Abstract
Mismatch repair (MMR) deficiency occurs in 10–20% of colorectal cancer (CRC) cases, leading to microsatellite instability (MSI). Although MSI/MMR testing is critical for CRC management, high costs and long turnaround times limit testing rates and clinical utility, highlighting the need for more accessible, cost-effective alternatives. PANProfiler Colorectal (PPC) is an artificial intelligence (AI)-based biomarker test that determines MSI/MMR status directly from haematoxylin and eosin (H&E)-stained slides. We conducted a blinded, multi-centred validation to assess PPC’s performance against standard testing. The study included 3,576 whole slide images from 1,243 CRC patients across three United Kingdom institutions. PPC produced definitive results for 86.55% of slides, achieving an overall agreement of 93.83%, positive agreement of 92.54%, and negative agreement of 94.02%. PPC accurately determined MSI/MMR status from routine H&E slides, offering a rapid, scalable alternative to conventional diagnostic methods.
Similar content being viewed by others
Introduction
The DNA mismatch repair (MMR) system is essential for maintaining DNA integrity by rectifying errors that occur during replication, such as base-base mismatches and insertion-deletion loops, to maintain genomic stability. Four key proteins, MLH1, MSH2, MSH6, and PMS2, are included in this process. If the expression of any of the corresponding genes is impaired, the MMR mechanism can become dysfunctional1. When MMR is disrupted, it leads to microsatellite instability (MSI), which is characterised by changes in these repetitive DNA sequences. As such, MSI is a phenotypic indicator of abnormal MMR function1. Deficient MMR (dMMR) is evident in 10-20% of colorectal cancers (CRCs)2,3,4. MSI/MMR status can inform the clinical management of CRC patients, with major diagnostic, prognostic, and therapeutic implications, as well as highlighting patients for Lynch syndrome testing5. Importantly, MSI-high (MSI-H) and dMMR serve as predictive biomarkers of response to immune checkpoint inhibitors. The pioneering work of Allison and Honjo demonstrated that immunotherapy is particularly effective in CRC with MSI-H or dMMR, with response rates of approximately 50% in metastatic CRC and up to 100% in early-stage cases6,7. Therefore, accurate and timely MSI/MMR testing is critical to guide treatment decisions and improve patient outcomes.
Testing for MSI or dMMR is recommended for all CRC patients by the National Institute for Health and Care Excellence (NICE)8, the European Society for Medical Oncology (ESMO)9, and jointly by the American Society of Clinical Oncology (ASCO) and the College of American Pathologists (CAP)10. However, universal MSI/MMR testing is not yet standard practice, with testing rates varying significantly across countries, healthcare settings, and patient demographics11,12,13. Current clinical practice relies on two primary methods for MSI/MMR testing: immunohistochemistry (IHC), which detects loss of nuclear expression of the MMR proteins MLH1, MSH2, MSH6, and PMS2, and polymerase chain reaction (PCR)-based MSI testing, which identifies instability in specific microsatellite loci14. Both approaches demonstrate good sensitivity (91–93%) and moderate specificity (79–83%)15, yet are limited by practical challenges. While IHC is generally highly concordant, its interpretation can be subject to inter-observer variability in challenging cases16, whereas PCR can be affected by poor DNA quality, especially from formalin-fixed, paraffin-embedded (FFPE) samples17,18. Both require additional tumour tissue beyond that used for preparing the haematoxylin and eosin (H&E)-stained specimen, specialised infrastructure, and may incur high costs. Combined with pathology workforce shortages and variable turnaround times13,19,20, these limitations highlight the need for a rapid, reliable, cost-effective, and widely accessible alternative for MSI/dMMR detection in all CRC patients.
In routine H&E-stained CRC tissue, MSI and dMMR tumours are associated with certain morphological patterns21, such as the presence of prominent tumour-infiltrating lymphocytes (TILs) or peritumoral “Crohn’s-like” lymphoid reaction, mucinous differentiation and/or poorly-differentiated morphology21. This suggests that MSI/MMR status has distinct histomorphological features identifiable from H&E-stained specimens, which could be leveraged in image-driven algorithms using artificial intelligence. This potential has been demonstrated in multiple studies where AI models were successfully used to determine MSI or MMR status from whole slide images (WSIs) of H&E-stained tissue21,22,23,24.
PANProfiler Colorectal (PPC) is an AI-based biomarker test for determining MSI/MMR status from H&E-stained WSIs (Fig. 1). In this paper, we report the performance characteristics of PPC based on a retrospective validation performed in a blinded setting. Data were collected from three UK institutions, ensuring a diverse cohort that enabled assessment of PPC’s robustness and effectiveness across different clinical settings. The results were generated centrally and analysed at St James’s University Hospital (SJUH) in Leeds, UK, without PPC having access to the reference test results at any time of the study. Variability in scanning equipment, image formats, patient demographics, and disease characteristics was considered to reflect real-world conditions. In addition, we discuss PPC’s utility as an AI-based diagnostic test within routine CRC workflows and its potential to streamline the diagnostic processes to enable faster turnaround times.
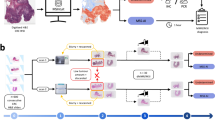
A The standard diagnostic pathway for CRC begins with the collection of tissue samples (biopsy or resection). Tissue sections are cut from FFPE blocks, mounted onto glass slides and stained with H&E. These slides are examined by a pathologist, who assesses the presence of tumour and, where present, its grade. Upon confirmation of malignancy, additional testing is performed to assess MSI/MMR status. IHC is commonly used to evaluate the expression of MMR proteins (MLH1, PMS2, MSH2, and MSH6), while PCR/NGS can be used to detect changes in the number of repeats in microsatellite loci, characteristic of MSI. These biomarkers are important for guiding treatment decisions, including the use of immunotherapy. Integrating PPC, a digital test, into routine pathological workflow has the potential to streamline the diagnostic process, reducing the turnaround times from days or weeks to minutes (see also Supplementary Fig. S3), potentially reducing the pressure on histopathology services, and enabling timely treatment decisions. B Overview of PPC’s end-to-end deep learning pipeline, including data preprocessing steps, background removal, detection of relevant tissue, tiling and colour normalisation, and selection of tiles for biomarker profiling. PPC’s deep learning architecture consists of a feature extractor and proprietary aggregation, attention and classification modules.
Results
Blinded validation results of PANProfiler colorectal
The key performance characteristics from the blinded validation are given in Table 1. The results demonstrate consistently strong performance of the PPC model across all three independent cohorts. High overall percent agreement (OPA), positive percent agreement (PPA), and negative percent agreement (NPA) were observed, with C-statistics exceeding 0.92 in all cases. The model delivered particularly robust results on the largest cohort (L1-UK-CRC-SVS-1-BLIND). The test replacement rate (TRR) varied across datasets but remained high overall with only 13.45% of tested samples returning an Indeterminate result. These findings support the generalisability and clinical potential of PPC across different institutions and regions in the UK.
Performance of PANProfiler Colorectal across diverse subpopulations
To demonstrate the generalisability of PPC across different regions within the UK, results from all institutions were analysed together. To this end, we included the I1-UK-CRC-CZI-1-DEV cohort as an additional site to represent Scotland. Since this dataset was validated using 5-fold cross-validation rather than a blind evaluation, it is not included in the main results. The I1-UK-CRC-CZI-1-DEV cohort, consisting of 1,125 WSIs with a prevalence of 13.6%, demonstrated strong performance, achieving an OPA of 86.49% (95% CI: 83.80-88.89%), PPA of 97.41% (95% CI: 92.63-99.46%), NPA of 84.44% (95% CI: 81.34-87.21%), a C-statistic of 0.928, and a TRR of 65.2% (95% CI: 62.29-67.94%). These metrics are within the range of the blind performance results (shown in Table 1), which assessed performance in England, Wales, and Northern Ireland.
Cohort characteristics
The breakdown of resections and biopsies for development and blind cohorts are provided in Supplementary Table S1. Study population demographics, including age and sex distribution, are detailed in Supplementary Table S2. The mean age across the cohorts ranged from 65.6 to 68.9 years (standard deviation: 11.6 to 12.8 years). Males comprised 52-57% of patients across cohorts, while females accounted for 43-48%, consistent with previously reported figures in the literature2. Tumour site distribution is presented in Supplementary Tables S3, with the colon being the most frequently affected site, which aligns with published data25,26. Supplementary Table S4 shows the breakdown of histological subtypes across different cohorts. Adenocarcinoma was the predominant histological subtype, observed in 73.24–94.4% of cases, in agreement with prior studies27. It was followed by mucinous adenocarcinoma, with a prevalence of 5.56–20.42% across cohorts.
Tumour grade and pathological stage data are summarised in Supplementary Tables S5-S7. These tables also provide the distribution of MSI and dMMR status within each cohort. Moderately differentiated was the most prevalent histological grade, consistent with the literature27. Poorly differentiated carcinoma was more frequently observed in patients with MSI-H/dMMR compared to those with non-MSI-H or proficient mismatch repair (pMMR), a finding also supported by earlier studies21. Tumour stage varied across cohorts, with stage II and III being the most common. MSI-H/dMMR prevalence was highest in stage II cases. Compared to the published literature, this study observed a lower proportion of stage IV diagnoses and a higher proportion of stage II cases26,28. However, the association between lower tumour stage and increased MSI-H/dMMR prevalence is consistent with previous findings29.
Impact of backbone model on performance
The self-supervised learning (SSL)-pretrained backbone outperformed the ImageNet backbone across all metrics in the five-fold cross-validation on the L1-UK-CRC-SVS-1-DEV cohort (see Supplementary Table S8 and Supplementary Fig. S1). It achieved a higher C-statistic (0.96 vs. 0.93), OPA (95.25% vs. 91.48%), PPA (95.51% vs. 87.78%), and NPA (95.19% vs. 92.39%). Additionally, the TRR was slightly higher for SSL (84.03% vs. 83.12%). Consequently, the SSL-based model, demonstrating optimal performance, was selected for the blinded validation study.
Explainability and interpretability of deep learning models
We unblinded and analysed 61 images from the L1-UK-CRC-SVS-1-BLIND cohort, with a breakdown of 16 true positives (TP), 18 true negatives (TN), 7 false positives (FP), 6 false negatives (FN), 8 indeterminate negatives (IN), and 6 indeterminate positives (IP). In Figs. 2, 3, and Supplementary Fig. S2, example images demonstrate how our models direct their attention, by overlaying the “attention heatmaps” (B) onto the original WSIs (A). The same panel also highlights the regions of interest selected based on the model’s attention.
A Original WSI and two regions of interest (ROIs) beneath it (selected from different regions indicated by the red boxes). B Attention scores with darker hues of red indicating high attention and blue indicating low attention. C Annotated regions based on a classification of the tissue, including adipose, background, debris, lymphocytes, mucus, smooth muscle, normal colon mucosa, stroma, and tumour.
A Original WSI and two ROIs beneath it (selected from different regions indicated by the red boxes). B Attention scores with darker hues of red indicating high attention and blue indicating low attention. C Annotated regions based on a classification of the tissue, including adipose, background, debris, lymphocytes, mucus, smooth muscle, normal colon mucosa, stroma, and tumour.
To analyse the predictions correctly classified by the model, we visualised the heatmaps for TP and TN cases. For TP cases, regions with high attention scores predominantly aligned with tumour tissue, such as poorly/moderately differentiated areas with medullary features, TILs, and adenocarcinoma with mucinous features (Fig. 2, A-B). In TN cases (Fig. 2, right), the model’s attention was directed toward tumour regions featuring moderately differentiated adenocarcinomas.
To analyse the predictions incorrectly classified by the model, we visualised the heatmaps for FP and FN cases (Fig. 3). FP predictions were focused on poorly differentiated regions, which could explain the misclassification, as poor differentiation is often linked to MSI-H/dMMR tumours (Fig. 3, left). For a FN case (Fig. 3, right), the model concentrated on regions of moderately differentiated adenocarcinoma, likely contributing to the misclassification.
To investigate the model’s indeterminate predictions, we visualised heatmaps for the IN and IP cases (Supplementary Fig. S2). Our analysis suggests that indeterminate predictions may arise from conflicting or ambiguous morphological signals. For instance, some non-MSI-H/pMMR (IN) cases presented with features typically associated with MSI-H/dMMR status (e.g., poor differentiation, TILs, mucinous features), while others were confounded by factors like limited tumour tissue or contained non-specific features such as moderate differentiation. Similarly, most indeterminate MSI-H/dMMR (IP) cases exhibited a substantial degree of well or moderate differentiation.
Overall, increased attention is observed on biologically meaningful morphological features, such as poor differentiation, TILs, and mucinous characteristics, which are well-documented as being associated with MSI-H/dMMR. By contrast, well-differentiated tumours are commonly linked to non-MSI-H/pMMR. Moderate differentiation, on the other hand, is not a reliable discriminator of MSI/MMR status, as it is observed in both MSI-H/dMMR and non-MSI-H/pMMR tumours23,24,30,31,32,33.
The model’s embedding space was also evaluated to determine whether MSI-H/dMMR and non-MSI-H/pMMR cases could be separated. Embedding spaces were visualised at both the slide level (Fig. 4A) and patch level (Fig. 4B). The slide-level (classification) embedding space (Fig. 4A) demonstrated clear separation between the MSI-H/dMMR and non-MSI-H/pMMR classes. At the patch level, distinct regions for each class were observed, with patches exhibiting similar morphology positioned closer together in the embedding space. For example, MSI-H/dMMR patches in one region formed clusters characterised by poorly differentiated adenocarcinoma, while another non-MSI-H/pMMR cluster of patches were associated with moderately differentiated adenocarcinoma.
The embedding space of 61 whole slide images (WSIs) from the L1-UK-CRC-SVS-1-BLIND cohort is visualised at both slide (A) and patch (B) levels. MSI-H/dMMR and non-MSI-H/pMMR images and patches are shown in red and blue, respectively. The slide level embeddings (A) are visualised using 2D t-SNE, while the top 100 scoring attention tiles per WSI (B) are visualised using 3D t-SNE. An example of three closest MSI-H/dMMR and non-MSI-H/pMMR patches (based on cosine distance) is shown, taken from regions indicated by red and blue boxes, respectively.
Discussion
This study evaluated the performance of AI-based PPC compared to standard pathology MSI/MMR testing in a retrospective, blinded setting. High PPA and NPA demonstrated that PPC accurately identified both MSI-high/dMMR (Unstable) and non-MSI-high/pMMR (Stable) samples. Additionally, of 3576 WSIs, PPC returned definite results (TRR) for 86.55% of images, indicating a potential significant reduction in the number of standard pathology tests required.
Many other studies have evaluated AI-driven methods for determining MSI/MMR status from WSIs34,35,36,37,38,39. These demonstrate the feasibility of AI-derived image features for MSI/MMR detection. However, most have focused on screening, where the primary target was identifying cases not requiring further molecular or IHC testing. For instance, one study33 conducted a blinded validation on 600 WSIs, reporting an area under the receiver operating characteristic curve (AUC, i.e. the C-statistic) of 88% and a sensitivity (i.e. PPA) of 96-98% with a TRR of 46-47%. Other studies reported comparable or worse performance. For example, one study40 achieved TRRs of 44.12% for resections and 52.73% for biopsies with a sensitivity of 95%. Moreover, another approach for MSI screening41 introduced a deep learning (DL)-based classifier capable of replacing testing for only 40% of CRC cases.
Other AI-based approaches have addressed the problem from a diagnostic perspective, aiming to identify both MSI-high/dMMR and non-MSI-high/pMMR cases. One diagnostic approach42 validated on 355 resections and 341 biopsies, reported AUCs of 84.57% and 76.79%, sensitivity scores of 90.91% and 92.31%, specificity (i.e. NPA) scores of 95.13% and 95.36%, and TRRs of 56.06% and 51.03%, respectively. Finally, a recent large-scale study23 by Wagner et al. reported sensitivity scores of 98-99% and specificity scores of 44-56%. While there exist key differences in model architecture and training data characteristics, our study’s higher specificity fundamentally stems from different clinical goals and model designs. The Wagner et al. model was developed for pre-screening, where a trade-off with specificity is accepted to maximise sensitivity. In contrast, our tool is designed for diagnostic use, requiring both high sensitivity and specificity. We achieve this by implementing a three-tiered classification system (‘Stable’, ‘Unstable’, ‘Indeterminate’), which allows the model to make definitive calls with higher confidence by classifying ambiguous cases as ‘Indeterminate’. This methodological choice is the primary driver of our higher specificity. Consequently, in contrast to previous methods, PPC demonstrates remarkable diagnostic utility by maintaining both high sensitivity and specificity. This also translates to great practical application, with an overall TRR of 86.55%.
We evaluated the safety of PPC by comparing its performance to standard MSI/MMR detection tests, including PCR for MSI testing and IHC for MMR deficiency, based on results from clinical studies in the literature. This is critical to show that PPC yields similar risk in the context of false negative and false positive results, as such there would be no greater risk of adverse effects compared to the current standard of care. Aggregated data from multiple studies primarily across the US and Europe15 indicate that MSI testing (n = 3,476) has a sensitivity (PPA) of 93% (95% CI: 87-96%) and a specificity (NPA) of 79% (95% CI: 70-86%), while MMR testing (n = 3,091) shows a sensitivity of 91% (95% CI: 85-95%), and a specificity of 83% (95% CI: 77-88%). The National Institute for Health and Care Excellence (NICE), England, reported similar figures8 for MSI testing (sensitivity: 91.3% [95% CI: 42.6-99.3%], specificity: 83.7% [95% CI: 63.8-93.7%]) and IHC-based MMR testing (sensitivity: 96.2% [95% CI: 69.4-99.6%], specificity: 88.4% [95% CI: 79.0-94.0%]). Recent studies report that concordance between MSI testing and MMR IHC is increasing, especially when sufficient tumour DNA is present and unusual IHC patterns are taken into account during MMR testing43. PPC’s performance appears comparable to standard pathology tests, however, direct head-to-head comparisons with other studies are often challenging due to variations in patient cohorts, experimental design, preanalytical protocols and the specific definitions of outcomes. Therefore, any comparison of our results to other published data should be interpreted with caution.
The integration of PPC into pathological workflows could offer significant advantages, particularly in its ability to streamline diagnostic processes by minimising reliance on time-intensive assays currently used as the standard for biomarker testing. This reduction in testing could assist users, such as pathologists and laboratory professionals, in managing growing workloads and addressing staffing shortages19,20. By contrast, PPC can generate biomarker results within minutes (Supplementary Fig. S3), enabling pathology reports to be finalised simultaneously during the histological assessment. Other potential benefits of PPC include lowering the diagnostic burden on pathologists and lab professionals, allowing them to dedicate more attention to complex cases. A digital test based on existing H&E-stained slides offers positive environmental benefits by reducing the need for additional wet laboratory procedures, minimising the use of toxic chemicals and consumables common in conventional diagnostics. By streamlining workflows and reducing the reliance on standard laboratory testing, PPC also has the potential to deliver timely results and greater cost efficiency for healthcare providers. Future work will include a health economics analysis to evaluate its cost-effectiveness in real-world clinical environments. In parallel, usability studies with target users will assess the impact of PPC on user experience and workflow integration.
While PPC’s overall performance demonstrated significant promise, certain aspects warrant additional exploration to improve the AI-based test. Approximately 14% of samples returned an Indeterminate result, which may be partially due to preanalytical factors such as suboptimal tissue processing, staining or slide digitisation. Standardising and optimising these workflows will be key to increasing the proportion of assessable cases and improving clinical usability. Indeterminate outcomes may also reflect model uncertainty driven by morphological inconsistencies. Our visual analysis of indeterminate cases showed they often contained conflicting features, such as non-MSI cases presenting with MSI-like features (e.g., TILs and poor differentiation), non-specific features like moderate differentiation, or confounding factors like limited tumour tissue, leading to an indeterminate result (Supplementary Fig. S2). Expanding the training dataset to include more diverse, representative cases could improve the model’s ability to resolve such differences. In addition, a deeper investigation into the variability of validation metrics such as TRR is needed to better understand and refine PPC’s performance across different cohorts. All institutions in the study were based in the UK, providing a strong foundation within a well-characterised healthcare system. To build on these findings and support broader applicability, future validation will benefit from including samples from other regions and more diverse demographics. While this retrospective study offers valuable insights, prospective studies in clinical environments will be key to further demonstrating PPC’s performance and utility in real-world clinical settings. Such studies would provide a critical understanding of PPC’s integration into routine pathology workflows and assess its impact on patient outcomes. Finally, this study utilised mostly resected tissue samples (Supplementary Table S1). Since biopsies are more commonly used in routine clinical practice, future validation efforts will expand to include biopsy samples, further supporting the test’s real-world applicability.
Taken together, these findings point to the potential of PPC as a proficient diagnostic tool capable of comparable levels of performance to current standard-of-care testing. This paves the way towards improved diagnostics, ensuring more accurate results and leading to the timely establishment of MSI/MMR status for CRC cases.
Methods
Cohort characteristics for blinded validation
A total of 3576 WSIs from three different UK cohorts were used for the multi-centred, blinded validation of PPC (Table 2). Only WSIs derived from FFPE blocks of primary CRC were included in the analysis. WSIs of fresh frozen samples, metastatic tumours from non-colorectal sites, and those with less than 10% tumour content were excluded. Additionally, WSIs failing visual assessment and quality control by pathologists due to excessive tissue folds, air bubbles, pen marks, adhesive tape, out-of-focus regions, pixel artefacts or other digital distortions were also excluded. The number of excluded images (n = 1234) accounted for 25.65% of the images originally acquired from the three institutions (n = 4,810). The WSIs were acquired using different scanners and stored in multiple image formats. MSI-H/dMMR prevalence ranged from 12.52 to 20.60%. This is comparable to global estimates2,3,4, which serve as the primary reference for clinical practice in the UK. The make-up of each independent cohort was as follows:
L1-UK-CRC-SVS-1-BLIND
This cohort comprised 3,884 archival CRC images, sourced from St James’s University Hospital (SJUH) in Leeds, UK, of which 3,124 had MSI/MMR status (391 Unstable and 2,733 Stable). Tissue samples were scanned at 20/40x magnification using Leica Aperio GT 450 DX, AT2 or Scanscope CS scanners and stored in SVS format.
W1-UK-CRC-CZI-1-BLIND
This cohort was composed of a total of 510 WSIs, sourced from Wales Cancer Biobank (WCB) in Cardiff, UK, of which 54 had MSI/MMR status (11 Unstable and 43 Stable). Tissue samples were scanned at 40x magnification and stored in CZI format.
N1-UK-CRC-SVS-1-BLIND
This cohort comprised 398 WSIs collected from Northern Ireland Biobank (NIB) in Belfast, UK (82 Unstable, 316 Stable). Tissue samples were scanned at 20/40x magnification using a Leica Aperio AT2 scanner and stored in SVS format.
Cohort characteristics for development and calibration
A total of 2502 WSIs of FFPE CRC specimens from four cohorts were used for development, internal validation, and calibration of PPC (Table 3). While samples from the blinded validation and development/calibration cohorts may originate from the same institution, there is no case overlap between these distinct cohorts. Each case included in the blinded validation cohort is unique and has not been used in the development or calibration phases of the model. The make-up of each cohort was as follows:
T1-US-CRC-SVS-1-DEV
This cohort consisted of 563 WSIs (78 Unstable and 485 Stable), acquired from The Cancer Genome Atlas (TCGA) Colon (COAD) and rectal (READ) adenocarcinoma studies (https://portal.gdc.cancer.gov/). It was used for model development and internal validation. Tissue samples were scanned at 20/40x magnification and stored in SVS format.
I1-UK-CRC-CZI-1-DEV
This cohort consisted of 1,156 WSIs (159 Unstable and 997 Stable), sourced from the Industrial Centre for Artificial Intelligence Research in Digital Diagnostics (iCAIRD) in Scotland, UK. It was used for model development and internal validation. Tissue samples were scanned using Zeiss Axioscan at 40x magnification and stored in CZI format.
L1-UK-CRC-SVS-1-DEV
This cohort consisted of 551 WSIs (109 Unstable and 442 Stable) collected from SJUH in Leeds. It was used for internal validation and calibration. Tissue samples were scanned at 20/40x magnification using Leica Aperio GT 450 DX, AT2 or Scanscope CS scanners and stored in SVS format.
N1-UK-CRC-SVS-1-DEV
This cohort consisted of 130 WSIs (27 Unstable and 103 Stable) collected from NIB. It was used for internal validation and calibration. Tissue samples were scanned at 20/40x magnification using a Leica Aperio AT2 scanner and stored in SVS format.
Overview of PANProfiler colorectal
PPC is an AI-driven biomarker test developed to determine MSI/MMR status from digitally scanned H&E-stained slides of FFPE primary CRC specimens. PPC classifies MSI/MMR status as ‘Unstable’, Stable’ or ‘Indeterminate’. ‘Unstable’ was defined as MSI-high/dMMR (i.e. positive class), while ‘Stable’ corresponded to pMMR/non-MSI-high (i.e. negative class). An ‘Indeterminate’ result was returned when the test could not confidently determine MSI/MMR status.
Performance characteristics and statistical procedures
PPC’s performance was evaluated through a comprehensive agreement analysis with standard pathology tests, employing four key metrics: overall percent agreement (OPA), positive percent agreement (PPA), negative percent agreement (NPA) and test replacement rate (TRR), which measured the percentage of cases for which PPC was able to provide a definitive result. In addition, the C-statistic, equivalent to the area under the receiver operating characteristic curve (AUC), was reported44,45. Formal definitions of all performance metrics are provided in Supplementary Tables S9 and S10.
Based on the expected prevalence of MSI-H/dMMR in colorectal cancer (10-20%), a power analysis determined that a minimum of 140-245 cases would be required to achieve a 95% confidence level with a 5% margin of error46. The sample size used in this study therefore exceeds the threshold necessary to ensure adequate statistical power.
Artificial intelligence for establishing MSI/MMR status
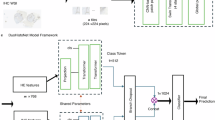
PPC is powered by AI and digital pathology. The platform relies on a series of DL methods, a subset of AI that enables computers to learn patterns from large datasets. Specifically, it can identify complex histomorphological features in relevant parts of H&E-stained WSIs of CRC and use them to determine MSI/MMR status47. Figure 1 illustrates the core preprocessing and DL components that underlie PPC and how they are connected in an end-to-end pipeline.
A DL model was trained to predict MSI/MMR status as follows. In the first step, input WSIs were subdivided into image patches (i.e. tiles) of 256×256 pixels at a resolution of 1.0 microns per pixel (MPP). Proprietary filtering algorithms were used to eliminate the background tiles and detect tissue. A WSI was discarded from analysis if it contained fewer than 10 tiles after the filtering process. Colour normalisation48 was applied to the remaining tiles before they were assigned with a reference biomarker status.
The selected pre-processed tiles were then used in an end-to-end DL pipeline. This pipeline consisted of the following components: (1) a smart sampling module used for extracting groups of tiles that are spatially adjacent (referred to as windows); (2) a feature extractor CNN (i.e., an encoder) for learning unique feature representations for each tile; (3) an attention module that aggregated the tile embedding into a slide level embedding, weighting the features based on their relevance (using the attention scores) for the target biomarker. The attention module was finally connected to a classification layer that made the final diagnostic call on the biomarker status.
Explainability and interpretability of deep learning models
Understanding the internal mechanisms of DL models can provide insights into how they detect visual patterns in tissue WSIs that may be correlated with biological signals. Visualising the “black-box” nature of these models offers valuable insights into the explainability of their outputs. A common approach to visualising the spatial regions critical for determining biomarker status is overlaying tile-level attention on WSIs to create spatial heatmaps. These heatmaps highlight the areas of the input image that have the strongest influence on the biomarker result. DL models can identify morphological patterns or visual features that are not immediately visible to the human eyes and correlate them with the existence (or absence) of biological signals, such as biomarker expression. We visualise the heatmaps by overlaying the model attention onto the WSI in Figs. 2 and 3.
In addition, to enhance the interpretability of the models, a DL model was developed and trained to classify various tissue types in CRC samples using the NCT-CRC-HE-100K dataset49. The model categorised tissues into the following classes: adipose, background, debris, lymphocytes, mucus, smooth muscle, normal colon mucosa, stroma, and tumour. This model was used to confirm whether the attention of the primary models was directed toward relevant morphological regions by overlaying the classification maps onto the WSIs and comparing them with the attention heatmaps.
To gain deeper insights into the PPC model, its embedding space, a lower-dimensional vector representation of image patches or whole slides, was visualised. Patch-level embeddings were obtained by processing each WSI through the model’s feature extractor, and the resulting vectors for individual patches were stored. These patch embeddings were then aggregated into slide-level embeddings using the model’s attention layer and saved for each WSI. The embeddings were visualised in TensorBoard using t-distributed Stochastic Neighbour Embedding (t-SNE), a technique for visualising high-dimensional data in two or three dimensions, with data points organised by their respective classes (Fig. 4). To measure similarity between embeddings, the cosine distance, a metric based on the angle between two vectors, was used. For this analysis, a subset of 61 WSIs from the L1-UK-CRC-SVS-1-BLIND cohort was unblinded, comprising 28 samples with MSI-H/dMMR and 33 with non-MSI-H/pMMR.
Development and validation strategies
We utilised an SSL framework (Mocov2) for pre-training base models, allowing them to learn feature representations from unlabelled data and develop robustness for diverse tasks50. Using the pre-trained SSL model as a backbone, a model was further fine-tuned through supervised learning with labels derived from reference MSI/MMR results. The SSL training datasets consisted of 588 WSIs from the T1-US-CRC-SVS-1-DEV cohort. The development datasets (Table 3) consisted of 1,719 WSIs (237 Unstable, 1,482 Stable WSIs), including T1-US-CRC-SVS-1-DEV (n = 563, with 78 Unstable and 485 Stable WSIs), and I1-UK-CRC-CZI-1-DEV (n = 1,156, with 159 Unstable, and 997 Stable WSIs).
To determine the best model to be deployed for blinded validation, different models were evaluated on the L1-UK-CRC-SVS-1-DEV cohort using cross-validation. To this end, this cohort was divided into five equal-sized folds. A model was fine-tuned, validated, and tested five times, cycling through a few fold combinations. This ensured that the models were thoroughly evaluated over the entirety of a cohort while minimising the risk of overfitting and providing a robust assessment of model performance.
Calibration refers to finding the most optimal decision thresholds for a model. These thresholds help the model operate safely by allowing an ‘Indeterminate’ output when no definitive result can be provided. PPC’s safety and effectiveness were maintained across different sites by calibrating cohort-specific thresholds for each biomarker. The decision boundaries identified during the calibration process were applied to compute the final model outputs, which were then used to calculate performance metrics on the test set for each cohort (see Performance characteristics for details).
After determining the optimal configuration, PPC underwent separate calibrations on two cohorts before the blinded validation phase. Specifically, calibration was performed on the L1-UK-CRC-SVS-1-DEV cohort, consisting of 551 WSIs (109 Unstable and 442 Stable images), and the N1-UK-CRC-SVS-1-DEV cohort, which included 130 WSIs (27 Unstable and 103 Stable). No calibration was performed for the WCB cohort (W1-UK-CRC-CZI-1-BLIND), due to a low sample size.
Blinded validation
Authors from the University of Leeds conducted the blinded validation across all three cohorts. MSI/MMR status associated with blinded cohorts was not disclosed at any stage to the analysts. WSIs were analysed using PPC, generating test results classified as Stable, Unstable, or Indeterminate. These results were then shared with the clinical research team performing the blinded validation, who constructed confusion matrices and calculated performance metrics.
Data availability
The Cancer Genome Atlas (TCGA) used for training is publicly available from the following link: [https://portal.gdc.cancer.gov/]. The additional datasets used for training and blinded validation were obtained through agreements between Panakeia Technologies Limited and the respective data providers and are not publicly available.
Code availability
The software and AI algorithms used in the current study are Panakeia Technologies Limited proprietary IP and cannot be shared.
References
Pećina-Šlaus, N., Kafka, A., Salamon, I. & Bukovac, A. Mismatch Repair Pathway, Genome Stability and Cancer. Front. Mol. Biosci. 7, (2020).
Koopman, M. et al. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br. J. Cancer 100, 266–273 (2009).
Kang, Y.-J. et al. A scoping review and meta-analysis on the prevalence of pan-tumour biomarkers (dMMR, MSI, high TMB) in different solid tumours. Sci. Rep. 12, 20495 (2022).
Lorenzi, M., Amonkar, M., Zhang, J., Mehta, S. & Liaw, K.-L. Epidemiology of Microsatellite Instability High (MSI-H) and Deficient Mismatch Repair (dMMR) in Solid Tumors: A Structured Literature Review. J. Oncol. 2020, 1807929 (2020).
Ma, J., Setton, J., Lee, N. Y., Riaz, N. & Powell, S. N. The therapeutic significance of mutational signatures from DNA repair deficiency in cancer. Nat. Commun. 9, 3292 (2018).
Cercek, A. et al. PD-1 blockade in mismatch repair–deficient, locally advanced rectal cancer. N. Engl. J. Med. 386, 2363–2376 (2022).
Morse, M. A., Hochster, H. & Benson, A. Perspectives on treatment of metastatic colorectal cancer with immune checkpoint inhibitor therapy. Oncologist 25, 33–45 (2020).
Molecular testing strategies for Lynch syndrome in people with colorectal cancer. Nice https://www.nice.org.uk/guidance/dg27.
Stjepanovic, N. et al. Hereditary gastrointestinal cancers: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 30, 1558–1571 (2019).
Vikas, P. et al. Mismatch repair and microsatellite instability testing for immune checkpoint inhibitor therapy: ASCO Endorsement of College of American Pathologists Guideline. J. Clin. Oncol. 41, 1943–1948 (2023).
Garcia-Carbonero, R. et al. Real-world study on microsatellite instability and mismatch repair deficiency testing patterns among patients with metastatic colorectal cancer in Spain. Clin. Transl. Oncol. 26, 864–871 (2024).
Papke, D. J., Lindeman, N. I., Schrag, D. & Iorgulescu, J. B. Underutilization of guideline-recommended mismatch repair/microsatellite instability biomarker testing in advanced colorectal cancer. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 31, 1746–1751 (2022).
Echle, A. et al. Clinical-grade detection of microsatellite instability in colorectal tumors by deep learning. Gastroenterology 159, 1406–1416.e11 (2020).
Chen, J. et al. Microsatellite status detection of colorectal cancer: evaluation of inconsistency between PCR and IHC. J. Cancer 14, 1132–1140 (2023).
Ladabaum, U., Ford, J. M., Martel, M. & Barkun, A. N. American Gastroenterological Association technical review on the diagnosis and management of lynch syndrome. Gastroenterology 149, 783–813.e20 (2015).
Yu, F., Makrigiorgos, A., Leong, K. W. & Makrigiorgos, G. M. Sensitive detection of microsatellite instability in tissues and liquid biopsies: Recent developments and updates. Comput. Struct. Biotechnol. J. 19, 4931–4940 (2021).
Sieben, N. L. G., Ter Haar, N. T., Cornelisse, C. J., Jan Fleuren, G. & Cleton-Jansensen, A.-M. PCR artifacts in LOH and MSI analysis of microdissected tumor cells. Hum. Pathol. 31, 1414–1419 (2000).
Monument, M. J., Lessnick, S. L., Schiffman, J. D. & Randall, R. L. Microsatellite Instability in Sarcoma: Fact or Fiction?. Int. Sch. Res. Not. 2012, 473146 (2012).
Walsh, E. & Orsi, N. M. The current troubled state of the global pathology workforce: a concise review. Diagn. Pathol. 19, 163 (2024).
Robboy, S. J. et al. The Pathologist Workforce in the United States: II. An Interactive Modeling Tool for Analyzing Future Qualitative and Quantitative Staffing Demands for Services. Arch. Pathol. Lab. Med. 139, 1413–1430 (2015).
Greenson, J. K. et al. Pathologic predictors of microsatellite instability in colorectal cancer. Am. J. Surg. Pathol. 33, 126–133 (2009).
Chang, X. et al. Predicting colorectal cancer microsatellite instability with a self-attention-enabled convolutional neural network. Cell Rep. Med. 4, 100914 (2023).
Wagner, S. J. et al. Transformer-based biomarker prediction from colorectal cancer histology: A large-scale multicentric study. Cancer Cell 41, 1650–1661.e4 (2023).
Kather, J. N. et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 25, 1054–1056 (2019).
White, A. et al. A review of sex-related differences in colorectal cancer incidence, screening uptake, routes to diagnosis, cancer stage and survival in the UK. BMC Cancer 18, 906 (2018).
Himbert, C. et al. Clinical characteristics and outcomes of colorectal cancer in the colocare study: differences by age of onset. Cancers 13, 3817 (2021).
Fleming, M., Ravula, S., Tatishchev, S. F. & Wang, H. L. Colorectal carcinoma: Pathologic aspects. J. Gastrointest. Oncol. 3, 153–173 (2012).
McPhail, S., Johnson, S., Greenberg, D., Peake, M. & Rous, B. Stage at diagnosis and early mortality from cancer in England. Br. J. Cancer 112, S108–S115 (2015).
Gutierrez, C., Ogino, S., Meyerhardt, J. A. & Iorgulescu, J. B. The Prevalence and Prognosis of Microsatellite Instability-High/Mismatch Repair-Deficient Colorectal Adenocarcinomas in the United States. JCO Precis. Oncol. e2200179 https://doi.org/10.1200/PO.22.00179. (2023).
Cao, R. et al. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics 10, 11080–11091 (2020).
Nguyen, H.-G. et al. Image-based assessment of extracellular mucin-to-tumor area predicts consensus molecular subtypes (CMS) in colorectal cancer. Mod. Pathol. 35, 240–248 (2022).
Adib, E. et al. Deep learning–powered analysis of tumor-infiltrating lymphocytes (TILs) in colorectal cancer. J. Clin. Oncol. 43, 288–288 (2025).
Saillard, C. et al. Validation of MSIntuit as an AI-based pre-screening tool for MSI detection from colorectal cancer histology slides. Nat. Commun. 14, 6695 (2023).
Hezi, H., Gelber, M., Balabanov, A., Maruvka, Y. E. & Freiman, M. CIMIL-CRC: A clinically-informed multiple instance learning framework for patient-level colorectal cancer molecular subtypes classification from H&E stained images. Comput. Methods Prog. Biomed. 259, 108513 (2025).
Gustav, M. et al. Deep learning for dual detection of microsatellite instability and POLE mutations in colorectal cancer histopathology | npj Precision Oncology. Npj Precis. Oncol. 8, (2024).
He, B. et al. Development of a Multimodal Deep Learning Model for Predicting Microsatellite Instability in Colorectal Cancer by Integrating Histopathological Images and Clinical Data. Preprint at https://doi.org/10.21203/rs.3.rs-4200523/v1 (2024).
Raza, M., Awan, R., Bashir, R. M. S., Qaiser, T. & Rajpoot, N. M. Dual attention model with reinforcement learning for classification of histology whole-slide images. Comput. Med. Imaging Graph. 118, 102466 (2024).
Nowak, M. et al. Single-cell AI-based detection and prognostic and predictive value of DNA mismatch repair deficiency in colorectal cancer. Cell Rep. Med. 5, 101727 (2024).
Lo, C.-M., Jiang, J.-K. & Lin, C.-C. Detecting microsatellite instability in colorectal cancer using Transformer-based colonoscopy image classification and retrieval. PloS One 19, e0292277 (2024).
Echle, A. et al. Artificial intelligence for detection of microsatellite instability in colorectal cancer—a multicentric analysis of a pre-screening tool for clinical application. ESMO Open 7, 100400 (2022).
Lee, S. H., Song, I. H. & Jang, H.-J. Feasibility of deep learning-based fully automated classification of microsatellite instability in tissue slides of colorectal cancer. Int. J. Cancer 149, 728–740 (2021).
Clinical actionability of triaging DNA mismatch repair deficient colorectal cancer from biopsy samples using deep learning - eBioMedicine. The Lancet 81, (2022).
Loughrey, M. B. et al. Identifying mismatch repair-deficient colon cancer: near-perfect concordance between immunohistochemistry and microsatellite instability testing in a large, population-based series. Histopathology 78, 401–413 (2021).
Harrell, F. E. Regression Modeling Strategies: With Applications to Linear Models, Logistic Regression, and Survival Analysis. (Springer, New York, NY, https://doi.org/10.1007/978-1-4757-3462-1 (2001).
Hanley, J. A. & McNeil, B. J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143, 29–36 (1982).
Naing, L., Nordin, R. B., Abdul Rahman, H. & Naing, Y. T. Sample size calculation for prevalence studies using Scalex and ScalaR calculators. BMC Med. Res. Methodol. 22, 209 (2022).
Arslan, S. et al. A systematic pan-cancer study on deep learning-based prediction of multi-omic biomarkers from routine pathology images. Commun. Med. 4, 48 (2024).
Macenko, M. et al. A method for normalizing histology slides for quantitative analysis. in 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro 1107–1110 https://doi.org/10.1109/ISBI.2009.5193250 (2009).
Kather, J. N., Halama, N. & Marx, A. 100,000 histological images of human colorectal cancer and healthy tissue. Zenodo https://doi.org/10.5281/zenodo.1214456 (2018).
Chen, X., Fan, H., Girshick, R. & He, K. Improved Baselines with Momentum Contrastive Learning. Preprint at https://doi.org/10.48550/arXiv.2003.04297 (2020).
Acknowledgements
This project was (partially) funded by Innovate UK Smart Grant 10054824. The results shown in this study are partially based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. We acknowledge the support of iCAIRD (Industrial Centre for Artificial Intelligence Research in Digital Diagnostics), funded by Innovate UK, for the provision of the I1-UK-CRC-CZI-1-DEV cohort. Biosamples in the W1-UK-CRC-CZI-1-BLIND cohort were obtained from the Wales Cancer Biobank (https://openbioresources.metajnl.com/articles/10.5334/ojb.46) which is funded by Health and Care Research Wales. Other investigators may have received specimens from the same subjects. Biosamples in the N1-UK-CRC-SVS-1-BLIND cohort were obtained from the Northern Ireland Biobank (https://openbioresources.metajnl.com/articles/10.5334/ojb.47 which received core funding from the Health and Social Care Research and Development Division of the Public Health Agency in Northern Ireland. O.M is funded by Barts Charity (MGU0504).
Author information
Authors and Affiliations
Contributions
S.A. led the study. S.W., S.A., F.N., C.B., and A.G. designed the study. S.W., F.N., and C.B., performed the study experiments and analysed the results. S.W., C.B., F.N., A.G., S.A., and J.S. developed the software and the core AI algorithms and models as well as the computational approaches used in the study. A.G., J.S., C.B., F.N., S.W., and S.A. developed the platform to run the study experiments. D.J.H., I.H.U., N.B., M.H., B.M., G.R., M.C., C.F., E.W., M.L., M.S.T., R.C., and N.M.O. provisioned data for the study. D.J.H., I.H.U., and J.B. facilitated access to a part of the data. N.B. carried out blinded validation analysis for the results. P.P., J.N.K, N.K., N.M.O., E.W., D.M., O.M., and S.S. provided clinical and biological inputs for the study. D.M. and S.S. evaluated the images and performed data quality controls. S.W., S.A., and C.B. created the visualisations and illustrations for the manuscript. S.A., S.W., C.B., and F.N. verified the study results. S.A. and C.B. prepared the manuscript. J.C.B. provided suggestions on the contextual positioning of the manuscript and contributed editorial feedback. All authors discussed the findings and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interests: S.W., N.K., D.M., S.S., P.P., J.S., F.N., A.G., C.B., A.M., and S.A are employees of Panakeia Technologies Limited, UK. S.W., N.K., P.P., J.S., F.N., A.G., C.B., and S.A declare stock ownership interests with Panakeia Technologies Limited, UK. P.P. holds a leadership position at Panakeia Technologies Limited, UK. O.M. provides consulting services for Panakeia Technologies Limited, UK. J.B. holds leadership positions at Panakeia Technologies Limited and Bering Limited, UK. J.B. declares travel, accommodations, and expenses for QURE AI, UK, and stock ownership interests with Orli Health, UK. He has received honoraria from Bayer, Germany, and research funding from QURE AI, UK, and IBEX Medical Analytics, Israel. M.S.T. is a scientific advisor to Mindpeak and Sonrai Analytics, and has received honoraria recently from BMS, MSD, Roche, Sanofi and Incyte. He has received grant support from Phillips, Roche, MSD and Akoya. None of these disclosures are related to this work. J.N.K. declares consulting services for Bioptimus, France; Owkin, France; DoMore Diagnostics, Norway; Panakeia Technologies Limited, UK; AstraZeneca, UK; Mindpeak, Germany; and MultiplexDx, Slovakia. Furthermore, he holds shares in StratifAI GmbH, Germany, and Synagen GmbH, Germany, and has received a research grant from GSK. He has also received honoraria by AstraZeneca, Bayer, Daiichi Sankyo, Janssen, Merck, MSD, BMS, Roche, Pfizer, and Fresenius. D.J.H. is an employee of and holds a leadership position at Nucana PLC, UK, and ILC Therapeutics Limited, UK, with which he also declares stock ownership interests. He has received research funding from Nucana PLC, UK. N.M.O. is a stock owner at 4D Path. He also previously received research funding from, and had a consultancy role in, 4D Path. He currently receives research funding from Tristar Technologies, with whom he also has a consultancy role. M.C and C.F. have a consultancy role in TriStar Technologies. E.W. has previously been in receipt of a 4D Path PhD studentship, and is currently in receipt of funding from Panakeia Technologies Limited, UK. J.C.B. is currently employed by Precede Bio. He holds a leadership position on the Board of Directors at Saga Diagnostics. He holds stock or ownership interests in AstraZeneca, Precede Bio, Corista, and Nexosomes. He has provided consulting or advisory services to Akoya, Leica, Agilent, Multiplex, Bain Capital, and ExAI. Other authors declare no other competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bass, C., Ntelemis, F., Schmidt, J. et al. H&E-based MSI/MMR testing with AI in colorectal cancer: a multi-centred blinded evaluation. npj Digit. Med. 9, 44 (2026). https://doi.org/10.1038/s41746-025-02218-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-02218-5