Abstract
This study focuses on refining growth-rate-based drug response metrics for patient-derived tumor organoid screening using brightfield live-cell imaging. Traditional metrics like Normalized Growth Rate Inhibition (GR) and Normalized Drug Response (NDR) have been used to assess organoid responses to anticancer treatments but face limitations in accurately quantifying cytostatic and cytotoxic effects across varying growth rates. Here, we introduce the Normalized Organoid Growth Rate (NOGR) metric, specifically developed for brightfield imaging-based assays. A label-free image analysis model was applied to segment organoids precisely, track their growth rates over time, and classify viable and dead organoids. Testing eleven phenotypically distinct pancreatic cancer organoid models with five chemotherapeutics demonstrates that the NOGR metric more effectively captures cytostatic and cytotoxic drug effects compared to existing methods. This approach enhances the biological relevance of drug sensitivity assessments on organoids and offers a valuable tool for advancing personalized cancer treatment strategies.
Similar content being viewed by others
Introduction
Patient-derived tumor organoids are revolutionizing cancer drug screening, heralding a new era of drug development and personalized oncology treatments. These advanced three-dimensional cultures, developed from a patient’s tumor cells, faithfully replicate the intricate structural and genetic features of cancer tissues1. The relative ease of establishing tumor organoids from a diverse array of tumor types has led to a significant expansion in tumor organoid biobanks2. These repositories, housing a vast and varied collection of tumor organoids, represent a critical resource in cancer research and drug development1. This advancement allows researchers to evaluate the efficacy and toxicity of potential drugs on a patient-specific basis, vastly improving the predictive accuracy of preclinical drug screening.
Drug screening using patient-derived organoids often relies on basic ATP-based assays, a method that, despite its utility, fails to capture the full complexity of these sophisticated models3,4. Such assays predominantly offer a bulk readout, which significantly limits the ability to analyze the heterogeneity in drug responses within the organoid population. Moreover, this approach does not facilitate the detailed investigation of drug mechanisms of action, particularly in distinguishing between cytostatic (inhibiting cell growth) and cytotoxic (killing cells) effects. This limitation is a notable concern, as it oversimplifies the intricate interactions and responses within tumor organoids, thereby potentially overlooking nuanced but crucial aspects of drug efficacy and behavior. Imaging offers a more refined approach to drug screening, overcoming all the limitations of ATP-based assays. Widefield live-cell imaging especially, is becoming more widely available to researchers because of its cost-effectiveness and ease of use compared to other imaging systems, such as confocal microscopes.
To support the implementation of live-cell imaging for organoid drug screening, we have developed a high-throughput compatible method, called OrBITS (Organoid Brightfield Identification-based Therapy Screening), which utilizes label-free bulk and single-organoid deep learning-based segmentation from brightfield images5,6. Similarly, other tools, such as deepOrganoid, OrgaExtractor, OrganoID, and OrgaSegment, have also been developed for label-free organoid segmentation and are becoming more widely available7,8,9,10.
Since then, we have generated thousands of images of organoids treated with a variety of anticancer drugs. This repository allowed us to further refine our label-free organoid detection algorithm to obtain a universally applicable image analysis module. Additionally, we used this repository to develop a new drug response metric, the Normalized Organoid Growth Rate (NOGR), specifically designed to capture key aspects of organoid reactions to treatments in a single metric. Conventionally, relative viability (RV) is used, referring to the proportion of living cells after exposure to a treatment compared to a 0% control group (usually the drug’s solvent) with or without at 100% control group (maximal response)11. This is often used in luminescence-based drug screenings of organoids12,13. A dose-response curve is plotted using RV measurements obtained from a range of drug concentrations, known as a drug titration range. Several key parameters such as IC50, AUC and Emax values are then derived from this curve to assess drug sensitivity14. However, these metrics fail to capture the complexity of a drug response because they cannot distinguish between cytostatic and cytotoxic response and RV is sensitive to variations in seeding density. More importantly, Hafner et al. showed that these traditional drug response metrics are highly sensitive to the number of cell divisions during the assay15. This means that the observed drug response may be more a reflection of the cell’s division rate rather than its actual genetic susceptibility or resistance to the drug. This led to the development and adoption of Normalized Growth Rate Inhibition (GR) metrics like GR50, AOC, and GRmax, which are designed to be insensitive to the number of cell divisions and thus offer a more consistent and biologically relevant measure of drug sensitivity and resistance by comparing dynamic changes in growth rate15,16. Gupta et al. further refined the GR-based metrics and propose the use of the Normalized Drug Response (NDR) which make use of both positive and negative control conditions while the GR only accounts for a negative or vehicle control17. The NDR was originally tested in 2D drug screening assays using both luminescence and image-based readouts. These assays employed a fluorescence marker for cell death in addition to a viability marker. GR metrics have already been used in endpoint ATP-based organoid drug screening, in which one or more reference wells are measured at the initial timepoint as a reference point for all conditions4,18,19,20. Without the ability to perform intra-well normalization, it becomes challenging to accurately calculate and compare growth rates, as variations in initial seeding density across wells can skew results. This is where the major advantage of live-cell imaging comes into play. With live-cell imaging, growth rates are calculated for each well individually at multiple timepoints, allowing for precise normalization that accounts for any differences in seeding density. This method ensures that the growth rate measurements are specific and relevant to each unique condition, providing a more accurate and representative analysis of how organoids respond to different anticancer drugs over time. We have already established that the NDR outperforms RV and GR when applied to live-cell imaging based organoid drug screening5,6,21. In this study we applied the knowledge we gained from high-throughput drug screenings to evaluate available metrics and develop a new and improved Normalized Organoid Growth Rate (NOGR) metric for live-cell imaging-based organoid drug screenings. In addition, we tested the accuracy of our label-free organoid detection model OrBITS on an independent selected panel of eleven PDAC organoids which were identified from a large organoid cohort in order to capture a maximum of phenotypic differences between them. We utilized this set of organoids to critically assess the limitations of the GR and NDR metrics previously employed in organoid drug screenings, both by our team and others5,6,21. Additionally, we aimed to validate the NOGR metric, demonstrating its effectiveness as a more accurate and reliable method for evaluating drug responses to five standard-of-care chemotherapeutics used for the treatment of pancreatic cancer22.
Results
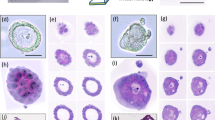
Phenotypic characterization of PDAC organoids
Our PDAC organoid panel, derived from from The PaCaOmics clinical trial cohort that includes 220 patients (ClinicalTrials.gov ID N° NCT01692873), was selected based on their transcriptomic differences in order to obtain a large panel of phenotypic differences represented by a significant variation in morphology. This selected subset of eleven organoid models was systematically classified into four distinct phenotypic classes (Figs. 1, 2). (i) The most prevalent class comprised of thick-walled cystic organoids, as exemplified by PC_003, PC_079, PC_083, PC_089, and PC_104. These organoids were characterized by their robust outer walls and sizable cystic spaces. (ii) Following this, we identified a group of organoids exhibiting mixed morphologies, PC_009, PC_076, and PC_379, combining both solid and thick-walled cystic features. (iii) A notable exception within our panel was PC_028, which exhibited a unique, dense, and entirely solid morphology. (iv) Finally, we observed a less common phenotype in PC_117 and PC_372, characterized by their thin-walled cystic structure.
Our label-free image analysis algorithm excelled in identifying various morphological subtypes within our PDAC organoid panel, underscoring the algorithm’s overall effectiveness (Fig. 2). Large, thin-walled cystic organoids like PC_372 at 120 h highlight an area for future refinement (Fig. 2, bottom right).
Besides distinct morphologies, the organoid panel also demonstrated a large variation in their growth rate (Fig. 3A). Timelapse images (0 h, 72 h, 120 h) for all organoid models are available in Supplementary Fig. 1. Interestingly, PC_379 did not grow over time, but instead, these organoids were highly migratory and with a tendency to merge (Supplementary fig. 1). This distinct behavior could indicate a more aggressive or advanced stage of tumor development, which warrants further investigation. Altogether, our characterizations underscore the need for more refined, growth-rate based drug response metrics.
Image-based quantification of organoid growth rate based on the Viability (V) metric, normalized to timepoint 0 (mean ± SD, n = 3) for A vehicle control and (B) positive control staurosporine. C Representative images of dying organoids. The upper panel displays the overlap of the raw brightfield and green fluorescence images that mark dead cells using cytotox green. The lower panel displays the overlay with the following segmentation and masking: Magenta = label-free organoid segmentation; Green = green fluorescence; Yellow = label-free death organoid segmentation. Scale bar: 75 μm.
Label-free classification of dying organoids
In developing new growth-rate drug response metrics, it was crucial to exclude dead cells or organoids to ensure accurate growth-rate calculations. Our initial approach involved calculating the Viability (V) metric by subtracting the total area showing cytotoxic green fluorescence, which indicates regions of cell death, from the total brightfield area that represents the overall organoid area. For detailed methodology, refer to the Materials and Methods section. However, we observed that a complete overlap of the fluorescent signal with the cytotoxic regions was not consistently achieved (Fig. 3C). This discrepancy underscores the importance of normalizing these measurements against a full cell death control, as described in the NDR method by Gupta et al.17. To enhance the cytotoxic dynamic range, which we define as the range of observable growth rates from 1 (no growth) to 0 (completely inhibited) (Fig. 3B), we refined our methodology by incorporating label-free detection of dead organoids. Dying organoids are characterized by a distinct phenotype which exhibit a dark and granulated appearance (Fig. 3C). Importantly, these images also visualize the limitation of fluorescent cell death markers to mark dead cells in organoids, especially at earlier timepoints (e.g., 72 h). The label-free cell death segmentation accurately classified dying organoids at various stages and sizes, although detection of smaller dead organoids needs further refinement in future versions. Moreover, using label-free cell death detection improved the dynamic range of growth rate. By using fluorescence area as an indicator of cell death, the growth-rate ranged from 1 to 0.5, but with label-free cell death detection, the growth -rate ranged from 1 to 0.2 (Fig. 3B)5.
Taken together, it is clear that individual growth-rates of organoid can vary drastically and must be comprehensively captured when reporting the effects of drugs in a screening. Label-free organoid and cell death detection can more accurately reflect the growth-rates and mitigates the limitations of fluorescence detection.
Comparison of growth rate drug response metrics
For further analysis, we have excluded PC_379 due to a lack of growth (growth rate = 1) and selected timepoint 72 h due to a more accurate label-free segmentation of thin-walled cystic organoids compared to timepoint 120 h (Supplementary fig. 1). At this timepoint, we identified PC_009 as the slowest growing organoid in our panel with a growth rate of approximately 1.5, and PC_117 as the fastest with a rate of 8 (Fig. 3A). These values were used as the maximum benchmarks for slow-growing and fast-growing organoids, respectively. We then calculated the GR, NDR, and NOGR growth rate metrics values across growth rate ranges from 0 to 1.5 to mimic drug-induced growth rate inhibition in slow-growing organoids (Fig. 4A), and from 0 to 8 for fast-growing organoids (Fig. 4B). Importantly, GR is normalized to a vehicle control and NDR and NOGR are normalized to both the vehicle and positive control. The lower growth rate limit was set at 0.3, indicated by the vertical red dotted line, corresponding with full cell death induced by the positive control as discussed earlier. For slow-growing organoids, the NDR metric strongly overestimates the cytotoxic effect (NDR < 0) of a given drug, since a value of -1 is already achieved at a growth rate of 0.75 instead of 0.3 (Fig. 4A). The cytostatic range (0 < growth rate metric < 1) yields similar results for all three metrics (Fig. 4A). For fast-growing organoids, the GR metric fails to accurately capture cytotoxic responses and underestimates the effect due to the lack of a positive control normalization (Fig. 4B). Similarly, the NDR metric underestimates the cytostatic effect of a given drug due to its logarithmic fit, exemplified by an NDR value of approximately 0.75 instead of 0.5 at 50% growth rate inhibition (GR = 4). The NOGR metric combines the strengths of the GR and NDR metrics and accurately captures the cytostatic and cytotoxic drug response of both slow and fast-growing organoids (Fig. 4A, B). This is clearly visualized in the data extracted from our drug screening with five standard-of-care chemotherapeutics (raw data in Supplementary Data 1). For example, the NDR overestimates the cytotoxic effect in the slow-growing PC_009 and underestimates the cytostatic effect in the fast-growing PC_117 compared to the NOGR metric (Fig. 4C). The GR metric strongly correlates with the NOGR in the cytostatic range, but strongly underestimates the cytotoxic response in the fast-growing organoids such as PC_117 (Fig. 4D).
A Simulated growth rate response metrics (GR, NDR, and NOGR) plotted against growth rate ranges from 0 to 1.5 to mimic drug response (i.e., reduction in growth rate) in slow-growing organoids. The red and green vertical dashed lines represent the growth rates of the positive and negative controls, respectively. The vertical blue dashed line marks a growth rate of 1, the cutoff between cytostatic (> 1) and cytotoxic (< 1) drug responses. The NDR metric overestimates cytotoxicity (NDR < 0), as an NDR value of -1 is already obtained at a growth rate of around 0.75 instead of 0.3 (positive control). B Growth rate metrics plotted against growth rates ranging from 0 to 8, representing fast-growing organoids. The GR metric underestimates cytotoxicity (GR < 0) due to the lack of positive control normalization, while the NDR metric underestimates the cytostatic effect (1 > NDR > 0) due to its logarithmic fit. C Comparison of NOGR vs. NDR values for a panel of 10 PDAC organoids treated with a concentration range of five chemotherapeutics for 72 h. Each data point represents an individual concentration, chemotherapeutic, and organoid, with the color legend indicating the corresponding organoid line and its growth rate. D Comparison of NOGR vs. GR values for the same panel of PDAC organoids at 72 h. The scatter plot shows the relationship between the two metrics for different concentrations of the tested chemotherapeutics and organoid lines.
Altogether, we illustrated the strengths and limitations of the different metrics, particularly in the context of fast- and slow-growing organoids (Table 1). Moreover, we demonstrated that the NOGR combines the advantages of both GR and NDR to more accurately portray cytostatic and cytotoxic effects for organoids of various growth rates.
The NOGR accurately captures the drug response that is visualized on microscopic images
A significant benefit of employing live-cell imaging lies in its capacity to serve as a reliable reference for confirming the precision and accuracy of various measured and calculated metrics, including GR, NDR, and NOGR. For this experiment, we selected paclitaxel (1, 4.6, 21.5, 100 nM) treated organoids, due to the wide variability in response between the organoids (Fig. 5A). Figure 5B–F show the corresponding image of untreated (left) and treated (right) organoids for selected conditions. The NOGR accurately captures the drug response for proliferative organoids (NOGR ≥ 1, Fig. 5B), a half cytostatic effect (NOGR = 0.44, Fig. 5C), a full cytostatic effect (NOGR = 0.06, Fig. 5D), a half cytotoxic effect (NOGR = −0.40, Fig. 5E) and a nearly full cytotoxic effect (NOGR = −0.93, Fig. 5F). The underestimation of cytostatic effects by the NDR metric (Fig. 5C, D) and the underestimation of the cytotoxic effect by the GR metric (Fig. 5E, F) are also evidenced.
A Dose-response curve of paclitaxel treated organoids (1, 4.6, 21.5, 100 nM) for 72 h (mean ± SD, n = 3). The circles represent the conditions that were selected for representative images for (B) proliferative organoids; (C) a half cytostatic effect; (D) a full cytostatic effect; (E) a half cytotoxic effect; (F) a nearly full cytotoxic effect. The left images represent untreated organoids and the right image represents treated organoids at 72 h. GR Normalized growth rate inhibition, NDR Normalized Drug Response, NOGR Normalized Organoid Growth Rate. The circles indicate the corresponding conditions of the images on the dose-response curve. Image label concentrations were rounded to the nearest whole number. Scale bar: 200 μm.
Drug screening results
Having established the more accurate representation of growth-rate with the NOGR metric, we aimed to derive relevant drug metrics. Figure 6A visualizes an example of the curve fitting and outlier detection to obtain the NOGR50 metric, the concentration at which the growth of the organoid is inhibited by 50%, following treatment with Gemcitabine. Fitted curves of all therapeutics are presented in Supplementary fig. 2 and all drug response data is available in Supplementary Data 1. In addition, we also calculated the Area Over the Curve (AOC), visualized by the blue area on Fig. 6A, and normalized it to the maximal area. This ratio provided a value between 0 and 1 to reflect no effect and complete effect of the drug, respectively (Fig. 6B). Derivates of this metric are the NOGR_AOC_0 which is the area over the curve up to y = 0 as a selective measure for cytotoxicity and the NOGR_AOC_Ratio which represents the fraction of the total AOC below < 0 (Fig. 6C, D). A value closer to one indicates a higher cytotoxicity across the titration range. When applying these different metrics, we observed that paclitaxel was the most potent drug with a variable response in our organoid panel. Interestingly, PC_079 and PC_083 were uniquely sensitive to paclitaxel compared to the other chemotherapeutics that were tested, while PC_028 was resistant to all therapies. Neither 5FU nor oxaliplatin induced any cytotoxic effect as evidenced by the AOC_0 and AOC_Ratio values in the tested range (0.1 μM–80 μM) (Fig. 6C, D). Drug sensitivity of organoids is ideally quantified in relation to each other. For this, percentile-based grouping was applied to categorize organoids as resistant, intermediate, and sensitive to easily interpret the results and guide therapy selection. From the classification of gemcitabine (Fig. 6E, F) and SN38 (Fig. 6G, H) we could see a high degree of overlap in the sensitivity of organoids to treatment, except for PC_372 which was more sensitive to SN38.
A Example of a fitted dose-response curve for gemcitabine, highlighting automatically determined outliers and the NOGR50 values. Overview of the (B) normalized AOC_1 of the fitted NOGR curves up to y = 1; (C) normalized AOC_0 of the fitted NOGR curves up to y = 0; and (D) AOC ratio of AOC_0 and AOC_1 for gemcitabine, paclitaxel and SN38 (1–100 nM) and 5FU and oxaliplatin (0.1 μM–80 μM) (72 h, n = 3). Fitted dose response curve for (E) gemcitabine and (G) SN38 with the corresponding percentile-based classification for each drug based on AOC_1 values (F–H).
Overall, our analysis demonstrates that the application of NOGR metric and its derivatives provides a more nuanced understanding of drug sensitivity in organoids, highlighting variable responses to chemotherapeutics in our panel of PDAC.
Discussion
The development and implementation of the Normalized Organoid Growth Rate (NOGR) metric, as highlighted in our study, represents a significant methodological advancement for brightfield imaging-based organoid drug screening. This metric, designed to capture organoid responses to treatments in a more holistic manner, addresses the limitations of traditional metrics like relative viability, which often fail to distinguish between cytostatic and cytotoxic effects. The NOGR metric builds upon the GR and NDR metrics that have been previously described for 2D screening assays by Hafner et al. and Gupta et al., respectively, and combines the strengths of both models15,17. Importantly, we demonstrated that the NOGR metric is less sensitive to variations in seeding density and growth rate and offers a more consistent and biologically relevant measure of drug sensitivity of different therapy classes as evidenced by our recent publication on a high-throughput drug combination screening using the NOGR metric23.
Our simulations indicated that the choice of growth-rate metric can significantly impact the interpretation of drug responses derived from widefield imaging in organoids, especially considering the growth rate (slow vs. fast-growing) and the accuracy of classifying death organoids. For the latter, we have significantly enhanced the label-free segmentation analysis of brightfield images by incorporating label-free cell death detection, a substantial improvement over previous approaches with fluorescent detection of cell death5. This innovative approach has proven effective in accurately classifying late-stage dying and dead organoids across various sizes and broadened the dynamic growth rate range that our analysis can capture, now ranging from 1 to −0.2. This is a notable improvement from the previous range of 1 to −0.5, which did not utilize label-free cell death detection5.
Concerning the growth rates, we observed that the GR metric was less reliable for the accurately quantifying the cytotoxic response of fast-growing organoids, which was drastically improved by the NDR metric via normalization to a positive control. However, both our simulations and screening data highlighted the limitation of the NDR to capture the cytotoxic response of slow-growing organoids and the cytostatic response of the fast-growing organoids. Contrastingly, the NOGR metric demonstrated a more robust performance in these scenarios. The NOGR effectively addressed these limitations, providing a more accurate representation of both cytotoxic and cytostatic drug responses across organoids with varying growth rates. The application of its derivatives, such as NOGR50, AOC_1, and AOC_0, allowed for more detailed classification of drug sensitivity in organoids and underlines the potential in guiding personalized therapy.
As organoid research is maturing for greater adoption in cancer drug screening, drug development, and even personalized therapy, the analysis methods for organoids have drastically fallen short of the biological advancements and culturing methods. Despite uncovering better ways to grow organoids, their analysis often relies on translating methods traditionally used for 2D cell cultures, which do not accurately represent this sophisticated model3,4. Therefore, in this manuscript we focused on the technical and methodological aspects of utilizing growth-rate based metrics for tumor organoid research. In the future, it will be of critical importance to correlate these in vitro findings with actual clinical outcomes in patients in order to further progress organoid research towards its full potential to recapitulate patient in the lab. In our previous research, we have already established a significant link between the results obtained from growth-rate based metrics (NDR) and single organoid analysis with the clinical responses of PDAC patients to standard-of-care chemotherapeutics21. In that retrospective study, we observed a strong correlation between the organoid responses and the actual patient outcomes, suggesting that the methodologies we are refining and employing can indeed serve as reliable predictors of how patients might respond to specific treatments. Since the NOGR developed here is a much more accurate representation of the true growth-rate organoids following therapy screening, we believe the correlation with patient outcomes will be further improved, which we plan to investigate in future work.
A secondary objective of our study was to study the robustness of the Orbits® label-free organoid segmentation model described previously to detect organoids with various sizes and morphologies that were not included in the training data5. Live-cell imaging has long been dominated by the use of fluorescent markers for viability, cell death and a range of other biological processes or targets. Additionally, confocal imaging is often required when working with 3D models to extract reliable data, and repeated excitation of fluorochromes can affect therapy response and induce phototoxicity through oxidative stress24. However, deep learning now allows for development of algorithms, such as Orbits®, deepOrganoid, OrgaExtractor, OrganoID, and OrgaSegment that can detect organoids label-free from brightfield and phase-contrast images7,8,9,10. As evidenced by our study, this approach can be extended to label-free classification of dying organoids and potentially other phenotypes of interest. Consequently, we can already extract a large amount of information about a drug response using widefield live-cell imaging systems that are more widely available and cost-effective, especially in combination with advanced drug metrics such as the NOGR. This approach significantly reduces imaging time and data storage needs compared to multicolor confocal imaging, while maintaining the flexibility to complement these more traditional methods. The complexity of deep learning algorithms often hinders their adoption by wet-lab scientists. User-friendly interfaces, coupled with effective segmentation capabilities, can make these tools more accessible and less intimidating, thus appealing to a wider range of biologists25. In our study, we validate the accuracy of the Orbits® label-free organoid segmentation module, which effectively segments various organoid morphologies, leveraging extensively pretrained deep learning models to streamline label-free organoid analysis. Furthermore, we are currently developing an integrated image and data analysis platform, aiming to further facilitate the adoption and application of these advanced tools in biological research in an automated and user-friendly way.
Conclusions
The introduction of the NOGR metric, alongside advancements in label-free live-cell imaging, represents a methodological improvement in organoid-based drug screening. This metric effectively captures a complete spectrum of drug responses, addressing the limitations of traditional methods and enabling more nuanced and accurate image-based analyses. The successful validation the Orbits® label-free segmentation models further enhances the potential of widefield live-cell imaging in organoid research. These innovations not only streamline the drug screening process but also open new avenues for personalized treatment strategies. This study lays the technical and methodological foundation of advanced organoid analysis with implications extend to clinical applications. In fact, as evidenced by our previous research correlating in vitro findings with patient outcomes, we believe that using the newly-defined NOGR could lead to even more accurate predictions of patient therapy response, which is currently under investigation in ongoing studies.
Materials and methods
Patient Material
Pancreatic ductal adenocarcinoma (PDAC) samples were obtained from three expert clinical centers under the PaCaOmics clinical trial (number 2011-A01439-32) after receiving ethics review board approval (Comité de protection des personnes Sud Méditerranée I). Consent forms of informed patients were collected and registered in a central database. All ethical regulations relevant to human research participants were followed. PDOs were established using endoscopic ultrasound-guided fine-needle aspirations (EUS-FNA) from PDAC patients. Briefly, PDAC cells were obtained from the biopsies through slight digestion with the Tumor Dissociation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) at 37 °C for 5 min. Isolated cells were placed into 12-well plates (Corning, #3513) coated with 150 µl growth factor reduced matrigel (Corning, Wiesbaden, Germany) and cultured with advanced DMEM/F12 supplemented with hydroxyethyl-piperazineethanesulfonic acid (HEPES) (10 mmol/L Thermo Fisher, Waltham, MA, USA), Wnt family member 3 A (WNT3A)-conditioned medium (30% v/v), R-spondin-1 (RSPO1)-conditioned medium (10% v/v); human recombinant fibroblast growth factor 10 (FGF10) (100 ng/mL; PeproTech, Rocky Hill, CT, USA), human recombinant epidermal growth factor (EGF) (50 ng/mL; PeproTech), human recombinant Noggin (100 ng/mL; Bio-Techne, Minneapolis, MN, USA), human Gastrin 1 (10 nmol/L; Sigma-Aldrich), Nicotinamide (10 mmol/L; Sigma-Aldrich), N-acetylcysteine (1.25 mmol/L; Sigma-Aldrich), B27 (1x; Thermo Fisher), A83-01 (500 nmol/L; Bio-Techne), and Y27632 (10.5 µmol/L; Bio-Techne). The plates were incubated at 37 °C in a 5% CO2 incubator, and the media changed every 3 to 4 days. Organoids were biobanked in liquid nitrogen and shipped to the University of Antwerp, Belgium.
Organoid cultures
Basic medium consisted of Ad-DF + ++ (Advanced DMEM/F12 (GIBCO, #12634028), with 1% GlutaMAX (GIBCO, 35050-061), 1% HEPES (GIBCO, #15630080), 1% penicillin/streptomycin (GIBCO, #15140122) supplemented with 2% Primocin (Invivogen, #ant-pm-05). For PDAC organoids, Ad-DF + ++ was supplemented with 0.5 nM WNT Surrogate-Fc-Fusion protein (ImmunoPrecise, #N001), 4% Noggin-Fc Fusion Protein conditioned medium (ImmunoPrecise, #N002), 4% Rspo3-Fc Fusion Protein conditioned medium (ImmunoPrecise, #R001), 1x B27 (Gibco, #17504044), 10 mM nicotinamide (Sigma-Aldrich, #N0636), 1.25 mM N-acetylcysteine (Sigma-Aldrich, #A9165), 100 ng/ml FGF-10 (Peprotech, #100-26), 500 nM A83-01 (Tocris, #2939), 10 nM gastrin (R&D Systems, #G9145) and 10 μM Y-27632 after passaging (Selleck Chemicals, #S1049). For passaging, the organoids were digested to single cells with TrypLE Express (GIBCO, #12604021) and resuspended in > 80% ice cold Cultrex growth factor reduced BME type 2 (R&D Systems, #3533-010-02) in full organoid medium. Small droplets of 20 µL were plated and incubated inverted for 30 min at 37 °C to allow them to solidify after which the drops were covered with full organoid medium.
Drug screening
Drug screening on 3D organoids was performed at the DrugVision.AI automated screening facility of the University of Antwerp, Belgium, using a prevalidated drug screening pipeline for which a detailed protocol is available in the Journal of Visualized Experiments6.
Briefly, established organoid lines were expanded in ECM domes (Cultrex type 2, R&D Systems, #3533-010-02). Next, 3-day-old organoids were harvested from ECM drops using the Cultrex Organoid Harvesting Solution (R&D Systems, #3700-100-01), collected in a 15 mL tube coated with 0.1% BSA/PBS, washed, and resuspended in medium. Next, the number of organoids was quantified by adding 5 µL of the organoid solution to 45 µL of medium in a 384-microplate well. A whole-well brightfield image was captured using the Tecan Spark Cyto and the number of organoids was counted label-free using Orbits. Next, the organoid solution was diluted in full medium supplemented with 4% Cultrex at a concentration of 4000 organoids / mL. Next, 50 µL (200 organoids) of this solution was dispensed into each well of a 384-well ultra-low attachment microplate (Corning, #4588) using the OT-2 pipetting robot (Opentrons) in a cooled environment. Thereafter, the plate was centrifuged (100 rcf, 30 s, 4 °C) to ensure that all organoids are in the same z-plane and incubated overnight at 37 °C. When the use of ECM domes is preferred, we recommend using low volume domes of 5 µL and low organoid density to avoid overlap of organoids in different z-planes. The following therapeutics were used: 5-fluoro-uracil (5FU, #HY-90006), oxaliplatin (#HY-17371), SN38 (active metabolite of irinotecan, #HY-13704)), gemcitabine (#HY-17026) and paclitaxel (#HY-B0015) (MedChemExpress). All drugs and fluorescent reagents were added to the plate using the Tecan D300e Digital Dispenser and dissolved in either DMSO or 0.3% Tween-20 (oxaliplatin). Cytotox Green (60 nM/well, Sartorius, #4633, DMSO) was used as a fluorescent cell death marker and Staurosporine (2 µM, Tocris Bioscience, #1285, DMSO) as a positive control. For each drug, a 7-point logarithmic titration was dispensed with the following concentration ranges: (i) SN38, gemcitabine, paclitaxel: 1 nM, 4.6 nM, 21.5 nM, 100.0 nM, 464.2 nM, 2154.4 nM, and 10,000 nM. (ii) 5FU, oxaliplatin: 100 nM, 304.7 nM, 928.2 nM, 2828.4 nM, 8617.7 nM, 26256.8 nM, and 80,000 nM. DMSO concentrations were normalized to the same level in each well ( < 1%). Brightfield and green fluorescence (Green channel: 461–487 nm/500–530 nm (excitation/emission)) whole-well images (4x objective) were taken at 0, 72 and 120 h with the Tecan Spark Cyto set at 37 °C/5% CO2.
Image and data analysis
Images and data were analysed with the Orbits® label-free organoid detection module (Orbits Oncology) as previously described in ref. 5. Viability (V) was quantified as Total Brightfield Organoid Area – Total Green Area and excluding organoids that were classified as death by the label-free cell death detection module. V was used to calculate the growth-rate based drug response metrics. For accurate growth rate metrics, we recommend a minimal growth rate of 1.2 for the negative control. Organoids with growth rates lower than this threshold may result in inaccurate metrics and are therefore excluded from further analysis.
Normalized growth rate inhibition (GR)15:
Normalized drug response (NDR)17:
where the growthRate is the ratio of the V at timepoint x and the V at timepoint 0, medPos is the median growthRate of the positive control and medNeg is the median growthRate of the vehicle control.
Normalized organoid growth rate (NOGR):
where V(0) is the viability at timepoint 0, V(x) is the viability at timepoint x, Gdrug is the G corresponding to the drug treated condition, GmedPos is the median G of the positive control and GmedNeg is the median G of the vehicle control.
Based on the GR, NDR or NOGR, the drug effects can be classified as: > 1, proliferative effect; = 1, normal growth as in vehicle control; = 0, complete growth inhibition; = −1, complete killing as in positive control.
The dose-response relationship was modeled using the Growth Rate (GR) equation:
Where GRinf is the response at infinite concentration, GEC50 is the concentration that produces half the maximum possible effect, h_GR is the Hill coefficient, determining the steepness of the curve, and c is the concentration. Next, the Python SciPy library’s ‘curve-fit’ function was employed to fit the GR model to the observed data for each biological replicate. Initial guesses for GRinf, GEC50, and h_GR were set to 0.1, median concentration and 2, respectively. Residual errors between observed and predicted responses were calculated for each data point using the Root Mean Square Error approach. Points exhibiting an error greater than 2.5 times the mean error and an absolute error greater than 0.25 were deemed outliers and the model was refitted to this refined dataset. The following metrics were derived from the fitted curve: NOGR50 as the concentration at which the response is 0.5; NOGR_AOC_1_fitted_n as the area over the curve (AOC) up to y = 1, normalized to the maximum area; NOGR_AOC_0_fitted_n as the AOC up to y = 0, normalized to the maximum area; and NOGR_AOC_Ratio as NOGR_AOC_0_fitted_n / NOGR_AOC_1_fitted_n.
A percentile-based approach was employed to classify the cell lines into distinct response groups based on their NOGR_AOC_1_fitted_n values. The dataset was divided into three groups based on the distribution of the values: (1) Resistant: Organoid lines falling in the lower 33% of NOGR_AOC_1_fitted_n values (below the 33rd percentile). (2) Intermediate: Organoid lines with NOGR_AOC_1_fitted_n values between the 33rd and 66th percentiles (mid-range responses). (3) Sensitive: Organoid lines in the top 33% of NOGR_AOC_1_fitted_n values (above the 66th percentile).
Finally, we simulated growth rate data to compare the performance of the GR, NDR, and NOGR metrics under controlled conditions, independent of experimental variability. Theoretical growth rates were generated for slow-growing organoids (0–1.5) and fast-growing organoids (0–8). GR, NDR, and NOGR were then calculated using the formulas provided in this section and plotted. This simulation allowed us to systematically assess the strengths and limitations of each metric across a broader range of growth rates, providing a clear comparison of their behavior without the influence of experimental noise.
Statistics and Reproducibility
A total of 11 patient-derived tumor organoid lines were used. Drug screenings were performed in technical triplicate on the same plate. No statistical analysis was performed.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available in Supplementary Data 1. Image data is available from the corresponding author upon reasonable request.
Code availability
All algorithms to calculate the drug response metrics used in this manuscript are detailed in the materials & methods section. The Orbits® label-free organoid segmentation module is available through Orbits® Oncology.
References
Weeber, F., Ooft, S. N., Dijkstra, K. K. & Voest, E. E. Tumor Organoids as a Pre-clinical Cancer Model for Drug Discovery. Cell Chem. Biol. 24, 1092–1100 (2017).
Thorel, L. et al. Patient-derived tumor organoids: a new avenue for preclinical research and precision medicine in oncology. Exp. Mol. Med. 56, 1531–1551 (2024).
Driehuis, E., Kretzschmar, K. & Clevers, H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 15, 3380–3409 (2020).
Ooft, S. N. et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 11, https://doi.org/10.1126/scitranslmed.aay2574 (2019).
Deben, C. et al. OrBITS: label-free and time-lapse monitoring of patient derived organoids for advanced drug screening. Cell Oncol. (Dordr.) 46, 299–314 (2023).
Le Compte, M. et al. Multiparametric Tumor Organoid Drug Screening Using Widefield Live-Cell Imaging for Bulk and Single-Organoid Analysis. J. Vis. Exp. https://doi.org/10.3791/64434 (2022).
Park, T. et al. Development of a deep learning based image processing tool for enhanced organoid analysis. Sci. Rep. 13, 19841 (2023).
Matthews, J. M. et al. OrganoID: A versatile deep learning platform for tracking and analysis of single-organoid dynamics. PLoS Comput Biol. 18, e1010584 (2022).
Lefferts, J. W. et al. OrgaSegment: deep-learning based organoid segmentation to quantify CFTR dependent fluid secretion. Commun. Biol. 7, 319 (2024).
Powell, R. T. et al. deepOrganoid: A brightfield cell viability model for screening matrix-embedded organoids. SLAS Discov. 27, 175–184 (2022).
Sebaugh, J. L. Guidelines for accurate EC50/IC50 estimation. Pharm. Stat. 10, 128–134 (2011).
Farin, H. F. et al. Colorectal cancer organoid-stroma biobank allows subtype-specific assessment of individualized therapy responses. Cancer Discov. 13, 2192–2211 (2023).
Seppälä, T. T. et al. Precision Medicine in Pancreatic Cancer: Patient-Derived Organoid Pharmacotyping Is a Predictive Biomarker of Clinical Treatment Response. Clin. Cancer Res. : Off. J. Am. Assoc. Cancer Res. 28, 3296–3307 (2022).
Fallahi-Sichani, M., Honarnejad, S., Heiser, L. M., Gray, J. W. & Sorger, P. K. Metrics other than potency reveal systematic variation in responses to cancer drugs. Nat. Chem. Biol. 9, 708–714 (2013).
Hafner, M., Niepel, M., Chung, M. & Sorger, P. K. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat. Methods 13, 521–527 (2016).
Clark, N. A. et al. GRcalculator: an online tool for calculating and mining dose-response data. BMC Cancer 17, 698 (2017).
Gupta, A., Gautam, P., Wennerberg, K. & Aittokallio, T. A normalized drug response metric improves accuracy and consistency of anticancer drug sensitivity quantification in cell-based screening. Commun. Biol. 3, 42 (2020).
Ooft, S. N. et al. Prospective experimental treatment of colorectal cancer patients based on organoid drug responses. ESMO Open 6, 100103 (2021).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Millen, R. et al. Patient-derived head and neck cancer organoids allow treatment stratification and serve as a tool for biomarker validation and identification. Med. 4, 290–310.e212 (2023).
Le Compte, M. et al. Single-organoid analysis reveals clinically relevant treatment-resistant and invasive subclones in pancreatic cancer. NPJ Precis Oncol. 7, 128 (2023).
Garajova, I., Peroni, M., Gelsomino, F. & Leonardi, F. A Simple Overview of Pancreatic Cancer Treatment for Clinical Oncologists. Curr. Oncol. 30, 9587–9601 (2023).
Deben, C. et al. Auranofin repurposing for lung and pancreatic cancer: low CA12 expression as a marker of sensitivity in patient-derived organoids, with potentiated efficacy by AKT inhibition. J. Exp. Clin. cancer Res. : CR 43, 88 (2024).
Icha, J., Weber, M., Waters, J. C. & Norden, C. Phototoxicity in live fluorescence microscopy, and how to avoid it. Bioessays 39, https://doi.org/10.1002/bies.201700003 (2017).
Eisenstein, M. AI under the microscope: the algorithms powering the search for cells. Nature 623, 1095–1097 (2023).
Acknowledgements
We acknowledge financial support from the Fund for Scientific Research (FWO-Vlaanderen; M.L.C. and A.L.) and the Research Fund of UAntwerp (BOF; S.S.). We would also like to thank several patrons, as part of this research was funded by donations from different donors, including Dedert Schilde vzw, Mr. Willy Floren and the Vereycken family.
Author information
Authors and Affiliations
Contributions
Conceptualization and design of the study, C.D.; data collection, data analysis, investigation and interpretation, C.D., E.C.D.L.H, S.S., F.R.F.; sample collection, N.D.; writing—original draft, C.D.; writing—review and editing, C.D., E.C.D.L.H., F.R.F., M.L.C., S.S., S.V., H.V., B.V.D.B., N.D., A.L., G.R., M.P., H.P., F.L., E.S.; visualization, C.D., E.C.D.L.H.; supervision, C.D., F.L., E.S.; project administration: C.D. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interests: C.D., M.L.C., E.C.D.L.H., and A.L. are co-founders of Orbits® Oncology, an image and data analysis platform that was used in this study.
Ethical approval
Pancreatic ductal adenocarcinoma (PDAC) samples were obtained from three expert clinical centers under the PaCaOmics clinical trial (number 2011-A01439-32) after receiving ethics review board approval. Consent forms of informed patients were collected and registered in a central database.
Peer review
Peer review information
Communications Biology thanks Tero Aittokallio, Maite Calucho and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Christina Karlsson Rosenthal. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Deben, C., Cardenas De La Hoz, E., Rodrigues Fortes, F. et al. Development and validation of the Normalized Organoid Growth Rate (NOGR) metric in brightfield imaging-based assays. Commun Biol 7, 1612 (2024). https://doi.org/10.1038/s42003-024-07329-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07329-5
This article is cited by
-
Organoids in translation: a bench-to-bedside framework for pancreatic cancer precision medicine
Journal of Translational Medicine (2026)
-
Towards a quality control framework for cerebral cortical organoids
Scientific Reports (2025)