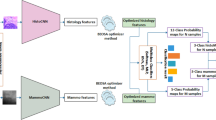
Abstract
Reliable interpretation of clinical imaging requires integrating complementary evidence across modalities, yet most AI systems remain limited by single-modality analysis and poor generalization across institutions. We propose a unified cross-modal framework that bridges mammography and histopathology for breast cancer diagnosis through: (1) a shared vision transformer encoder with lightweight modality-specific adapters, (2) a weakly supervised patient-level contrastive alignment module that learns cross-modal correspondences without pixel-level supervision, (3) domain generalization strategies combining MixStyle augmentation and invariant risk minimization, and (4) causal test-time adaptation for unseen target domains. The model jointly addresses classification, lesion localization, and pathological grading while generating reasoning-guided attention maps that explicitly link suspicious mammographic regions with corresponding histopathological evidence. Evaluated on four public benchmarks (CBIS-DDSM, INbreast, BACH, CAMELYON16/17), the framework consistently outperforms state-of-the-art unimodal, multimodal, and domain generalization baselines, achieving mean AUC of 0.90 under rigorous leave-one-domain-out evaluation and substantially smaller domain gaps (0.03 vs. 0.06–0.10). Visualization and interpretability analyses further confirm that predictions align with clinically meaningful features, supporting transparency and trust. By advancing multimodal integration, cross-institutional robustness, and explainability, this study represents a step toward clinically deployable AI systems for diagnostic decision support.
Similar content being viewed by others
Data availability
All imaging data analyzed in this study were obtained from publicly accessible biomedical databases: CBIS-DDSM (Curated Breast Imaging Subset of DDSM), accessible via The Cancer Imaging Archive: https://www.cancerimagingarchive.net/collection/cbis-ddsm/; INbreast dataset, available on Mendeley Data: https://data.mendeley.com/datasets/3w8hnz2wff/1; BACH (Grand Challenge on Breast Cancer Histology images) dataset, available via Zenodo: https://zenodo.org/records/3632035 CAMELYON16/17 datasets are publicly available through the Grand Challenge website: https://camelyon17.grand-challenge.org/Data/, and mirrored on AWS Open Data: https://registry.opendata.aws/camelyon/. Processed or derived data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
The implementation of the proposed cross-modal breast cancer diagnosis framework, including all training scripts, evaluation pipelines, and model architectures, is publicly available at the following repository: https://anonymous.4open.science/r/ruxian-6A03/README.md (for review purposes). Upon publication, the code will be made permanently available under an open-source license. The codebase is implemented in Python 3.8+ using PyTorch 1.10.0 or higher. Key parameters used to generate the results reported in this study are as follows: image size 224 × 224 pixels, patch size 16 × 16 pixels, embedding dimension 768, transformer depth 12 layers, 12 attention heads, batch size 8–16, learning rate 1 × 10−4, weight decay 1 × 10−4, trained for 50–100 epochs using the Adam optimizer. Mammography images (DICOM format, single-channel grayscale) were normalized to [ − 1, 1] range, and histopathology images (PNG/JPEG format, RGB channels) underwent Macenko stain normalization. Cross-modal pairing was performed at the patient level (same patient ID for mammography and histopathology pairs). All random seeds were set to 42 for reproducibility. A complete list of dependencies with specific version requirements, detailed usage instructions, and configuration files are provided in the repository.
References
Lee, R. S. et al. A curated mammography data set for use in computer-aided detection and diagnosis research. Sci. Data 4, 1–9 (2017).
Moreira, I. C. et al. Inbreast: toward a full-field digital mammographic database. Academic Radiol. 19, 236–248 (2012).
Aresta, G. et al. Bach: Grand challenge on breast cancer histology images. Med. image Anal. 56, 122–139 (2019).
Litjens, G. et al. 1399 h&e-stained sentinel lymph node sections of breast cancer patients: the camelyon dataset. GigaScience 7, giy065 (2018).
Huang, Y. et al. Nomogram for predicting neoadjuvant chemotherapy response in breast cancer using mri-based intratumoral heterogeneity quantification. Radiology 315, e241805 (2025).
Schwarzhans, F. et al. Image normalization techniques and their effect on the robustness and predictive power of breast MRI radiomics. Eur. J. Radiol. 187, 112086 (2025).
Braman, N. et al. Novel radiomic measurements of tumor-associated vasculature morphology on clinical imaging as a biomarker of treatment response in multiple cancers. Clin. Cancer Res. 28, 4410–4424 (2022).
Shubeitah, M., Hasasneh, A. & Albarqouni, S. Two-steps approach for breast cancer detection and classification using convolutional neural networks. Int. J. Eng. Appl. 12, (2024).
Wei, X. et al. Vikl: A mammography interpretation framework via multimodal aggregation of visual-knowledge-linguistic features. arXiv preprint arXiv:2409.15744 (2024).
Hou, J. et al. Self-explainable ai for medical image analysis: A survey and new outlooks. arXiv preprint arXiv:2410.02331 (2024).
Wang, A. Q. et al. A framework for interpretability in machine learning for medical imaging. IEEE Access 12, 53277–53292 (2024)..
Musa, A., Prasad, R. & Hernandez, M. Addressing cross-population domain shift in chest x-ray classification through supervised adversarial domain adaptation. Sci. Rep. 15, 11383 (2025)..
Sethi, S. et al. ProtoECGNet: Case-Based Interpretable Deep Learning for Multi-Label ECG Classification with Contrastive Learning. In Proc. of the 10th Machine Learning for Healthcare Conference (eds Agrawal, M. et al) Vol. 298 https://proceedings.mlr.press/v298/sethi25a.html (PMLR, 2025).
Mayilvahanan, P. et al. In Search of Forgotten DomainGeneralization. International Conference on Learning Representations (ICLR), (Spotlight) (2025).
Tian, Y. et al. Learning vision from models rivals learning vision from data. In Proc. of the IEEE/CVF conference on computer vision and pattern recognition, 15887–15898 (2024).
Wang, Y., Wu, Y. & Zhang, H. Lost domain generalization is a natural consequence of lack of training domains. In Proc. of the AAAI Conference on Artificial Intelligence Vol. 38, 15689–15697 (2024).
Tan, Z., Yang, X. & Huang, K. Rethinking multi-domain generalization with a general learning objective. In Proc. of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 23512–23522 (2024).
Addepalli, S., Asokan, A. R., Sharma, L. & Babu, R. V. Leveraging vision-language models for improving domain generalization in image classification. In Proc. of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 23922–23932 (2024).
Zhou, K., Yang, Y., Qiao, Y. & Xiang, T. Mixstyle neural networks for domain generalization and adaptation. Int. J. Computer Vis. 132, 822–836 (2024).
Khoee, A. G., Yu, Y. & Feldt, R. Domain generalization through meta-learning: a survey. Artif. Intell. Rev. 57, 285 (2024).
Bai, S. et al. Diprompt: Disentangled prompt tuning for multiple latent domain generalization in federated learning. In Proc. of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 27284–27293 (2024).
Li, Y. et al. Federated domain generalization: A survey. In Proc. of the IEEE (IEEE, 2025).
Yan, S. et al. Prompt-Driven Latent Domain Generalization for Medical Image Classification. IEEE Transac. Med. Imaging 44, 348–360 (2025).
Tian, F. et al. Prediction of tumor origin in cancers of unknown primary origin with cytology-based deep learning. Nat. Med. 30, 1309–1319 (2024).
Li, H., Wang, S., Zhang, Y. & Li, W. A new paradigm for cytology-based artificial intelligence-assisted prediction for cancers of unknown primary origins. Innov. Life 2, 100086 (2024).
Ghani, H. et al. Gpsai: A clinically validated ai tool for tissue of origin prediction during routine tumor profiling. Cancer Res. Commun. 5, 1477–1489 (2025).
Xin, H. et al. Automatic origin prediction of liver metastases via hierarchical artificial-intelligence system trained on multiphasic ct data: a retrospective, multicentre study. EClin. Med. 69, (2024).
Wang, X. et al. A pathology foundation model for cancer diagnosis and prognosis prediction. Nature 634, 970–978 (2024).
Ma, W. et al. New techniques to identify the tissue of origin for cancer of unknown primary in the era of precision medicine: progress and challenges. Brief. Bioinforma. 25, bbae028 (2024).
Wang, H. et al. Clap: learning transferable binary code representations with natural language supervision. In Proceedings of the 33rd ACM SIGSOFT International Symposium on Software Testing and Analysis, 503–515 (2024).
Zhang, J., Huang, J., Jin, S. & Lu, S. Vision-language models for vision tasks: A survey. IEEE Trans. Pattern Anal. Mach. Intell. 46, 5625–5644 (2024).
Zhang, Y. et al. Exploring the transferability of visual prompting for multimodal large language models. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 26562–26572 (2024).
Rezaei, R. et al. Learning visual prompts for guiding the attention of vision transformers. In Proc. of TV4 Workshop (ICLR, 2025).
Ndir, T. C., Schirrmeister, R. T. & Ball, T. EEG-CLIP: Learning EEG representations from natural language descriptions. Front. Robot. AI 12, 1625731 (2025).
Yuan, K., Navab, N., Padoy, N. et al. Procedure-aware surgical video-language pretraining with hierarchical knowledge augmentation. Adv. Neural Inf. Process. Syst. 37, 122952–122983 (2024).
Jiang, X. et al. Supervised fine-tuning in turn improves visual foundation models. arXiv preprint arXiv:2401.10222 (2024).
Zheng, F. et al. Exploring low-resource medical image classification with weakly supervised prompt learning. Pattern Recognit. 149, 110250 (2024).
Acknowledgements
The authors gratefully acknowledge the institutional support from their affiliated hospitals and research institutes, which provided the necessary infrastructure and collaborative environment for this study. We also thank the open-access biomedical imaging databases, whose publicly available resources enabled the reproducibility and validation of our findings. Funding for this project was provided by Suzhou Gusu talent plan for Health Technical Personnel project (Grant No. GSWS2021024) and the Natural Science Foundation of Jiangsu Province (Grant No. BK20250383) and Nanjing Medical University Gusu School Youth Talent Development Program (Grant No. GSKY20250523) and Postgraduate Research & Practice Innovation Program of Jiangsu Province (Grant No. SJCX25_1793).
Author information
Authors and Affiliations
Contributions
X.Z. had full access to all study data and assumed responsibility for the integrity and accuracy of the analyses (Validation, Formal analysis). Z.G. and MS conceptualized the study, designed the methodology, and participated in securing research funding (Conceptualization, Methodology, Funding acquisition). M.L. and M.D. carried out data acquisition, curation, and investigation (Investigation, Data curation) and provided key resources, instruments, and technical support (Resources, Software). G.J., H.S., and Q.C. drafted the initial manuscript and generated visualizations (Writing – Original Draft, Visualization). M.D., G.J., H.S., and Q.C. supervised the project, coordinated collaborations, and ensured administrative support (Supervision, Project administration). All authors contributed to reviewing and revising the manuscript critically for important intellectual content (Writing – Review \& Editing) and approved the final version for submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhong, X., Gu, Z., Shanmuganathan, M. et al. Bridging radiology and pathology: domain-generalized cross-modal learning for clinical. npj Digit. Med. (2026). https://doi.org/10.1038/s41746-026-02423-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41746-026-02423-w