Abstract
Fear, while crucial for survival, is a component of a myriad of psychiatric illnesses in its extreme. Persistent fear memories can form through processes such as second-order conditioning (SOC), during which a second-order conditioned stimulus (CS2) acquires significance by associating with a first-order conditioned stimulus (CS1). The neural circuitry underlying SOC, particularly the roles of sensory cortices, remains poorly understood. Here we explore the mechanisms of olfactory SOC in rats, focusing on the basolateral amygdala (BLA) and posterior piriform cortex (pPC). Our results demonstrate that NMDAR-dependent plasticity in both regions is essential for SOC. The BLA mediates the CS2-CS1 association, while the pPC, receiving inputs from the locus coeruleus and BLA, is critical for memory acquisition and retrieval. Single-nucleus multiomics analysis of Fos+ ensembles in both regions reveals distinct yet overlapping gene activation profiles in excitatory neurons, accompanied by global chromatin remodeling. These findings highlight the specific yet coordinated roles of these structures in supporting learning and memory.

Similar content being viewed by others
Introduction
Pavlovian threat conditioning, or first-order threat conditioning (FOC), is a widely recognized model of associative learning in which a neutral stimulus (conditioned stimulus, CS) becomes associated with an aversive event (unconditioned stimulus, US). This model has been instrumental in deepening our understanding of learning, memory, and various psychiatric disorders, including pathological anxiety and post-traumatic stress disorder (PTSD)1,2,3. However, in the natural environment, organisms rarely encounter direct threats or primary CS repeatedly in isolation. Instead, defensive behaviors often arise from complex associative chains, involving higher-order conditioning such as second-order conditioning (SOC). In SOC, a stimulus acquires significance indirectly by associating with a primary CS, rather than the US, thereby influencing behavior through more elaborate associative networks4,5. This capacity for complex associative learning contributes to the persistence of fear memories, which are often more resistant to extinction when rooted in second-order associations6. This presents significant challenges for therapeutic interventions that only target the primary CS6.
The amygdala, particularly its basal and lateral nuclei, collectively referred to as the basolateral amygdala (BLA), is central to the study of threat learning5,7,8,9. Several molecular mechanisms have been implicated in threat learning within the BLA, including NMDA receptor (NMDAR) activation, which is essential for synaptic plasticity10,11,12, as well as downstream signaling cascades that lead to gene transcription and long-term changes in neuronal function13,14,15. In addition, epigenetic modifications within the BLA have been shown to play a crucial role in the consolidation and maintenance of threat memories16,17,18.
While the BLA is indispensable for the initial encoding of threat memories, a growing body of evidence suggests that long-term storage and expression of these memories involve a distributed network of brain regions, including both multimodal and primary sensory cortices5,19,20,21,22,23. In particular, the auditory cortex (Aud) and the olfactory piriform cortex (PC) have emerged as key sites in which threat-related sensory information is processed and stored19,21,24. These regions exhibit strong reciprocal connections with the BLA25,26,27,28, facilitating the integration of sensory and affective information29,30,31,32,33,34,35. The functional connectivity between the BLA and sensory cortices is enhanced following FOC, with evidence of increased synaptic plasticity in these pathways19,21,24. Moreover, recent evidence indicates that the posterior PC (pPC) plays a critical role in the extinction of olfactory threat memories22.
Despite significant advances in our understanding of FOC, the specific networks and molecular mechanisms underlying SOC remain poorly understood. The question of how sensory cortices, particularly the PC and Aud, contribute to the formation and retrieval of SOC memories has not been explored. In this study, we sought to address this gap by investigating the role of NMDAR-dependent plasticity in the BLA, pPC, and Aud during SOC. We employed an auditory-olfactory conditioning model to systematically examine the involvement of these regions in SOC at both anatomical and molecular levels.
We employed event-labeling techniques (cFos-tTA), retrograde tracing, and optogenetic manipulation to map the functional circuits involved in SOC. In addition, we conducted chemogenetic silencing and pharmacological interventions to assess the necessity of specific pathways for SOC memory formation and retrieval. Finally, we performed a combined single-nucleus RNA sequencing (snRNA-seq) and assay for transposase-accessible chromatin using sequencing (snATAC-seq) on tissues from the BLA and pPC following SOC, with the aim of identifying gene expression changes and chromatin dynamics associated with SOC memory. Our findings provide new insights into the neural circuits and molecular mechanisms underlying SOC, with potential implications for our understanding of the persistence of fear memories and the development of more effective therapeutic strategies for fear-related disorders.
Results
NMDAR-dependent plasticity is engaged in the BLA and pPC during SOC
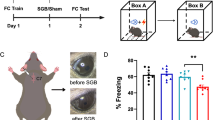
To investigate the network activation during SOC (Fig. 1A), we first established FOC by pairing a tone (CS1) with a foot shock. Following this, SOC was induced by pairing the CS1 with an odor (CS2), creating a second-order association. In a control group, CS1 was presented 30 min prior to CS2 to prevent associative learning. SOC memory was assessed 24 h later by measuring freezing behavior in response to the odor (CS2). Rats in the paired group exhibited significantly higher freezing responses to the odor, indicating successful SOC (Fig. 1B).
A Schematics illustrating the SOC training flow and cFos IHC. Sampling areas for cFos are denoted in the images on the right. LA: lateral amygdala, BA: basal amygdala, aPC: anterior piriform cortex, pPC: posterior piriform cortex, Aud: auditory cortex. B Percentage of freezing responses to CS1 tone during SOC and to CS2 odor during memory retrieval (F1,13 = 15.850, p = 0.002). Both unpaired (UP; n = 4 F/2 M) and paired (P; n = 3 F/4 M) groups exhibited freezing responses to the CS1 tone after the first-order conditioning, whereas only the tone/odor paired group manifested SOC memory for odor (t = 8.954, p < 0.001). C Comparisons of cFos cell counts across various structures in the UP and P groups (n = 6-7). LA (t = − 2.648, p = 0.024), BA (t = − 7.605, p < 0.001), pPC (t = − 3.198, p = 0.008) and Aud (t = − 4.004, p = 0.002) displayed higher cFos activation in the paired group. D Schematics depicting SOC training with drug or vehicle infusion. E Percentage of freezing to the CS1 tone and CS2 odor when D-APV (n = 3 F/4 M) or vehicle (n = 4 F/5 M) was infused into the BLA (F1,14 = 51.397, p < 0.001). The D-APV-infused group exhibited significantly reduced freezing to the odor (p < 0.01). F Percentage of freezing responses to CS1 tone and CS2 odor when D-APV (n = 4 F/4 M) or vehicle (n = 3 F/3 M) was infused to the pPC (F1,14 = 13.902, p = 0.002). The D-APV-infused group displayed significantly reduced freezing to the odor (p < 0.05). G Percentage of freezing responses to CS1 tone and CS2 odor when D-APV (n = 2 F/4 M) or vehicle (n = 3 F/4 M) was infused to the Aud. No difference was observed between the two groups (p > 0.05). Upper panels in E-G show example cannular targeting in the three structures. Red arrows indicate the cannular tip locations. Scale bars, 500 µm. *p < 0.05, **p < 0.01. Error bars, SEM.
We next examined the cFos protein expression as a marker of neuronal activation following SOC memory retrieval. Ninety minutes after exposure to CS2, we observed significantly elevated cFos expression in the paired group across several brain regions, including the lateral amygdala, basal amygdala, pPC, and Aud (Fig. 1C and Supplementary Fig. 1). These findings suggest that SOC engages a distributed network of brain regions, including both the BLA and sensory cortices.
To further explore the role of NMDAR-dependent plasticity in these regions, we infused the NMDAR antagonist D-APV into the BLA, pPC, or Aud immediately following SOC training and assessed memory retrieval the next day (Fig. 1D and Supplementary Fig. 2). In the BLA, D-APV infusion resulted in a significant reduction in freezing behavior in response to CS2, indicating that NMDAR activity in the BLA is necessary for SOC memory consolidation (Fig. 1E). Similarly, D-APV infusion in the pPC significantly reduced freezing behavior, suggesting that NMDAR-dependent plasticity in the PC is also critical for SOC memory (Fig. 1F). In contrast, D-APV infusion in the Aud did not produce a significant effect on freezing behavior, indicating that the Aud may play a less direct role in SOC memory retrieval (Fig. 1G).
CS1 and CS2 converge in the BLA to undergo plasticity during SOC
Given the importance of the BLA in threat learning, we hypothesized that CS1 and CS2 converge in the BLA to initiate the plasticity that supports SOC memory. To test this hypothesis, we employed a cFos-tTA/TRE-GFP tagging approach36, in which neurons responsive to the tone (CS1) were labeled with GFP, and their co-expression with odor-responsive (CS2) neurons was examined following SOC (Fig. 2A). Rats in the paired group showed significantly higher overlap of CS1 and CS2 neuronal ensembles in the BLA compared to the unpaired group, as measured by co-expression of GFP and cFos (Fig. 2B, C and Supplementary Fig. 3). This higher overlap was paralleled by heightened freezing behavior in response to CS2 (Fig. 2D), suggesting that the convergence of CS1 and CS2 in the BLA may underlie SOC memory. In contrast, no enhancement of overlap between the CS1 and CS2 ensembles was observed in the Aud (Supplementary Fig. 4).
A Schematic representation of CS1 tone labeling using the cFos-tTA-TRE method, SOC training, and subsequent CS2 odor labeling via cFos IHC following odor memory retrieval. B Example images displaying GFP+ cells (representing the tone CS1 ensemble, white arrow heads), cFos+ cells (magenta, representing the odor CS2 ensemble; yellow arrow heads), and co-labeled cells (representing the CS1 + CS2 ensemble; white arrows) in the BLA. C Percentage overlap between tone and odor ensembles in the BA in the UP (n = 4 F/3 M) and P (n = 5 F/6 M) groups. The paired group exhibited higher overlap of CS1 tone and CS2 odor ensembles (t = − 2.528, p = 0.022). D Percentage of freezing responses to CS1 tone and CS2 odor in rats transduced with cFos-tTA-TRE (F1,16 = 54.448, p < 0.001). E Flowchart depicting the optogenetic activation of the CS1 + CS2 ensembles in the BLA. F Significant freezing responses were induced in ChR2-expressing rats (n = 3 F/2 M) during light activation of CS1 + CS2 ensembles in the BLA (t = − 2.899, p = 0.044; baseline vs. light), a subset of which (cyan colored lines) were subsequently reversed following an optical depotentiation protocol (t = − 0.933, p = 0.420 compared to baseline). Gray-shaded bars in the background represent freezing behavior binned by minute. Black bars indicate the average freezing during the 3 min baseline period prior to light stimulation and the 3 min period during light stimulation. G Schematic illustrating the combined CTB and cFos-labeling approach. H Percentage of freezing responses to CS1 tone during SOC and to CS2 odor during memory retrieval (UP: n = 2 F/3 M; P: n = 4 F/1 M) (F1,8 = 22.131, p = 0.002). I Example image showing CTB (magenta) infusion in the BLA. J1 Example images of CTB-expressing cells in the pPC. cFos+ cells are indexed by GFP. Yellow arrows indicate cFos- CTB cells. Arrow heads indicate cFos+ CTB cells. J2 Comparable numbers of BLA-projecting cells in the pPC were observed in the UP and P groups. J3 A larger portion of BLA-projecting cells in the pPC were cFos+ (t = − 2.511, p = 0.036). K1 Example images of CTB-expressing cells in the Aud layer II/III (left) and V (right). Yellow arrows indicate cFos- CTB cells. Arrow heads indicate cFos+ CTB cells. K2 Comparable numbers of BLA-projecting cells in the Aud were observed in the paired and unpaired groups. K3 A larger portion of BLA-projecting cells in Aud layer II/III were cFos+ (t = − 2.756, p = 0.025). Scale bars, 50 µm. *p < 0.05, **p < 0.01. Error bars, SEM.
To further confirm the role of these convergent ensembles in SOC memory, we induced the expression of the light-sensitive protein channelrhodopsin-2 (ChR2) in the CS1 + CS2 ensemble in the BLA during SOC and optically stimulated these neurons following SOC (Fig. 2E). In the absence of the odor CS2, optical activation of the CS1 + CS2 ensemble at 20 Hz induced a robust freezing response, which persisted minutes beyond the period of light stimulation, indicating that activation of this ensemble is sufficient to drive SOC memory recall (Fig. 2F and Supplementary Fig. 5). Importantly, a subsequent optical depotentiation protocol, designed to weaken synaptic connections within the CS1 + CS2 ensemble, significantly reduced the freezing response, suggesting that synaptic potentiation within this ensemble is necessary for SOC memory expression (Fig. 2F).
In addition, we examined cFos activation of BLA-projecting neurons within the pPC and Aud following SOC retrieval, using retrograde dye cholera toxin subunit B (CTB) infusion in the BLA prior to SOC (Fig. 2G–I). Rats subjected to SOC exhibited a heightened freezing response to the CS2 odor (Fig. 2H), which was paralleled by a greater activation of BLA-projecting neurons in the pPC (Fig. 2J1–J3). Moreover, increased activation was observed in BLA-projection neurons in layer II/III of the Aud (Fig. 2K1–K3). These results suggest that inputs from the pPC to the BLA, as well as from layer II/III neurons in the Aud to the BLA, are potentiated following SOC.
Interestingly, CTB infusion in the Aud revealed a higher proportion of Aud-projecting neurons in the BLA exhibiting cFos expression after SOC (Supplementary Fig. 6). This enhanced connectivity from the BLA to the Aud is consistent with recent findings on amygdalo-cortical remodeling following auditory FOC25. However, this potentiation is not sufficient to generate overlapping representations of CS1 and CS2, likely because the auditory cortex also receives indirect olfactory inputs37 that do not converge with the auditory representation of CS1. Consistent with previous studies showing that the PC does not directly project to the Aud37,38, no CTB labeling was observed in the PC.
Adrenergic locus coeruleus (LC)-pPC and BLA-pPC projections support learning and retrieval of SOC respectively
We next investigated the specific inputs to the pPC that contribute to SOC. In addition to the reciprocal projections between the PC and BLA27, the PC also receives direct input from the Aud37. In addition, previous research has underscored the importance of LC adrenergic inputs to the PC in olfactory learning39. Our objective was to elucidate the contributions of these inputs to the PC in the context of olfactory SOC (Fig. 3A).
A Simplified circuitry diagram depicting the information flow triggered by tone and odor. LC: locus coeruleus, Aud: auditory cortex, BLA: basolateral amygdala, pPC: posterior piriform cortex. Red lines represent the inputs to the pPC. Odor CS2 directly activates pPC, while tone CS2 can modulate pPC neuronal activity through the LC, Aud or BLA inputs. B Schematic illustrating the combined CTB and cFos-labeling approach. C Example image demonstrating CTB infusion in the pPC. D1 Example images of CTB-expressing cells in the BLA. Arrow heads indicate cFos+ CTB cells. D2 Comparable numbers of pPC-projecting cells in the BLA were observed in the UP (n = 3 F/3 M) and P (n = 3 F/2 M) groups. D3 A larger portion of pPC-projecting cells in the BLA were cFos+ (t = − 4.796, p < 0.001). E1 Example images of CTB-expressing cells in the Aud layer II/III (left) and V (right). Yellow arrows indicate cFos- CTB cells. White arrow heads indicate cFos+ CTB cells. E2 Comparable numbers of pPC-projecting cells in the Aud were observed in the paired and unpaired groups. E3 Similar portions of pPC-projecting cells in Aud between the two groups were cFos+ (t = − 0.167, p = 0.871). F1 Example images of CTB-expressing cells in the LC. The LC region is defined by dopamine β-hydroxylase (DBH) staining (left panel). Yellow arrows indicate cFos- CTB cells. White arrow heads indicate cFos+ CTB cells. F2 Comparable numbers of pPC-projecting cells in the LC were observed in the paired and unpaired groups. F3 A larger portion of pPC-projecting cells in the LC were cFos+ (t = − 2.527, p = 0.035). G Percentage of freezing responses to CS1 tone during SOC and to CS2 odor during memory retrieval (F1,9 = 15.234, p = 0.004). H Flowchart illustrating the chemogenetic silencing of the BLA-pPC pathway in SOC. Bottom panels show example images of GFP expression in the BLA and projection fibers in the pPC. Scale bars, 50 µm. I Activation of DREAAD in the pPC by clozapine N-oxide (CNO) did not affect odor SOC acquisition (CNO: n = 3 F/3 M; Vehicle: n = 2 F/3 M) (t = 0.871, p = 0.406), but impaired memory retrieval (t = 3.564, p = 0.016), compared to the vehicle (Veh) group (n = 3 F/3 M). A control group with GFP AAV infusion showed no effect on memory recall when CNO was infused before retrieval (t = − 1.368, p = 0.305). J Schematics depicting SOC training with adrenoceptor (AR) antagonists (n = 3 F/5 M) or vehicle (n = 4 F/3 M) infusion. K Post-SOC acquisition infusion of AR antagonists impaired odor-induced freezing responses during memory retrieval (F2,16 = 6.580, p = 0.008). Scale bars, 50 µm. *p < 0.05, **p < 0.01. Error bars, SEM.
To determine which inputs are potentiated following SOC, we infused CTB into the pPC and analyzed the activation of pPC-projecting neurons in the BLA, Aud and LC following SOC retrieval (Fig. 3B, C). SOC significantly increased the activation of pPC-projecting neurons in the BLA, indicated by a higher proportion of cFos+ neurons in the paired compared to the unpaired group (Fig. 3D1–D3). In contrast, the activation of pPC-projecting neurons in the Aud did not differ significantly between the paired and unpaired groups (Fig. 3E1–E3). In addition, the projection from the LC to the pPC was enhanced in the paired group (Fig. 3F1–F3 and Supplementary Fig. 7). These CTB tracing and cFos mapping results, supported by a heightened freezing response to the odor in the paired group (Fig. 3G), suggest that the BLA-pPC and LC-pPC pathways play a more critical role in SOC than the Aud-pPC pathway.
To directly assess the functional significance of the BLA-pPC pathway in SOC memory, we employed a chemogenetic approach to selectively silence BLA projections to the pPC during SOC memory retrieval. AAV-SYN-hM4Di was infused bilaterally into the BLA, and clozapine-N-oxide (CNO) was infused into the pPC prior to SOC learning or retrieval (Fig. 3H). Silencing of the BLA-pPC pathway before memory retrieval, but not before SOC acquisition, resulted in a significant reduction in freezing behavior in response to CS2 (Fig. 3I and Supplementary Fig. 8). These findings indicate that the BLA-pPC pathway is specifically required for the retrieval, rather than the acquisition, of SOC memories.
Finally, we examined the role of adrenergic input to the pPC in SOC acquisition. A mixture of α1- and β-adrenoceptor antagonists was infused into the pPC immediately after SOC training to block adrenergic signaling (Fig. 3J). This intervention significantly impaired SOC memory formation, as evidenced by a reduction in freezing behavior during the memory test, suggesting that adrenergic input to the pPC facilitates SOC memory encoding (Fig. 3K). In addition, drug infusion in the BLA also impaired SOC memory retrieval (Fig. 3K). Together, these results suggest LC-derived norepinephrine broadly influences the SOC memory process.
Neuronal clustering and Fos + ensembles in the BLA and pPC
To explore the molecular mechanisms underlying SOC memory, we conducted a multiome assay, which combined snRNA-seq and snATAC-seq, on tissues from the BLA and pPC regions following SOC acquisition and retrieval. Interestingly, we did not observe any significant differences in freezing levels following SOC between male and female rats (Supplementary Fig. 9). In the sequencing study, we used male rats to minimize potential variability in gene expression related to hormonal fluctuations across the estrous cycle in females40. Our analysis focused on Fos+ neurons, which are likely to represent memory engram ensembles following SOC. Tissue samples were collected at two critical time points: 90 min post-SOC encoding to capture memory acquisition, including early consolidation, and 90 min post-memory retrieval (Fig. 4A). These time points were chosen to capture ensembles with sustained Fos mRNA activation and subsequent downstream gene activation41,42,43.
A Schematic representation of the SOC and snRNA-seq experimental workflow, with cell-type-specific clustering illustrated for the BLA and PC on the right. B Neuron-specific clusters identified in the BLA, comprising 2 excitatory (VGLUT1, VGLUT2), 5 inhibitory (SST, MSN, NPY, PV, VIP) and 2 mixed clusters labeled unknown. C Distinct top gene markers identified for the 9 neuronal clusters (0–8) of the BLA. D Fos+ ensembles in the BLA activated following SOC acquisition. E Fos+ ensembles in the BLA activated following SOC retrieval. F Neuron-specific clusters identified in the PC, comprising 4 excitatory (VGLUT1a, VGLUT1b, VGLUT1c and VGLUT2), 3 inhibitory (VIP, PV and an unidentified inhibitory cluster labeled INu), and 2 mixed clusters labeled unknown. G Distinct top gene markers identified for the 9 neuronal clusters (0–8) in the PC. H Fos+ ensembles in the PC activated following SOC acquisition. I Fos+ ensembles in the PC activated following SOC retrieval.
snRNA-seq analysis identified nine distinct subclusters of excitatory and inhibitory neurons in the BLA region (Fig. 4B, C and Supplementary Figs. 10–12), with Fos+ cells sparsely distributed across these clusters (Fig. 4D, E and Supplementary Fig. 13). The major excitatory VGLUT1 (Slc17a7 +) cluster, and inhibitory neuron clusters including somatostatin (SST)-expressing neurons (Sst+/Pvalb-), medium spiny neurons (MSNs; Penk+/Npy-), neuropeptide Y (NPY)-expressing neurons (Npy+ also Penk+), parvalbumin (PV)-expressing neurons (Pvalb+/Erbb4+, also Npy+/Sst+), and vasoactive intestinal peptide (VIP)-expressing neurons (Vip+), were selected for further analysis. Genes known to be selectively expressed in the CeA but not the BLA, such as Prkcd44, Tac245, and Crh46, were not detected in any of the clusters, suggesting minimal, if any, contamination from CeA neurons in our tissue. However, Foxp2, a well-established marker for neurons in the intercalated cell masses47 (ITCs; inhibitory nuclei bordering the BLA), was identified within the NPY+ cluster. This suggests that our tissue collection likely included a portion of the ITCs.
Similarly, nine neuronal clusters were identified in the pPC, also showing sparse Fos expression (Fig. 4F–I and Supplementary Figs. 14–17). Among the major excitatory neuron clusters, VGLUT1a, VGLUT1b (Cux1+) and VGLUT1c (Reln+) likely represent superficial, deep pyramidal neurons and semilunar neurons48, respectively. Major inhibitory neuron clusters included PV-expressing (Pvalb+/Erbb4+, also Npy+/Sst+) neurons, VIP-expressing neurons (Vip+), and an unidentified cluster (GAD1+).
Gene activation in Fos + ensembles supports SOC memory in both the BLA and pPC
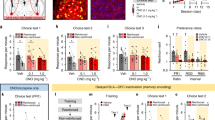
We analyzed differentially expressed genes (DEGs) in the Fos+ vs. Fos- ensembles within the same neuronal clusters using a pseudo-bulk approach. For excitatory BLA neurons, we focused on the VGLUT1 cluster (Fig. 5 and Supplementary Table 1). VGLUT1 neurons also form a major excitatory cluster in the mouse BLA in a recent report43. During SOC acquisition (Fig. 5A, C, D), 59 upregulated genes were identified in the paired group and 27 in the unpaired group, with Fos as the only overlap. During retrieval (Fig. 5B, C–E), 57 DEGs were detected in the unpaired group and 53 in the paired group, with eight shared (~ 8% overlap), including immediate early genes (Fos, Egr1, Egr4, Nr4a1), regulator of transcription factor (Tiparp49), G-protein coupled receptor (Gprc5a50), neurotrophic factors (Bdnf), and MAP kinase-related phosphatases (Dusp651) (Fig. 5E). Although the number of DEGs was similar across groups, Fos+ neurons showed minimal transcriptional overlap, suggesting engagement in distinct molecular programs depending on the training condition.
A Volcano plot displaying differentially expressed genes (DEGs) in the unpaired (U_A) and paired (P_A) groups following SOC acquisition. B Volcano plots displaying DEGs in the unpaired (U_R) and paired (P_R) groups following SOC retrieval. The top 20 DEGs are labeled, with non-significant genes indicated as NS. C Venn diagram illustrating the common DEGs across the four conditions (U_A, P_A, U_R, P_R). D Scatter plot showing DEGs in the U_A and P_A groups. Blue dots represent DEGs in the P_A group, green dots represent DEGs in the U_A group. E Scatter plot showing DEGs in the U_R and P_R groups. Blue dots represent DEGs in the P_R group, green dots represent DEGs in the U_R group, and red dots indicate common DEGs between the two groups. Only detected genes shared between both groups were included in the scatter plots. Note that Fos was not included in the scatter (D, E) plots due to high fold changes (out of range). F Bubble plot showing enriched pathways in the U_R and P_R groups of the VGLUT1 cluster. U unpaired, P paired, A acquisition, R retrieval. NS: not significant.
Gene ontology analysis during acquisition showed no significant enriched pathways, while retrieval in the paired group enriched pathways related to synaptic signaling and transmission (Bdnf, Ntrk2, Nr4a1, Snap25), neurotransmitter receptor regulation (Arc, Rab5al1, Snap25, Nptx2), synaptic plasticity (Arc, Bdnf, Egr1), regulation of peptidyl-serine phosphorylation (Bndf, Bdkrb2, Irgm, Ntrk2), cell adhesion (Bmp2, Ncam1), long-term memory (Arc, Ntrk2, Egr1, Snap25, Ncam1, Nptx2), and stress responses, including glucocorticoid signaling and norepinephrine (Bdnf, Ntrk2, Egr1) (Fig. 5F). The unpaired groups also showed enrichment in fewer pathways related to norepinephrine response and synaptic plasticity (Fig. 5F), potentially associated with non-specific defensive responding or general learning processes. In contrast, the paired group, in which associative learning occurred, exhibited enrichment of a significantly greater number of pathways, particularly those related to synaptic function and long-term memory. This suggests that while Fos expression may indicate general neuronal activation in both conditions, the transcriptional programs specifically supporting associative learning are more robustly engaged in the paired group.
In inhibitory neurons in the BLA region, DEGs were identified across five subclusters (SST, MSN, NPY, PV, and VIP) following SOC acquisition and retrieval (Fig. 6A, B and Supplementary Table 1). Each subcluster exhibited distinct DEG patterns with minimal overlap. Following SOC retrieval, SST and NPY neurons had more DEGs compared to other clusters, with a number of shared genes in SST (Fos, Trib1, Efcab8, Afap1/2, Vtcn1) and NPY (Fos, Tbx10, Man1a1) neurons between the paired and unpaired groups (Fig. 6C–F). Gene ontology analysis highlighted distinct enriched pathways in SST and NPY clusters compared to VGLUT1 following retrieval (Fig. 6G). NPY neurons was enriched in pathways related to stress responses, including norepinephrine transport (Crh, C5, Adora2a), cAMP-mediated signaling (Crh, Gpr3, Rxfp2, Pomc), regulation of corticotropin secretion (Crh, Avpr1a) and neuropeptide signaling (Pgr15l, Cysltr2, Pomc, Gpr83). In addition, pathways related to synaptic plasticity were also enriched (Arc, Egr1, Vgf, Egr2). In contrast, the SST cluster was enriched in pathways involved in extracellular structure and matrix organization (Plg, Ccn1, Serpinf2, Tie1, Nr2e1, Col27a1, Fmod, Grem1), cell maturation (Ednrb, Trip13, Cdkn1a, Nr4a2, Cspg4, C1ql1, Catsper3) and chemotaxis (Itga1, Ednrb, Nr4a1, Rab13, Il12a, Pdgfd, Gpr18, Grem1), consistent with the role of SST neurons in synaptic remodeling following threat conditioning52.
A Heat map showing distinct patters of DEGs in the five inhibitory clusters following SOC acquisition. B Heatmap showing distinct patters of DEGs in the five inhibitory clusters following SOC retrieval. C Scatter plot displaying DEGs in the U_R and P_R groups within the medium spinal neuron (MSN) clusters. D Scatter plot showing DEGs in the U_R and P_R groups within the parvalbumin neuron (PV) clusters. E Scatter plot showing DEGs in the U_R and P_R groups within the somatostatin neuron (SST) clusters. F Scatter plot showing DEGs in the U_R and P_R groups within the neuropeptide Y neuron (NPY) clusters. Blue dots represent DEGs in the P_R group, green dots represent DEGs in the U_R group, and red dots indicate common DEGs between the two groups. Only detected genes shared between both groups were included in the scatter plots. Note that Fos was not included in the scatter plots (C-F) due to high fold changes (out of range). G Bubble plot illustrating enriched pathways in the P_R groups of the NPY and SST clusters, compared to the VGLUT1 cluster. U unpaired, P paired, R retrieval. NS: not significant.
In excitatory pPC neurons, we focused on the VGLUT1a cluster (Fig. 7 and Supplementary Table 2). The VGLUT1c cluster had few DEGs during acquisition and retrieval, while VGLUT1b lacked sufficient Fos+ neurons during retrieval (Supplementary Fig. 18). During acquisition (Fig. 7A, C), 90 upregulated genes were found in the paired group and 89 in the unpaired group, with 27 shared ( ~ 18% overlap). Retrieval analysis (Fig. 7B, C) revealed 60 DEGs in the unpaired group and 108 in the paired group, with 15 shared (~ 10% overlap). Eleven genes, including Arc, Egr1, Nr4a1, and Bdnf, were consistently upregulated across all conditions. The overlap in DEGs suggests these genes are characteristic of Fos+ neurons, regardless of their specific roles in SOC.
A Volcano plot displaying differentially expressed genes (DEGs) in the unpaired (U_A) and paired (P_A) groups following SOC acquisition. B Volcano plots displaying DEGs in the unpaired (U_R) and paired (P_R) groups following SOC retrieval. The top 20 DEGs are labeled, with non-significant genes indicated as NS. C Venn diagram illustrating the common DEGs across the four conditions (U_A, P_A, U_R, P_R). D Scatter plot showing DEGs in the U_A and P_A groups. Blue dots represent DEGs in the P_A group, green dots represent DEGs in the U_A group, and red dots indicate common DEGs between the two groups. E Scatter plot showing DEGs in the U_R and P_R groups. Blue dots represent DEGs in the P_R group, green dots represent DEGs in the U_R group, and red dots indicate common DEGs between the two groups. Only detected genes shared between both groups were included in the scatter plots. Note that Fos was not included in the scatter (D, E) plots due to high fold changes (out of range). F Bubble plot showing enriched pathways in the U_A, P_A, U_R, and P_R groups of the VGLUT1a cluster. G Comparison of the enriched pathways in the P_R groups between the BLA VGLUT1 cluster and the PC VGLUT1a cluster. U unpaired, P paired, A acquisition, R retrieval. NS: not significant.
Scatter plots of log2 fold-changes highlighted shared DEGs (Fig. 7D, E), while gene ontology analysis revealed both distinct and shared enriched pathways (Fig. 7F). During acquisition, pathways related to norepinephrine response, synaptic transmission and plasticity, neuropeptide signaling, and cellular hormone responses were enriched in both groups, with the paired group showing additional enrichment in pathways critical for learning, such as synaptic assembly (Ntrk2, Bdnf, Adgrf1, Npas4, Numb, Prkca), regulation of protein phosphorylation (Ntrk2, Bdnf, Ptgs2, Camk4, Prkca), and signaling via MAPK (Ntrk2, Gadd45b, Lpar3, Epor, Scimp, Grm1, Alkal2, Igf1r, Rell2, Prkca), ERK (Dusp6, Epor, Scimp, Alkal2, Spred3, Prkca), calcium (Homer1, Gpr39, Ptgs2, Ms4a2, Lpar3, Lgals3, Pde4b), and G-proteins (Homer1, Grm1). During retrieval, fewer pathways were enriched, but key synaptic and learning-related pathways were conserved across both phases in the paired group. In addition, retrieval in the paired group was associated with enrichment in synaptic remodeling (Arc, Ntrk2, Bdnf, Homer1, Numb, Rheb, Hspa8, Frmpd4, Gpr158), lipid metabolism (Nr4a3, Egr1, Irs2, Sik1, Ccn1, Adgrf5, Sik2, Atp1a1, Ptgs2), and cellular response to chemical stress (Egr1, Nr4a2, Gch1, Hspa8, Atp1a1, Lrrc8c, Ptgs2, Fos).
An integrated UMAP analysis revealed molecular correlations between BLA and pPC subclusters (Supplementary Fig. 19), showing partial overlap between BLA VGLUT1 and PC VGLUT1a, and nearly complete overlap of VIP and PV subclusters. This aligns with findings that inhibitory neuronal types share similar molecular features across brain regions53. A comparison of VGLUT1 neurons in the BLA and VGLUT1a neurons in the pPC during SOC retrieval (Fig. 7G) revealed overlapping enriched pathways, including those involved in synaptic signaling, norepinephrine response, protein phosphorylation, and learning and memory processes. Notably, pPC pyramidal neurons displayed distinct enrichment in pathways related to the regulation of synaptic plasticity, synapse organization, lipid signaling and responses to chemical stress. In the pPC, most DEGs in the paired group, including shared genes, exhibited higher fold changes than in the unpaired group (Fig. 7D, E).
In inhibitory neurons of the pPC, distinct DEG patterns were observed. DEGs were identified across all three inhibitory neuron clusters during acquisition (Fig. 8A and Supplementary Table 2), and two clusters (PV and VIP) during retrieval (Fig. 8B). Two shared genes (Fos, Trim36) were identified in the PV cluster (Fig. 8C, D), though no significant enrichment was observed in gene ontology analysis.
A Heat map showing distinct patters of DEGs in the three inhibitory clusters following SOC acquisition. B Heat map showing distinct patters of DEGs in the inhibitory clusters following SOC retrieval. C Scatter plot displaying DEGs in the U_R and P_R groups within the PV clusters. Blue dots represent DEGs in the P_R group, green dots represent DEGs in the U_R group, and red dots indicate common DEGs between the two groups. D Scatter plot showing DEGs in the U_R and P_R groups within the vasoactive intestinal peptide (VIP) clusters. Only detected genes shared between both groups were included in the scatter plots. Note that Fos was not included in the scatter (C, D) plots due to high fold changes (out of range). U unpaired, P paired, R retrieval. NS: not significant.
SOC-driven chromatin remodeling in the BLA and pPC
The formation and retrieval of SOC memories require complex plasticity within both the BLA and pPC. To dissect the transcriptional regulation underlying these processes, we analyzed snATAC-seq on Fos+ and Fos- excitatory neuronal pseudo-bulk from these regions following SOC. Our findings reveal marked shifts in chromatin accessibility in Fos+ neurons, highlighting the significance of chromatin remodeling in SOC.
Fos+ neurons showed a global increase in chromatin accessibility compared to Fos- neurons. In BLA VGLUT1 neurons, this increase was more pronounced during acquisition (Fig. 9A) than retrieval (Fig. 9B), with a notable LOESS regression shift (Fig. 9C). Conversely, pPC VGLUT1a neurons exhibited consistently elevated chromatin accessibility across both acquisition (Fig. 9D) and retrieval (Fig. 9E), without a regression shift (Fig. 9F). These findings suggest substantial chromatin remodeling in the BLA during acquisition, which stabilizes by retrieval, while the pPC maintains chromatin modification throughout both phases. The global absolute shift in chromatin accessibility for Fos+ relative to Fos- peaks was 6.7 in the BLA during acquisition and 3.8 during retrieval. In the pPC, these shifts were 5.0 during acquisition and 5.2 during retrieval. These results suggest that the BLA undergoes extensive chromatin remodeling during memory acquisition, while the pPC maintains a relatively stable chromatin state across both acquisition and retrieval phases.
A Scatter plot showing Fos+ and Fos- snATAC-seq peaks with Fos motif weight in BLA VGLUT1 neurons during acquisition in the paired group (P_A). B Scatter plot of Fos+ and Fos- snATAC-seq peaks in the BLA during retrieval (P_R). C LOESS regression showing a shift in chromatin accessibility from acquisition (P_A) to retrieval (P_R) in the BLA. D Scatter plot displaying Fos+ and Fos- snATAC-seq peaks with Fos motif weight in pPC VGLUT1a during acquisition in the paired group (P_A). E Scatter plot of Fos+ and Fos- snATAC-seq peaks in the pPC during retrieval (P_R). F No observed LOESS regression shift in chromatin accessibility in the pPC between acquisition and retrieval phases. P paired, A acquisition, R retrieval.
When combining snATAC-seq peaks from all assays (BLA and pPC), motif analysis revealed that 36.8% (± 14.7%) of snATAC-seq peaks in Fos+ neurons across genomic regions contained Fos binding motifs associated with synaptic plasticity, memory consolidation, and stress response. Regional distribution of Fos motifs varied: 19.4% (± 0.3%) in promoters, 48.7% (± 0.7%) in introns, 29.5% (± 2.2%) in exons, 47.9% (± 0.5%) in intergenic regions, and 36.2% (± 2.5%) in other regions (Supplementary Fig. 20). These findings reinforce the role of Fos in chromatin remodeling54, which may be essential for gene expression changes underlying SOC. However, further research is needed to elucidate the specific mechanisms involved. Collectively, our results suggest that SOC triggers region-specific chromatin remodeling crucial for memory formation and retrieval, with distinct shifts in accessibility in Fos+ neurons of the BLA and pPC that may reflect their roles in different SOC phases.
Discussion
Our study provides compelling evidence that SOC is mediated by a coordinated set of circuits involving the BLA and sensory cortices, particularly the pPC. Both regions, along with the Aud, are engaged during SOC memory retrieval, as indicated by elevated cFos expression following exposure to the CS2. We show that NMDAR-dependent plasticity in both the BLA and the pPC is required for SOC learning, indicating that these regions act in concert to encode, consolidate, and retrieve higher-order associative memories.
Functionally, the BLA and pPC appear to contribute similarly to both FOC and SOC. The BLA integrates sensory cues and mediates associative plasticity, while the pPC interacts with the BLA to support memory formation and may serve as a site for long-term memory storage. These roles are supported by our findings and are consistent with prior literature, suggesting that the core circuit mechanisms are preserved across FOC and SOC. However, differences in molecular requirements likely exist between the two forms of learning. For example, previous studies have reported that SOC may depend on protein synthesis in the BLA that is initiated during FOC, effectively leveraging prior molecular changes to support new associations15. Furthermore, the LC noradrenergic signaling are essential for both FOC and SOC, suggesting that common neuromodulatory mechanisms facilitate the encoding and retrieval of associative memories across learning paradigms.
The BLA has long been recognized as a critical site for threat learning, integrating sensory inputs with aversive stimuli to form associative memories. Our findings extend this role to SOC, where the BLA links CS1 and CS2 to support higher-order associative learning. Neurons in the BLA that are recruited during SOC acquisition are likely reactivated during memory retrieval, as evidenced by the convergence of CS1 and CS2 neuronal ensembles in the BLA and the ability of these ensembles to drive defensive freezing behavior when optically activated. This convergence highlights the BLA’s role as a hub wherein sensory and affective information integrate to form complex associative memories. CREB upregulation and enhanced excitability in the amygdala is known to bias neurons towards incorporation into memory engram55,56. Here, it is likely that increased excitability of US and CS1 ensembles following FOC predisposes them to selection for the SOC engram. The protein synthesis required for FOC memory consolidation may also contribute to SOC memory acquisition15. Moreover, the fact that synaptic depotentiation of the CS1 + CS2 ensemble attenuates SOC memory expression supports the crucial role of synaptic plasticity within these ensembles for the maintenance of SOC memories. While the BLA is essential for encoding and consolidating associative memories through synaptic plasticity, recent findings show that Calca-expressing neurons in the parabrachial nucleus contribute to SOC by reactivating the US pathway, encoding the learned aversiveness of CS1 and enabling the transfer of aversive valence to CS257. These regions likely serve parallel but complementary functions: the BLA integrates sensory associations, whereas the PBN reinstates affective significance during higher-order learning.
In addition to the BLA, our study identifies the pPC as a critical site for SOC memory acquisition and retrieval. This is the first demonstration of a sensory cortex’s involvement in SOC, revealing that NMDAR-dependent plasticity in the pPC is essential for the encoding of second-order associations. The pPC’s role in SOC is likely mediated by its extensive connections with the BLA27, which allow it to integrate sensory information with affective valence and contribute to the formation of long-term memories. The specific involvement of the pPC in SOC is further supported by our chemogenetic experiments, which show that silencing the BLA-PC pathway during memory retrieval significantly impairs SOC memory, indicating that this pathway is essential for the expression of SOC memories.
In olfactory FOC, the pPC is particularly important for long-term memory storage31,32. Lesions in the pPC one month after olfactory FOC impair remote, but not recent, memory, highlighting its specific role in long-term memory consolidation29. Similarly, blocking NMDARs in the pPC impairs remote, but not recent, memory of FOC34. The pPC is known to exhibit associative plasticity over time31,32 and engage in sleep-dependent memory replay58, both of which are essential for consolidating threat memories. In this study, we also demonstrate that NMDAR-dependent plasticity in the pPC is critical for SOC encoding. It is possible that the pPC also functions as a repertoire for SOC memory storage, in addition to its role in encoding. Involvement of both the BLA and pPC may enhance memory fidelity, ensuring reliable responding to associated cues, and facilitating the transition of memory storage from the BLA to the pPC for long-term retention.
Using a stringent experimental design that includes CS1–CS2 unpaired controls to isolate gene expression changes specific to associative learning while accounting for non-associative effects of sensory stimulation, our gene expression and chromatin accessibility analyses of Fos+ ensembles in the BLA and pPC reveal critical insights into the molecular mechanisms supporting SOC. In the BLA, excitatory VGLUT1 neurons exhibited marked chromatin remodeling during acquisition and differential gene expression during retrieval, with significant enrichment in pathways related to synaptic transmission, receptor signaling, stress response, cell adhesion, and learning-related processes. These findings parallel molecular changes reported in FOC23,43,59, suggesting that SOC and FOC engage overlapping rather than fundamentally distinct molecular pathways. We also identified distinct transcriptional programs among molecularly defined inhibitory neuron subtypes in the BLA region, highlighting previously uncharacterized cell-type-specific contributions of inhibitory neurons to learning. For example, SST-expressing neurons, which are known to coordinate memory circuitry by modulating synaptic connections with excitatory engram neurons52, and NPY-expressing neurons, which have been linked to stress regulation60, both showed gene pathway activation consistent with their respective functional roles.
In the pPC, VGLUT1a excitatory neurons emerged as key players in SOC, showing gene enrichment during both acquisition and retrieval in pathways related to synaptic assembly, protein phosphorylation, and MAPK signaling. Notably, pathways involved in synaptic remodeling and long-term memory were particularly enriched during retrieval, suggesting the pPC supports memory stabilization in later phases of learning. These transcriptional patterns were corroborated by snATAC-seq analyses, which revealed sustained increases in chromatin accessibility in the pPC across both learning phases. In contrast, the BLA exhibited more dynamic chromatin remodeling during acquisition, suggesting a role for epigenetic priming in facilitating plasticity within engram neurons. Collectively, these results demonstrate that SOC engages coordinated gene regulatory programs and chromatin dynamics across the BLA and pPC, enabling the integration of sensory and emotional information and supporting the formation and stabilization of higher-order associative memories.
Furthermore, our findings highlight the LC adrenergic signaling as a critical modulator during SOC memory encoding and support its role in ensemble linking61,62. Blocking adrenergic input to the pPC or BLA during training impaired SOC formation, suggesting that LC-derived norepinephrine is required for associating CS1 and CS2 representations. Given that the LC is activated by novelty, salience, and prediction error54,55, its engagement during SOC likely enhances synaptic plasticity and facilitates the integration of CS2 into existing memory networks. This linking may occur through β-adrenoceptor-mediated plasticity in the BLA and pPC39,63, enabling the formation of distributed engrams. The observed potentiation of LC-pPC projections further supports a role for the LC in coordinating cortical-subcortical plasticity essential for SOC.
In conclusion, our study demonstrates that SOC is mediated by a dual system involving both the BLA and pPC, with significant contributions from adrenergic inputs that enhance synaptic plasticity and facilitate the formation of complex associative memories. This dual system, involving both the BLA and pPC, highlights the complexity and specificity of SOC memory encoding and retrieval. Our findings suggest that higher-order learning processes like SOC elaborate upon initial FOC, providing insight into the neural mechanisms underlying psychiatric conditions such as PTSD and their resistance to extinction therapy. By investigating SOC in animal models, we can better understand these mechanisms and, ultimately, pave the way for the development of more effective therapeutic strategies for fear-related disorders.
Materials and methods
Subjects
Adult Sprague-Dawley rats aged between 3 to 6 months, of both sexes, were randomly assigned for the behavioral experiments. The experimental groups were balanced in terms of sex. Only male rats were used for Multiomics snRNA-seq and snATAC-seq, with three replicates in each group. Rats were maintained on a standard 12 h light-dark cycle with ad libitum access to food and water. All experimental procedures were conducted following approval from the Institutional Animal Care Committee at Memorial University of Newfoundland and adhered to the guidelines set by the Canadian Council on Animal Care.
Cannula surgery
Animals were anesthetized using isoflurane in an induction chamber and subsequently placed in a stereotaxic frame. Bilateral implantation of 23-gauge metal guide cannulas was performed in the BLA (coordinates: AP: − 2.5 mm, ML: ± 4.9 mm from bregma, DV: − 7.6 mm from the brain surface), pPC (coordinates: AP: − 2.3 mm or −1.5 mm for chemogenetic experiment, ML: ±5.4 mm, DV: −8.4 mm), or Aud (coordinates: AP: −5 mm, ML: ± 6.7 mm, DV: −2 mm). The cannulas were secured to the skull using dental cement, with miniature self-tapping stainless-steel screws implanted on the skull. Following surgery, incisions were sutured, and rats were allowed to recover in their home cages. A minimum recovery period of 1 week was provided before commencing behavioral experiments. Subsequently, a subset of rats underwent perfusion to verify cannula targeting (Supplementary Fig. 2).
Virus and tracer injection
For retrograde tracing experiments, 0.25 μl of CTB conjugated to Alexa 647 (0.5% w/v in PBS; Invitrogen) was bilaterally infused into the BLA (coordinates: AP: −2.5 mm, ML: ± 4.9 mm from bregma, DV: − 7.8 mm from the brain surface), pPC (coordinates: AP: − 1.5 and − 3.0 mm, ML: ± 5.6 mm, DV: − 8.4 mm), or Aud (coordinates: AP: − 4.5 mm, ML: ± 6.7, DV: − 2.5). For tTA-cFos ensemble labeling experiments, a mixture of AAV2/DJ-cFos-tTA (E11 GC/ml) and AAV2/DJ-TRE-eGFP (2.6E12 GC/ml) in a 1:1 ratio, totaling 0.75 µl, was infused into the BLA and Aud. A similar mixture of AAV2/DJ-cFos-tTA and AAV2/DJ-TRE-ChR2-eGFP was infused into the BLA for optogenetic activation experiments. For the chemogenetic inhibition experiment, 1 µl of AAV2/8-SYN-hM4D(Gi) or AAV2/8-SYN-GFP (Neurophotonics, Laval; 1.8 × 10¹³ GC/mL) was injected into the BLA. An infusion cannula was then implanted in the pPC following the virus infusion. Infusions were conducted using either a 32-gauge beveled 1 μl Hamilton syringe (Neuros 7001 KH) connected to a vertical infusion pump (Pump 11 Elite; Harvard Apparatus) or a glass micropipette attached to a 10 μl Hamilton syringe via polyethylene tubing and the Pump 11 Elite (Harvard Apparatus). Each infusion lasted 5 min, followed by a 10 min wait before syringe withdrawal. Rats were allowed 1–2 weeks to recover before initiating behavioral experiments, followed by perfusion with 4% paraformaldehyde (PFA).
Optical fiber cannula implantation
Mono fiber-optic cannula (Doric, 300 μm, NA = 0.37, 12 mm) were implanted in the BLA immediately following virus injection. The tip of the optic fiber was placed 1 mm above the injection site. Then, the cannula was the secured with dental cement.
Behavioral studies
Odorant
Terpinene (6.63%) or benzaldehyde (0.05%) diluted with mineral oil to specific concentrations was utilized as the odorant. The chosen odorant was determined to be affectively neutral to adult rats, and concentrations were selected to achieve a vapor-phase partial pressure of 1 Pa64.
Apparatus
All behavioral training and testing were conducted in a custom-made olfactometer for air and odorant delivery, which was integrated with a shock chamber. The shock chamber comprises a Plexiglas chamber mounted on an electrified grid, linked to a shock generator (Muromachi Kikai Model SGS-003DX, Japan or San Diego Instruments, U.S.A). Odorant was stored in a polyvinyl carbonate bottle, connected to the olfactometer via C-flex tubing, and sealed when not in use. A fan with evacuation tubing was affixed to the top lid of the shock chamber to expedite odor clearance. A background white noise of 60 dB was maintained in the behavioral rooms throughout the experiments.
Second-order conditioning (SOC)
Rats underwent a habituation period of 30 min in the shock chamber over two (non-surgery rats) or four (surgery rats) consecutive days. On days 3 or 5, FOC training involved individual rat training with four separate exposures to a pairing of a pure tone conditioned stimulus (CS1: 2 kHz, 80 dB) and an unconditioned stimulus (US: foot shock). The shocks were administered at 5, 15, 20, and 30 min during a 30 min training session, coinciding with the last second of tone delivery (0.5 mA for 1 s). On either day 4 or day 6, the SOC procedure was conducted. In the paired group, the tone (CS1) and odor (CS2) were presented simultaneously for 5 min, while the unpaired group received the CS1 and CS2 presentations with a 30 min interval between them. Freezing behavior in the presence of the tone CS1 was recorded. On day 5 or 7 (SOC recall), rats were pre-exposed to the shock chamber without odor delivery for 5 min to establish baseline activity before receiving a 5 min exposure to CS2. Freezing behavior in the presence of CS2 was measured as the percentage of time spent freezing.
Drug administration
For the NMDAR blocker experiment, D-APV (1 μg/0.5 μl per hemisphere, dissolved in saline; Tocris Bioscience) was infused into the BLA, pPC, or Aud immediately after SOC training. For adrenoreceptor blocker experiments, a mixture of adrenoreceptor antagonists (2.5 µg propranolol and 2.0 µg prazosin in 0.5 µl per hemisphere, dissolved in saline; Sigma) was infused into the BLA and pPC immediately after SOC training. For chemogenetic inhibition experiments, Clozapine N-oxide (CNO) (Tocris) was prepared as a 3 mM stock solution in 0.9% sterile saline and diluted to a 3 µM working solution on test day. Rats received 0.5 µL of either CNO or saline per side in the pPC, administered either 30 min before CS1 + CS2 pairing during SOC training or 30 min before odor CS2 testing on SOC recall day. Infusions were delivered at a rate of 0.2 μl/min using a stainless steel 33-gauge internal cannula connected to a 10 μl Hamilton syringe via polyethylene tubing, driven by an 11 Elite Syringe pump (Harvard Apparatus). The internal cannula extended 0.5 mm below the guide cannula, with an additional 1 min duration allowed for the infusion cannula to remain in place following the infusion.
Neuronal ensemble labeling with the cFos-tTA/TRE-GFP transgenes
Rats were maintained on a doxycycline (DOX) diet (40 mg/kg DOX, Envigo, U.S.) for at least one week prior to surgery. Ten days after infusion with AAVs carrying transgenes AAV2/DJ-cFos-tTA and AAV2/DJ-TRE-eGFP, the rats were habituated to the conditioning context over a four-day period. Following habituation, the DOX diet was replaced with a regular diet in the home cage for two days, allowing for GFP neuronal labeling of CS1 during a 5 min exposure to a neutral tone. Immediately after the tone presentation, the rats were switched to a high-dose DOX diet (200 mg/kg DOX) for the remainder of the experiment to prevent further GFP neuronal labeling.
Two days later, the rats underwent paired or unpaired SOC. The following day, they were pre-exposed to the conditioning context for 5 min without odor delivery, followed by a 5 min exposure to the odor. Freezing behavior in the presence of the odor was measured as the percentage of time spent freezing. Ninety minutes after the odor test, the rats were euthanized via transcardiac perfusion. Brain sections were subsequently processed for immunohistochemistry to visualize CS2 cFos+ cells.
Optogenetic activation
Two weeks after virus injection (AAV2/DJ-cFos-tTA and AAV2/DJ-TRE-ChR2-eGFP) and optic cannula implantation, the rats were habituated to the experimental context for two days (30 min per day) with the optic fiber connected. Following habituation, the rats underwent US + CS1 conditioning as previously described. After conditioning, the rats were returned to their home cages, and the DOX diet was replaced with regular food. After two days off DOX, the rats underwent paired SOC, in which they were simultaneously exposed to a tone CS1 and an odor CS2 for 5 min, labeling neurons representing the CS1 + CS2 pairing. Immediately following this session, the rats were returned to a high-dose DOX diet (200 mg/kg DOX) to halt CHR2-GFP expression.
Two days later, the rats underwent odor testing in the conditioning context in the morning, followed by optical stimulation (20 Hz, 15 ms pulse width) in a different context in the afternoon. Bilateral photo-activation of the BLA was delivered via two laser light sources (LDFLS_450; Doric Lenses) at 450 nm (20 mW/mm² at fiber tip) through mono-fiber optic patch cords. The stimulation protocol consisted of a 3-minute baseline period with no light, followed by two episodes of optical stimulation, each lasting 3 min, with a 4 min interval between them. Freezing behavior was measured both before and during the second optical stimulation.
A subset of rats then underwent two sessions of low-frequency light stimulation (1 Hz, 15 min) the next day for optical depotentiation. This group was subsequently tested for odor in the morning and received optical stimulation in the afternoon of the following day. One animal was not included from the depotentiation group due to the accidental loss of the cannula cap prior to stimulation. Another animal that exhibited low freezing in response to light following the conditioning was not included in the depotentiation experiment. Ninety minutes following the optical stimulation, the rats were euthanized via transcardiac perfusion. Brain sections were then subjected to immunohistochemistry to measure cFos expression for validation.
Chemogenetic silencing of the BLA-pPC pathway
For chemogenetic inhibition experiments, rats received 0.5 µl of either CNO or saline per hemisphere in the pPC, administered either 30 min before CS1 + CS2 pairing during SOC training or 30 min before odor CS2 testing on SOC recall day. For the retrieval phase, a within-subjects control design was employed. Rats were randomly assigned to receive either CNO or saline on the first day of testing, with the treatments switched on the following day.
Immunohistochemistry
Rats were perfused 90 min following SOC conditioning or odor (CS2) exposure on recall day. The perfusion was performed transcardially with ice-cold saline (0.9%), followed by 4% PFA in 0.1 M phosphate buffer. After extraction, the brains were stored in 4% PFA for 24 h at 4 °C and then transferred to 0.1 M PBS. Coronal sections of 50 µm were cut using a Compresstome (VF-310-OZ, Greenville, NC) and transferred to 24-well plates containing a PVP solution. The free-floating sections were stored at 4 °C until further processing.
For fluorescent cFos staining, sections were washed three times for 5 min each in Tris buffer (0.1 M, pH 7.6) and incubated for 30 min in 0.03% H2O2 in Tris buffer. Following this, the sections were washed for 10 min in Tris A (0.1% Triton X in Tris buffer) and then in Tris B (0.1% Triton X and 0.005% BSA in Tris buffer) before blocking with 10% normal goat serum (Sigma-Aldrich) in Tris B for 1.5 h. The sections were then washed in Tris A and Tris B buffers for 10 min each. After washing, they were incubated in a primary monoclonal antibody solution prepared in Tris B. The antibodies used included cFos (1:1000, New England Biolabs) and Dopamine beta-hydroxylase (DBH, 1:2,000, Millipore-Sigma). This incubation was carried out over two nights.
After primary antibody incubation, sections were washed in Tris A and Tris B buffers and incubated with a goat anti-rabbit IgG secondary antibody conjugated with either Alexa Fluor 488 for CTB tracing or Alexa Fluor 647 for cFos-tTA-TRE co-allocation or optogenetic validation experiments (1:1,000; ThermoFisher), or with a mixture of Alexa Fluor 488 and Alexa Fluor 555 for the LC sections. The incubation was performed in Tris B buffer for 45 min. Following incubation, the sections were washed in Tris buffer for 5 min, mounted on slides, air-dried, and coverslipped with DAPI (Abcam).
For SG-gray cFos staining (1:10,000, New England Biolabs), the sections were incubated with a goat anti-rabbit biotinylated secondary antibody (1:1000; ThermoFisher) followed by a 10 min wash in Tris D (0.1% Triton X and 0.005% BSA in 0.5 M Tris buffer) and incubation in an avidin–biotin horseradish peroxidase complex (1:1000 in Tris D; Vector ABC kit, Vector Laboratories) for 1.5 h at room temperature (allowing the complex to form 30 min before use). After incubation, the sections were washed in Tris B, and immunostaining was performed with a peroxidase substrate (Vector SG), staining cFos-positive nuclei blue-gray. The sections were then washed three times in Tris buffer, dehydrated in ethanol solutions and xylene, and coverslipped with Permount (Fisher).
Imaging and analysis
Images were acquired using an EVOS 5000 (Thermo Fisher Scientific) for fluorescent imaging and a bright-field microscope (10x, BX53 Upright Microscope, Olympus) for bright-field imaging. Standardized light intensity and exposure parameters were consistently applied across all captured images. Image analysis was performed using ImageJ software. The number of cFos immunopositive nuclei was automatically counted and normalized to the regions of interest (per mm²). For fluorescent cFos counting, images underwent background subtraction before automatic cell counting using the Trainable Weka Segmentation plugin. In retrograde tracer experiments, double-labeled CTB and cFos+ cells were counted manually. For LC cFos expression, cFos+ cells were also counted manually. Three to six images per animal from each area of interest within the same rostral to caudal range were analyzed, with values from both hemispheres averaged. Mis-targeted brains were excluded from analysis. A subset of images was analyzed blindly by experimenters who were not aware of the experimental conditions.
snRNA sequencing and snATAC sequencing
Sample preparation
A total of 12 rats were used in this experiment, with three rats per group (paired or unpaired) at each time point (early consolidation or memory retrieval). Tissue samples were harvested from the BLA and pPC at two time points following SOC: 90 min post-conditioning and 90 min post-retrieval test. Tissue from the BLA region (AP: − 2.3 to − 3.8 mm to bregma) was extracted from frozen sections in the cryostat using a 1 mm diameter biopsy punch, while pPC tissue was collected using a 0.5 mm diameter punch at three lateral-to-medial locations (Fig. 4A). Single-nucleus suspensions were prepared following the Chromium Next GEM Single Cell Multiome ATAC + Gene Expression protocol (10x Genomics, User Guide Rev F, CG000338)65. Briefly, single nuclei from the tissues were isolated using the 10x Genomics Chromium Nuclei Isolation with RNAse Inhibitor Kit (PN-1000494). The tissues were immediately immersed in pre-chilled lysis buffer post-extraction to facilitate dissociation and nuclei release. Nuclei were isolated by passing the suspension through a 40 μm cell strainer (Bel-Art™) to remove clumps and aggregates. The quality and concentration of nuclei were assessed by staining with a Trypan blue and ethidium bromide mixture, using a hemocytometer (NanoEntek) and fluorescence microscopy (Invitrogen™ EVOS™ M5000). The final nuclei concentration was optimized by diluting the nuclei buffer.
Library preparation and sequencing
Single-nucleus ATAC and RNA-seq libraries were constructed using the 10X Chromium Next GEM Single Cell Multiome ATAC + Gene Expression Kit (10x Genomics, User Guide Rev F, CG000338). Approximately 2000 nuclei per sample were loaded onto the Chromium Next GEM Chip G. Libraries were prepared in parallel for snATAC-seq and snRNA-seq, comprising double-strand DNA with standard Illumina paired-end construction. Sequencing was performed on an Illumina NovaSeq 6000 platform (TCAG Facility at SickKids, Toronto) to achieve a target depth of ~ 20,000 read pairs per cell for snATAC-seq and ~ 25,000 read pairs per cell for RNA-seq.
Data processing and quality control
The raw sequencing files were demultiplexed using Cell Ranger ARC (10x Genomics, version 2.0.2). FASTQ files for both gene expression and chromatin accessibility were aligned against the rat reference genome (mRatBN7.2). Post-alignment libraries derived from the same brain region (BLA or pPC) were aggregated using Cell Ranger ARC with normalization for sequencing depth to generate cell clusters.
Creation of Seurat objects
Processed data were imported into R for analysis using the ‘Seurat’ and ‘Signac’ packages66. Quality control metrics, including total reads, unique molecular identifiers (UMIs), and the fraction of reads mapping to mitochondrial DNA, were evaluated. Gene expression (RNA) and chromatin accessibility (ATAC) data were integrated into Seurat objects corresponding to BLA and pPC. Initial quality control filtering was applied to exclude cells with low-quality metrics, including those with more than 5% mitochondrial content and read counts for both snRNAseq and snATACseq comprise between 200 and 100,000.
snRNA-seq and snATAC-seq data integration
snRNA-seq and snATAC-seq data from BLA and pPC were integrated using Seurat and Signac. snRNA-seq read counts underwent normalization and variance stabilization via SCTransform67, followed by dimensionality reduction using Principal Component Analysis (PCA) on the first 50 principal components. snATAC-seq data were processed using Term Frequency-Inverse Document Frequency (TF-IDF) normalization and Singular Value Decomposition (SVD) to identify the most variable genomic regions. A Weighted Nearest Neighbor (WNN) analysis was conducted to create a joint snRNA-seq-snATAC-seq UMAP for integrated visualization of the multi-omic data65,68.
Clustering, cell type identification and differential gene expression analysis
After library preparation, 25,098 UMIs were captured in the BLA and 30,759 UMIs in the PC using Gel Beads-in-emulsions (GEMs). Following quality control and filtering, 23,385 UMIs were retained in the BLA and 25,211 UMIs in the PC. Cell types were identified using a graph-based clustering approach, followed by UMAP visualization of both snRNA-seq and snATAC-seq datasets69. Marker genes for each cluster were determined based on significant over-expression genes with a log-fold change greater than 0.5 and an adjusted p-value (Bonferroni) less than 0.05. Neurons were further refined using canonical neuronal markers, including Slc17a7, Slc17a6, Gad1, Gad2, and Slc32a143,70. A total of 14,171 BLA neurons and 15,927 PC neurons were selected for further analysis. PCA and UMAP were then performed on these subsets, with the first 60 principal components computed.
In the BLA, the snRNAseq dataset was organized into 13 distinct clusters including neurons and non-neuronal cell types (Fig. 4A and Supplementary Fig. 10), with minimal batch effects observed (Supplementary Fig. 11). Neuronal clusters were defined using canonical gene markers for both excitatory (Slc17a7, Slc17a6) and inhibitory (Gad1, Gad2, Slc32a1)43,71 neurons, resulting in the identification of nine subclusters. These included the excitatory VGLUT1 (Slc17a7+; cluster 0) and VGLUT2 (Slc17a6+; cluster 7) subclusters, multiple inhibitory subclusters (clusters 1–5), and two mixed subclusters that expressed markers for cortical projection neurons (e.g., Satb272) and astrocytes (e.g., Ccn273), categorized as unknown (cluster 6, 8) (Fig. 4B, C and Supplementary Fig. 12). The inhibitory clusters were define as somatostatin (SST) (Sst+/Erbb4-; cluster 1), medium spiny (MSN) (Penk+74/Npy- ; cluster 2), neuropeptide Y (NPY) (Npy+; also Penk+; cluster 3), parvalbumin (PV) (Erbb4+75/Pvalb+; also Npy+/Sst+; cluster 4), and vasoactive intestinal peptide (VIP) (Vip+; cluster 5) neuron clusters (Supplementary Fig. 12). Clusters 6–8 contained insufficient cells and were excluded for further analysis.
In the PC, 17 clusters were identified (Fig. 4A). Neuronal population was further divided into three excitatory VGLUT1 (Slc17a7 +; clusters 0, 4, 6), one excitatory VGLUT2 (Slc17a6+; cluster 7), and three inhibitory neuron subclusters (1, 3, 5), along with two mixed clusters labeled as unknown (cluster 2, 8) (Fig. 4A and Supplementary Figs. 14–16). Cluster 2 did not significantly express any of the distinct excitatory and inhibitory markers used in our analysis, whereas cluster 8 expressed both Satb2 and Ccn2, along with Slc17a7 (Fig. 4F, G and Supplementary Fig. 16). These two clusters were labeled unknown. Clusters identified as VGLUT2 (due to insufficient cell numbers) and unknown were excluded from further analysis. The three VGLUT1 excitatory neuron types in the PC were designated VGLUT1a, VGLUT1b and VGLUT1c. VGLUT1a (cluster 0) and VGLUT1b (cluster 6) likely represent superficial and deep pyramidal neurons, respectively, as distinguished by Cux1 expression, characteristic of neurons in the deep layers IIb and III of the PC48. VGLUT1c (cluster 4) likely consists of semilunar neurons expressing Reln48. The inhibitory neurons were defined as PV (Erbb4+/Pvalb+; also Npy+/Sst+; cluster 5), VIP (Vip+; cluster 3), and one unidentified inhibitory neuron cluster labeled INu (Gad1+, cluster 1).
The Fos+ ensemble was identified using the AddModuleScore function in Seurat66, which normalized Fos expression by comparing it to control genes with similar expression levels. Genes were binned by average expression, and the Fos score in each cell was generated by subtracting the average expression of randomly selected control genes from Fos expression. Cells with a module score > 0 were classified as Fos+. Differential expression analysis between Fos+ and Fos− cells was performed using the Wilcoxon test. A log fold-change threshold of |0.5| and an adjusted p-value (Bonferroni) of <0.05 were used to define DEGs.
Gene ontology enrichment analysis
Gene ontology enrichment analysis was performed on the identified gene sets using the clusterProfiler76 package in R, along with org.Rn.eg.db, DOSE, stringr, and readxl packages. DEGs identified in each behavioral condition and subcluster were converted to Entrez IDs. Gene ontology analysis focused on Biological Processes with an adjusted p-value cutoff of 0.05. Results were visualized using ggplot2 for GO terms relevant to neuronal function and plasticity.
Integration of BLA and pPC Data
To compare molecular signatures between BLA and PC neurons, canonical correlation analysis (CCA) was performed on the Seurat objects from each region77. The integrated dataset was scaled, PCA was conducted, and UMAP was generated for cross-region visualization and comparison.
snATAC-seq data processing
For generating chromatin accessibility data, we first sub-set the objects based on behavioral and Fos status information, followed by further subsetting the snATAC-seq portion to create a chromatin assay. Peak information were extracted using a custom R script (cf. code availability section). In brief, the create_bed_file function extracts peak ranges from the ChromatinAssay object by accessing the genomic ranges (granges) and converting the counts matrix into a data frame. For each peak, the chromosome, start, and end coordinates are retrieved, with a “chr” prefix added to the chromosome name. The start coordinates are adjusted by subtracting one to conform to BED file formatting. Normalized read counts are calculated as scores by dividing the sum of read counts for each peak by the total reads across all cells (pseudo-bulk) for the given condition, then multiplying by one million. The final output, including chromosome, start, end, score, and counts, is saved as a BED-like data frame and written to a BED.
snATAC-seq data from Fos+ and Fos- neurons were processed to assess chromatin accessibility. The genomic coordinates and snATAC-seq signal intensities for both conditions were combined into a single data frame, providing a unified view of chromatin accessibility across the two conditions. Each row in the dataset represents a peak, labeled with its genomic coordinates. snATAC-seq peak annotations were obtained from HOMER78, including gene names, genomic features, and distances to the transcription start site (TSS).
To assess chromatin dynamics between Fos+ and Fos- cells across BLA acquisition, BLA retrieval, pPC acquisition, and pPC retrieval, we quantified a global shift as a mean of difference as follows:
where \({{Fos}}^{+}{score}\) and \({{Fos}}^{-}{score}\) represent the re-scaled Fos+ and Fos- pseudo-bulk snATAC-seq peak scores normalized to a uniform read count, and \(N\) is the total number of peaks. This global shift was computed separately for each dataset, serving as a dataset-specific threshold for significant signal intensity shifts.
Motif analysis was conducted using matrixscan from RSAT79,80 to identify potential Fos binding sites within each snATAC-seq peak, employing the Fos matrix (ID: MM1_HSA_IMR-90_FOS) sourced from MethMotif81,82.
Data visualization, statistical analysis and reproducibility
Sequencing data visualization was performed using ‘ggplot2’, ‘EnhancedVolcano’, and ‘ggrepel’. Statistical analyses were conducted using built-in functions within Seurat and base R, with significance defined at p <0.05.
All other data were analyzed using OriginPro 2022b software and presented as mean ± SEM. Behavioral data were subjected to two-way repeated-measures ANOVAs to assess CS and conditioning/treatment interactions, followed by post hoc Tukey tests. Unpaired two-tailed Student t tests were employed for comparisons between two groups. Normality of the data was assessed with the Shapiro-Wilk test and met before group comparisons. Significance was established at p <0.05. Rats were randomly assigned to the experimental groups. A subset of data collection and imaging analysis were conducted by experimenters blind to the experimental conditions. Figures were generated using OriginPro 2022b and visualized using CorelDRAW X7 software.
The number of rats is indicated by N. Measurements were taken from distinct samples. Statistical results are included in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All sequencing data generated in this study are publicly available through GEO (under accession number GSE280826. Other data (Figs. 1–3) is linked in Figshare (https://doi.org/10.6084/m9.figshare.28827782).
Code availability
Analysis scripts and computational workflows are available on GitHub (https://github.com/karmaout/SOC_Multiomic_sequencing) for reproducibility. Processed datasets are archived on Zenodo (https://doi.org/10.5281/zenodo.14014437) to support reanalysis and integration. Detailed documentation on data processing, analysis pipelines, and visualization methods is included.
References
Pitman, R. K. et al. Biological studies of post-traumatic stress disorder. Nat. Rev. Neurosci. 13, 769–787 (2012).
Kent, J. M., Sullivan, G. M. & Rauch, S. L. The neurobiology of fear: Relevance to panic disorder and posttraumatic stress. Psychiatr. Ann. 30, 733–742 (2000).
Pape, H. C. & Pare, D. Plastic synaptic networks of the amygdala for the acquisition, expression, and extinction of conditioned fear. Physiol. Rev. 90, 419–463 (2010).
Gewirtz, J. C. & Davis, M. Using pavlovian higher-order conditioning paradigms to investigate the neural substrates of emotional learning and memory. Learn Mem. 7, 257–266 (2000).
Sepahvand, T., Power, K. D., Qin, T. & Yuan, Q. The basolateral amygdala: The core of a network for threat conditioning, extinction, and second-order threat conditioning. Biology12, 1274 (2023).
Wessa, M. & Flor, H. Failure of extinction of fear responses in posttraumatic stress disorder: evidence from second-order conditioning. Am. J. Psychiatry 164, 1684–1692 (2007).
Lingawi, N. W., Laurent, V., Westbrook, R. F. & Holmes, N. M. Acquisition and extinction of second-order context conditioned fear: Role of the amygdala. Neurobiol. Learn. Mem. 183, 107485 (2021).
Bouton, M. E., Maren, S. & McNally, G. P. Behavioral and neurobiological mechanisms of pavlovian and instrumental extinction learning. Physiol. Rev. 101, 611–681 (2021).
Fanselow, M. S. & LeDoux, J. E. Why we think plasticity underlying pavlovian fear conditioning occurs in the basolateral amygdala. Neuron 23, 229–232 (1999).
Maren, S., Aharonov, G., Stote, D. L. & Fanselow, M. S. N-methyl-D-aspartate receptors in the basolateral amygdala are required for both acquisition and expression of conditional fear in rats. Behav. Neurosci. 110, 1365–1374 (1996).
Miserendino, M. J., Sananes, C. B., Melia, K. R. & Davis, M. Blocking of acquisition but not expression of conditioned fear-potentiated startle by NMDA antagonists in the amygdala. Nature 345, 716–718 (1990).
Rodrigues, S. M., Schafe, G. E. & LeDoux, J. E. Intra-amygdala blockade of the NR2B subunit of the NMDA receptor disrupts the acquisition but not the expression of fear conditioning. J. Neurosci. 21, 6889–6896 (2001).
Silva, A. J. Molecular and cellular cognitive studies of the role of synaptic plasticity in memory. J. Neurobiol. 54, 224–237 (2003).
Rodrigues, S. M., Farb, C. R., Bauer, E. P., LeDoux, J. E. & Schafe, G. E. Pavlovian fear conditioning regulates Thr286 autophosphorylation of Ca2+/calmodulin-dependent protein kinase II at lateral amygdala synapses. J. Neurosci. 24, 3281–3288 (2004).
Lay, B. P. P., Westbrook, R. F., Glanzman, D. L. & Holmes, N. M. Commonalities and differences in the substrates underlying consolidation of first- and second-order conditioned fear. J. Neurosci. 38, 1926–1941 (2018).
Monsey, M. S., Ota, K. T., Akingbade, I. F., Hong, E. S. & Schafe, G. E. Epigenetic alterations are critical for fear memory consolidation and synaptic plasticity in the lateral amygdala. PloS ONE 6, e19958 (2011).
Siddiqui, S. A. et al. Enhanced histone acetylation in the infralimbic prefrontal cortex is associated with fear extinction. Cell. Mol. Neurobiol. 37, 1287–1301 (2017).
Itzhak, Y., Anderson, K. L., Kelley, J. B. & Petkov, M. Histone acetylation rescues contextual fear conditioning in nNOS KO mice and accelerates extinction of cued fear conditioning in wild type mice. Neurobiol. Learn. Mem. 97, 409–417 (2012).
Li, W. & Wilson, D. A. Threat memory in the sensory cortex: Insights from Olfaction. Neuroscientist 3, 285–293 (2023).
Josselyn, S. A. & Tonegawa, S. Memory engrams: Recalling the past and imagining the future. Science 367, https://doi.org/10.1126/science.aaw4325 (2020).
Concina, G., Renna, A., Grosso, A. & Sacchetti, B. The auditory cortex and the emotional valence of sounds. Neurosci. Biobehav. Rev. 98, 256–264 (2019).
Sepahvand, T., Nazari, N., Qin, T., Rajani, V. & Yuan, Q. Olfactory threat extinction in the piriform cortex: An age-dependent employment of NMDA receptor-dependent long-term depression. Proc. Natl. Acad. Sci. USA 120, e2309986120 (2023).
Cho, J. H., Huang, B. S. & Gray, J. M. RNA sequencing from neural ensembles activated during fear conditioning in the mouse temporal association cortex. Sci. Rep. 6, 31753 (2016).
Headley, D. B., Kanta, V., Kyriazi, P. & Pare, D. Embracing complexity in defensive networks. Neuron 103, 189–201 (2019).
Yang, Y. et al. Selective synaptic remodeling of amygdalocortical connections associated with fear memory. Nat. Neurosci. 19, 1348–1355 (2016).
Tsukano, H. et al. Reciprocal connectivity between secondary auditory cortical field and amygdala in mice. Sci. Rep. 9, 19610 (2019).
Majak, K., Ronkko, S., Kemppainen, S. & Pitkanen, A. Projections from the amygdaloid complex to the piriform cortex: A PHA-L study in the rat. J. Comp. Neurol. 476, 414–428 (2004).
Romanski, L. M. & LeDoux, J. E. Information cascade from primary auditory cortex to the amygdala: corticocortical and corticoamygdaloid projections of temporal cortex in the rat. Cereb. Cortex 3, 515–532 (1993).
Sacco, T. & Sacchetti, B. Role of secondary sensory cortices in emotional memory storage and retrieval in rats. Science 329, 649–656 (2010).
Cambiaghi, M., Grosso, A., Renna, A. & Sacchetti, B. Differential recruitment of auditory cortices in the consolidation of recent auditory fearful memories. J. Neurosci. 36, 8586–8597 (2016).
Mouly, A. M. et al. PET Metabolic imaging of time-dependent reorganization of Olfactory cued fear memory networks in rats. Cereb. Cortex 32, 2717–2728 (2022).
You, Y., Novak, L. R., Clancy, K. J. & Li, W. Pattern differentiation and tuning shift in human sensory cortex underlie long-term threat memory. Curr. Biol. 32, 2067–2075.e2064 (2022).
East, B. S. et al. Basolateral amygdala to posterior piriform cortex connectivity ensures precision in learned odor threat. Sci. Rep. 11, 21746 (2021).
Hegoburu, C., Parrot, S., Ferreira, G. & Mouly, A. M. Differential involvement of amygdala and cortical NMDA receptors activation upon encoding in odor fear memory. Learn Mem. 21, 651–655 (2014).
Meissner-Bernard, C., Dembitskaya, Y., Venance, L. & Fleischmann, A. Encoding of Odor fear memories in the mouse Olfactory cortex. Curr. Biol. 29, 367–380.e364 (2019).
Liu, X. et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 484, 381–385 (2012).
Budinger, E. & Scheich, H. Anatomical connections suitable for the direct processing of neuronal information of different modalities the rodent primary auditory cortex. Hearing Res. 258, 16–27 (2009).
Budinger, E., Heil, P., Hess, A. & Scheich, H. Multisensory processing via early cortical stages: Connections of the primary auditory cortical field with other sensory systems. Neuroscience 143, 1065–1083 (2006).
Shakhawat, A. M. et al. Arc-expressing neuronal ensembles supporting pattern separation require adrenergic activity in anterior piriform cortex: An exploration of neural constraints on learning. J. Neurosci. 35, 14070–14075 (2015).
Duclot, F. & Kabbaj, M. The estrous cycle surpasses sex differences in regulating the transcriptome in the rat medial prefrontal cortex and reveals an underlying role of early growth response 1. Genome Biol. 16, 256 (2015).
Sharp, F. R., Sagar, S. M., Hicks, K., Lowenstein, D. & Hisanaga, K. c-fos mRNA, Fos, and Fos-related antigen induction by hypertonic saline and stress. J. Neurosci. 11, 2321–2331 (1991).
Tyssowski, K. M. et al. Different neuronal activity patterns induce different gene expression programs. Neuron 98, 530–546 (2018).
Hochgerner, H. et al. Neuronal types in the mouse amygdala and their transcriptional response to fear conditioning. Nat. Neurosci. 26, 2237–2249 (2023).
Haubensak, W. et al. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature 468, 270–276 (2010).
Kim, J., Zhang, X., Muralidhar, S., LeBlanc, S. A. & Tonegawa, S. Basolateral to central amygdala neural circuits for appetitive behaviors. Neuron 93, 1464–1479 (2017).
Pomrenze, M. B. et al. A transgenic rat for investigating the anatomy and function of corticotrophin releasing Factor Circuits. Front. Neurosci. 9, 487 (2015).
Busti, D. et al. Different fear states engage distinct networks within the intercalated cell clusters of the amygdala. J. Neurosci. 31, 5131–5144 (2011).
Diodato, A. et al. Molecular signatures of neural connectivity in the olfactory cortex. Nat. Commun. 7, 12238 (2016).
Zhang, L., Cao, J., Dong, L. & Lin, H. TiPARP forms nuclear condensates to degrade HIF-1alpha and suppress tumorigenesis. Proc. Natl. Acad. Sci. USA 117, 13447–13456 (2020).
Zhou, H. & Rigoutsos, I. The emerging roles of GPRC5A in diseases. Oncoscience 1, 765–776 (2014).
Ahmad, M. K., Abdollah, N. A., Shafie, N. H., Yusof, N. M. & Razak, S. R. A. Dual-specificity phosphatase 6 (DUSP6): a review of its molecular characteristics and clinical relevance in cancer. Cancer Biol. Med. 15, 14–28 (2018).
Kim, T. et al. Activated somatostatin interneurons orchestrate memory microcircuits. Neuron 112, 201–208.e204 (2024).
Yao, Z. et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241.e3226 (2021).
Phillips, R. A. et al. Temporally specific gene expression and chromatin remodeling programs regulate a conserved Pdyn enhancer. ELife 12, RP89993 (2023).
Han, J. H. et al. Neuronal competition and selection during memory formation. Science 316, 457–460 (2007).
Frankland, P. W. & Josselyn, S. A. Memory allocation. Neuropsychopharmacology 40, 243 (2015).
Park, S., Zhu, A., Cao, F. & Palmiter, R. D. Parabrachial Calca neurons mediate second-order conditioning. Nat. Commun. 15, 9721 (2024).
Barnes, D. C. & Wilson, D. A. Slow-wave sleep-imposed replay modulates both strength and precision of memory. J. Neurosci. 34, 5134–5142 (2014).
Chen, M. B., Jiang, X., Quake, S. R. & Sudhof, T. C. Persistent transcriptional programmes are associated with remote memory. Nature 587, 437–442 (2020).
Leitermann, R. J., Rostkowski, A. B. & Urban, J. H. Neuropeptide Y input to the rat basolateral amygdala complex and modulation by conditioned fear. J. Comp. Neurol. 524, 2418–2439 (2016).
Chowdhury, A. et al. A locus coeruleus-dorsal CA1 dopaminergic circuit modulates memory linking. Neuron 110, 3374–3388 (2022).
Gonzalez, M. C., Rossato, J. I., Radiske, A., Bevilaqua, L. R. M. & Cammarota, M. Dopamine controls whether new declarative information updates reactivated memories through reconsolidation. Proc. Natl. Acad. Sci. USA 118, e2025275118 (2021).
Johansen, J. P., Cain, C. K., Ostroff, L. E. & LeDoux, J. E. Molecular mechanisms of fear learning and memory. Cell 147, 509–524 (2011).
Devore, S., Lee, J. & Linster, C. Odor preferences shape discrimination learning in rats. Behav. Neurosci. 127, 498–504 (2013).
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. & Satija, R. Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Lause, J., Berens, P. & Kobak, D. Analytic Pearson residuals for normalization of single-cell RNA-seq UMI data. Genome Biol. 22, 258 (2021).
Stuart, T. et al. Comprehensive integration of single-cell Data. Cell 177, 1888–1902 (2019).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2018).
O’Leary, T. P. et al. Neuronal cell types, projections, and spatial organization of the central amygdala. iScience 25, 105497 (2022).
Jin, S. B., Maddern, X. J., Campbell, E. J. & Lawrence, A. J. Examining ventral subiculum and basolateral amygdala projections to the nucleus accumbens shell: Differential expression of VGLuT1, VGLuT2 and VGaT in the rat. Neurosci. Lett. 788, 136858 (2022).
Alcamo, E. A. et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron 57, 364–377 (2008).
Mendes, F. A. et al. Connective-tissue growth factor (CTGF/CCN2) induces astrogenesis and Fibronectin expression of embryonic neural cells. PloS ONE10, e0133689 (2015).
Phillips, R. A. et al. Distinct subpopulations of D1 medium spiny neurons exhibit unique transcriptional responsiveness to cocaine. Mol. Cell. Neurosci. 125, 103849 (2023).
Xu, Y. et al. ErbB4 in parvalbumin-positive interneurons mediates proactive interference in olfactory associative reversal learning. Neuropsychopharmacology 47, 1292–1303 (2022).
Xu, S. et al. Using clusterProfiler to characterize multiomics data. Nat. Protoc. 19, 3292–3320 (2024).
Jendoubi, T. & Strimmer, K. A whitening approach to probabilistic canonical correlation analysis for omics data integration. BMC Bioinform. 20, 15 (2019).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Turatsinze, J. V., Thomas-Chollier, M., Defrance, M. & van Helden, J. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat. Protoc. 3, 1578–1588 (2008).
Santana-Garcia, W. et al. RSAT 2022: regulatory sequence analysis tools. Nucleic Acids Res. 50, W670–W676 (2022).
Dyer, M., Lin, Q. X. X., Shapoval, S., Thieffry, D. & Benoukraf, T. MethMotif.Org 2024: a database integrating context-specific transcription factor-binding motifs with DNA methylation patterns. Nucleic Acids Res. 52, D222–D228 (2024).
Xuan Lin, Q. X. et al. MethMotif: an integrative cell specific database of transcription factor binding motifs coupled with DNA methylation profiles. Nucleic Acids Res. 47, D145–D154 (2019).
Acknowledgements
This work was supported by a Natural Sciences and Engineering Research Council of Canada discovery grant to Q.Y. (RGPIN-2018-04401), a Canadian Institutes of Health Research project grant to Q.Y. and T.B. (#PJT-169197), Memorial University Faculty of Medicine Dean’s support fund to Q.Y., and the Canada Research Chair program to T.B. (#CRC-2022-00285). We thank Riyana Afroze, Chelsea Crossley, Brandon Hannam, Tamunotonye Omoluabi, Sarah Torraville, Camila Reinhardt, Lara Blackburn, Usha Memoriya, Yodadge Amarasena, Juliana Alonso, Rehnuma Tanvee, Fatemeh Ghaedi, Donovan Francis, and Mina Ashbahi for their assistance in the project.
Author information
Authors and Affiliations
Contributions
Q.Y. conceived the project idea; Q.Y. and T.B. designed the research; T.S., T.Q., Z.H., K.D.P., S.J.C., N.N., A.M.J., A.S., and Y.X. conducted research; T.S., T.Q., S.J.C., N.N., A.M.J., Y.X., T.B., and Q.Y. analyzed data; T.S., T.Q., K.D.P., T.B., and Q.Y. wrote the manuscript. T.S., T.Q., K.D.P., S.J.C., T.B. and Q.Y. edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Henry L. Hallock and Richard D. Palmiter for their contribution to the peer review of this work. Primary Handling Editor: Benjamin Bessieres. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sepahvand, T., Qin, T., Hasan, Z. et al. Second-order threat conditioning in the amygdala-posterior piriform cortex network. Commun Biol 8, 846 (2025). https://doi.org/10.1038/s42003-025-08287-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08287-2