z., W.X., J.L., R.Z., Z.L. and N.L. contributed to discussions on data annotation, while S.Z. and R.Z. organized the annotation and human evaluation processes. W.X. and J.L. handled data collection, and M.S., L.W., C.E., X.M., Y.J., Z.X., Y.C., M.T., Y.X. and E.S. participated in data annotation and human evaluation. S.Z. developed the experimental design, and J.L., W.X., Z.Z., H.Y. and S.Z. were responsible for model development and experiments. S.Z., W.X. and J.L. drafted the initial manuscript, and R.Z., Z.L. and N.L. provided supervision throughout the study. All authors contributed to research discussions, critically revised the manuscript, and approved the final version for submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhou, S., Xie, W., Li, J. et al. Automating expert-level medical reasoning evaluation of large language models. npj Digit. Med. 9, 34 (2026). https://doi.org/10.1038/s41746-025-02208-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-02208-7