Abstract
Human trophoblast stem (TS) cells are informative in vitro models for the generation and testing of biologically meaningful hypotheses. The goal of this project was to derive patient-specific TS cell lines from clinically available chorionic villus biopsies. Cell outgrowths were captured from human chorionic villus tissue specimens cultured in modified human TS cell medium. Cell colonies emerged early during the culture and cell lines were established and passaged for several generations. Karyotypes of the newly established chorionic villus-derived trophoblast stem (TSCV) cell lines were determined and compared to initial genetic diagnoses from freshly isolated chorionic villi. Phenotypes of TSCV cells in the stem state and following differentiation were compared to cytotrophoblast-derived TS (TSCT) cells. TSCV and TSCT cells uniformly exhibited similarities in the stem state and following differentiation into syncytiotrophoblast and extravillous trophoblast cells. Chorionic villus tissue specimens provide a valuable source for TS cell derivation. They expand the genetic diversity of available TS cells and are associated with defined clinical outcomes. TSCV cell lines provide a new set of experimental tools for investigating trophoblast cell lineage development.
Similar content being viewed by others
Introduction
The placenta is a critical organ that allows the fetus to develop within the female reproductive tract1. Specialized functions attributed to the placenta are executed by trophoblast cells2,3,4. The trophoblast cell lineage arises from the initial differentiation event of the embryo5. In the human, trophoblast cells organize into villous and extravillous structures. A villous is comprised of trophoblast and non-trophoblast cell types and includes a self-renewing trophoblast cell population referred to as cytotrophoblast6,7. Cytotrophoblast are the progenitors for two differentiated cell populations: syncytiotrophoblast (STB) and extravillous trophoblast (EVT) cells2,3,4. STBs have a fundamental role in regulating nutrient and waste transfer between mother and fetus6, whereas EVT cells exit the placenta and transform the uterus into an environment supporting placental and fetal development2,3. Failures in placentation are the root cause of an assortment of disorders of pregnancy, including early pregnancy loss, preeclampsia, intrauterine growth restriction, and pre-term birth8,9. Regulatory mechanisms underlying human cytotrophoblast self-renewal and differentiation have largely remained elusive.
Recently, conditions for capturing and maintaining human trophoblast stem (TS) cells in vitro were described10. Human TS cells have the capacity for self-renewal and differentiation into STB or EVT cells. This in vitro model system has led to the generation of new insights into mechanisms regulating human trophoblast cell development11,12,13,14,15,16,17,18,19,20,21,22. Initial human TS cell lines were derived from blastocysts or first-trimester placental tissue obtained from pregnancy terminations. Establishment of culture conditions for human TS cells led to the derivation of TS cells from pluripotent stem cells23,24,25,26,27,28,29. These in vitro model systems have provided new insights regarding trophoblast cell development; however, it is unknown whether the origin of these TS cells was compatible with a healthy pregnancy outcome.
Chorionic villus sampling (CVS) represents a standard prenatal care procedure that is performed between 10 and 14 weeks of gestation30. Sampling involves the removal of a small amount of chorionic villus tissue for the purpose of genetic testing. Common indications for retrieving chorionic villus tissue include advanced maternal age, history of infertility, family history (e.g., sibling with genetic anomalies), or an abnormal noninvasive prenatal test result30,31. In addition to their use in genetic diagnosis, first-trimester chorionic villus tissue has become a robust platform for investigation of placental pathobiology31,32,33.
In this study, we derived patient-specific human TS cell lines from clinically available chorionic villus tissue. Modifications of the epigenome, which are required for the derivation of TS cells from pluripotent stem cells, are not required for the derivation of TS cells from chorionic villus tissue34. Furthermore, utilization of chorionic villus tissue as a source for deriving TS cell lines significantly expands the genetic diversity of available human TS cell lines and each chorionic villus-derived TS cell line is linked to clinical data describing pregnancy outcomes.
Results
Derivation of TS cells from chorionic villus biopsies
Chorionic villus biopsies were acquired with patient consent as part of standard medical care. Surplus tissue fragments not used for clinical genetic testing were placed in culture medium used for the expansion of human TS cells21 (Fig. 1A). Tissue pieces attached to type IV collagen-coated tissue culture-treated plates (Fig. 1B). Cell outgrowths were evident at sites of attachment and expanded over the first several weeks of culture (Fig. 1C). Cells and tissue fragments were passaged prior to reaching confluency and replated in 24 well plates. Cell colonies emerged after the first passage and steadily expanded with culture medium changes every 2 days. Colony morphology and growth rates were heterogeneous for the first few passages but became more homogenous after 5–6 passages. The morphology of chorionic villus-derived TS (TSCV) cells was consistent with the morphology of cytotrophoblast-derived TS cells (TSCT; Figs. 1B, 2A). TSCV cell line expansion was carried out slowly to reduce clonal pressure on derived cells. Cell lines were slowly transitioned into 6-well and 10 cm plate formats after passages 3–4 and 7–8, respectively (Fig. 1C). Newly established cell lines were cryopreserved beginning at passage 6. Importantly, TSCV cells tolerated cryopreservation. Revived cells survived, attached, and proliferated for further expansion. Overall, TSCV cell line derivation required approximately 3 months from sample acquisition to functional assessments of derived lines.
A Simplified schematic depicting the process of obtaining chorionic villi tissue fragments, derivation of TS cells, and then subsequent differentiation into STB and EVT cell lineages. Created with BioRender.com. B Chorionic villus tissue fragments attach and form cellular outgrowths within a few days of initial plating. Within 1–2 weeks, the outgrowths expand and proliferate across the well. Two to three weeks after plating, the cells were passaged, and colonies emerged. Colony clusters were initially small but proliferated and grew rapidly. Significant heterogeneity is present initially, but subsequent passaging selects for a TS cell population that displays a similar morphology to the original TS cell lines, which possess the ability to differentiate into STB and EVT cell lineages. Scale bars represent 250 μm in the first panel and cropped inset images. All other scale bars represent 500 μm. C An example timeline for TS cell line derivation and characterization.
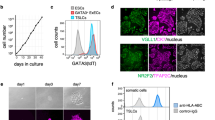
A Stem state phase contrast images of four chorionic villus-derived TS cell lines (TSCVK01, TSCVK09, TSCVK23, TSCVK24) alongside images of the reference cytotrophoblast-derived TS cell lines (TSCT27 and TSCT29) at different passage numbers (15–21). Scale bars represent 250 μm. B Bar graphs depicting expression of three microRNAs (miR) from the C19MC cluster (hsa-miR-517a-3p, has-miR-517-5p, and hsa-miR-526b-3p) in TSCT and TSCV cell lines relative to induced pluripotent stem (iPS) cells, measured by RT-qPCR. Data were normalized to the control miRNA, hsa-miR-103a-3p. Data are presented as the mean ± SEM. Each dot represents a biological replicate, n = 3 samples per group; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. C Plots representing DNA methylation levels in the ELF5 promoter at 11 sites in TSCT and TSCV cell lines compared to iPS cells. Methylated sites (black) and unmethylated sites (white) are shown for 10 replicates, and the average percent methylation is listed.
The success rate of TSCV line derivation was 42%, with ten TSCV lines (6 XY; 4 XX) successfully derived from 24 unique patient tissue specimens (15 XY; 9 XX). Success in cell line derivation may be impacted by the negative consequences of overnight shipping (Los Angeles to Kansas City) or the cellular contents of the tissue fragments but did not appear to be associated with the clinical karyotype of the CVS specimens. Characterization of four TSCV lines with normal karyotypes is presented in this report. Information about maternal age, karyotype, maternal and paternal ancestries, and gestational age at chorionic villus collection is presented in Supplementary Data 1.
Characterization of TSCV cells in the stem state
Karyotyping was repeated on TSCV cells following line derivation and expansion. Cell line karyotypes were largely consistent with the clinical karyotyping (Supplementary Data 1 and Supplementary Fig. 1). Karyotypes of TSCVK01 and TSCVK24 lines were normal and consistent with the clinical results. TSCVK09 and TSCVK23 cell lines exhibited mosaicism and were not consistent with the clinical results. A subset of cells karyotyped for TSCVK09 were 46, XY. The remaining cells analyzed displayed other genetic anomalies; however, each individual anomaly was restricted to 1–2 total cells. A subset of TSCVK23 cells was identified as mosaic for trisomy 20 (47, XX, +20) following cell line derivation (Supplementary Data 1 and Supplementary Fig. 1).
TSCT27 (XX) and TSCT29 (XY) served as reference standard human TS cell lines10 used for comparative characterization of the TSCV cell lines. TSCV cells were maintained in a stem/proliferative state and propagated beyond the Hayflick limit of 50 cell divisions for non-stem cells, which is consistent with TSCT cell proliferation10. TSCV cells in the stem state grew in discrete colonies and displayed a cobblestone morphology, consistent with the morphology of cytotrophoblast-derived cell lines, TSCT27 and TSCT29 (10; Fig. 2A). TSCV cells displayed additional characteristics consistent with their trophoblast cell identity35, including expression of microRNAs from the Chromosome 19 microRNA cluster (Fig. 2B; C19MC; hsa-miR-517a-3p, has-miR-517-5p, and hsa-miR-526b-3p) and hypomethylation of the E74 Like ETS Transcription Factor 5 (ELF5) promoter relative to induced pluripotent stem (iPS) cells (Fig. 2C). Additionally, cell proliferation rates were comparable between TSCT and TSCV cell lines (Supplementary Fig. 2A). All TS cell lines demonstrated the ability to survive and proliferate during single-cell cloning (Supplementary Fig. 2B). Overall, TSCV and TSCT cells cultured in the stem state displayed similar proliferative, morphologic, microRNA expression, and methylation properties.
Analysis of the differentiation capacity of TSCV cells
Comparisons of TSCV and TSCT cell capacities for differentiation into STB and EVT cell lineages were performed following cell line derivation (Fig. 1C). Assessments of cell differentiation were routinely performed following 10 passages. Differentiation was assessed at morphological and functional levels.
STB differentiation
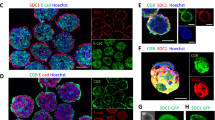
The ability of TSCV cells to differentiate into STB was assessed using previously described two-dimensional (ST2D) and three-dimensional (ST3D) protocols10. STB differentiation using ST3D conditions elicited significant morphological changes, including the formation of suspended spheroid cell clusters (Fig. 3A). Complementary to the morphological changes observed, TSCV-derived STB displayed downregulation of stem state transcripts TEA domain transcription factor 4 (TEAD4), LDL receptor related protein 2 (LRP2), and Lin-28 homolog A (LIN28A; Fig. 3B) and upregulation of STB lineage-specific transcripts, including cytochrome P450 Family 11 Subfamily 1 (CYP11A1), chorionic gonadotropin beta 7 (CGB7), and syndecan 1 (SDC1; Fig. 3C). STB differentiated from TSCV cells secreted chorionic gonadotropin (CG) at levels comparable to STB differentiated from TSCT cells as measured by ELISA (Fig. 3D). Additionally, E-cadherin (CDH1) immunostaining revealed comparable levels of syncytialization for TSCV- and TSCT-derived STB using the ST2D protocol (Supplementary Fig. 3). Overall, TSCV-derived STB had similar cell morphology, expression patterns of signature STB transcripts, CG production, and syncytialization that is observed in TSCT-derived STB.
A Representative phase contrast images of cytotrophoblast-derived TSCT27 and TSCT29 cells and four chorionic villus-derived TS cell lines possessing a normal karyotype, TSCVK01, TSCVK09, TSCVK23, and TSCVK24 cultured under STB differentiation conditions. Scale bars represent 250 μm. B, C Stem cell-associated transcripts (B; TEAD4, LRP2, and LIN28A) and STB cell-associated transcripts (C; CYP11A1, CGB7, and SDC1) were quantified by RT-qPCR in stem (dark gray) and STB differentiated (gray) TSCT27, TSCT29, TSCVK01, TSCVK09, TSCVK23, and TSCVK24 cells. D Chorionic gonadotropin (CG) protein levels (mlU/mg protein) were quantified by ELISA in cell culture supernatants collected from TSCT and TSCV cultured cells. Data are presented as the mean ± SEM. Each dot represents a biological replicate, n = 3 samples per group; **p < 0.01, ***p < 0.001, ****p < 0.0001.
EVT cell differentiation
Canonical features of EVT cell differentiation observed in TSCT cells were evident in TSCV cell lines with normal karyotypes (TSCVK01, TSCVK09, TSCVK23, and TSCVK24), including elongated cell morphology, cell migration (Fig. 4A; Supplementary Videos 1–3), and expression of major histocompatibility complex, class I, G (HLA-G) protein (Fig. 4B). EVT cells displayed downregulation of stem state transcripts TEAD4, LRP2, and LIN28A (Fig. 4C). Characteristic EVT transcripts were upregulated, including HLA-G, matrix metallopeptidase 2 (MMP2), and C-C motif chemokine receptor 1 (CCR1; Fig. 4D). Additionally, HLA-G and CCR1 protein levels were similar among TSCV and TSCT cell lines (Supplementary Fig. 4). Overall, these TSCV stem cell derived EVT cells were comparable to EVT cells derived from TSCT cells.
A Representative phase contrast images of cytotrophoblast-derived TSCT27 and TSCT29 cells and four chorionic villus-derived TS cell lines possessing a normal karyotype, TSCVK01, TSCVK09, TSCVK23, and TSCVK24 cells cultured under EVT cell differentiation conditions. Scale bars represent 250 μm. B Immunofluorescence detection of HLA-G (gray) by immunocytochemistry in TSCT and TSCV cells cultured in the stem state and on day 8 of EVT cell differentiation. DAPI (magenta) stains cell nuclei. Scale bars represent 100 μm. C, D Stem cell-associated transcripts (C; TEAD4, LRP2, and LIN28A) and EVT cell-associated transcripts (D; HLA-G, MMP2, and CCR1) were quantified by RT-qPCR in stem (dark gray) and EVT differentiated (gray) TSCT27, TSCT29, TSCVK01, TSCVK09, TSCVK23, and TSCVK24 cells. Data are presented as the mean ± SEM. Each dot represents a biological replicate, n = 3 samples per group; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Transcriptomic analysis of the developmental potential of TSCV cells
To obtain a broad comparative assessment of TSCV and TSCT in stem, STB, and EVT differentiated cell states, transcriptomes were captured using RNA-sequencing (RNA-seq).
STB differentiation from the stem state resulted in broad changes in gene expression in TSCT cells (TSCT27, TSCT29; Fig. 5A; Supplementary Data 2) and TSCV cells (TSCVK01, TSCVK09, TSCVK23, and TSCVK24; Fig. 5B; Supplementary Data 3), including downregulation of stem markers epithelial cell adhesion molecule (EPCAM), LIN28A, LRP2, paternally expressed 10 (PEG10), and TEAD4 and upregulation of STB markers chorionic gonadotropin beta 2 (CGB2), CGB7, CYP11A1, cytochrome P450 family 19 subfamily A member 1 (CYP19A1), and SDC1. STB differentiation-induced changes in gene expression were consistent between TSCT (TSCT27 and TSCT29) and TSCV (TSCVK01, TSCVK09, TSCVK23, and TSCVK24) cells (R = 0.87, p < 2.23-16; Fig. 5C).
A, B Volcano plots depicting significantly up- and downregulated genes based on transcripts measured by RNA-seq in STB versus stem cell states in TSCT (A) and TSCV (B) cells. Gene transcript levels unchanged between STB and stem state cells are depicted in gray (n = 3 per group; absolute Log2 fold change >1, adjusted p < 0.05). C Two-dimensional density plot comparing gene expression changes between STB and stem cell states in TSCT (TSCT27 and TSCT29) cells versus TSCV (TSCVK01, TSCVK09, TSCVK23, and TSCVK24) cells (Pearson correlation coefficient (R) = 0.87, p < 2.2e-16). D, E Volcano plots depicting significantly up- and downregulated genes based on transcripts measured by RNA-seq in EVT versus stem cell states in TSCT (D) and TSCV (E) cells. Gene transcript levels unchanged between EVT and stem cell states are depicted in gray (n = 3 per group; absolute Log2 fold change >1, adjusted p < 0.05). F Two-dimensional density plot comparing gene expression changes between EVT and stem cell states in TSCT (TSCT27 and TSCT29) cells versus TSCV (TSCVK01, TSCVK09, TSCVK23, and TSCVK24) cells. Pearson correlation coefficient (R) = 0.85, p < 2.2e-16. G Principal component analysis based on RNA-seq datasets generated from TSCT and TSCV cells cultured in the stem state or following differentiation into STB and EVT cell lineages. H Heat map showing scaled normalized read counts representing gene expression profiles of stem state, STB, and EVT differentiated cells across TSCT and TSCV cell lines.
EVT cells successfully differentiated from the stem state exhibited broad gene expression changes in TSCT (TSCT27, TSCT29; Fig. 5D; Supplementary Data 4) and TSCV cell lines with normal karyotypes (TSCVK01, TSCVK09, TSCVK23, and TSCVK24; Fig. 5E; Supplementary Data 5). These changes included the downregulation of stem markers EPCAM, LIN28A, LRP2, PEG10, and TEAD4 and upregulation of EVT cell markers CCR1, HLA-G, integrin subunit alpha1 (ITGA1), MMP2, and Notum, palmitoleoyl-protein carboxylesterase (NOTUM). Gene expression changes induced by EVT cell differentiation were consistent between TSCT (TSCT27 and TSCT29) and TSCV (TSCVK01, TSCVK09, TSCVK23, and TSCVK24) cells (R = 0.85, p < 2.23-16; Fig. 5F).
Principal component analysis of TSCT and TSCV cell lines identified three primary cell-state-specific clusters (Fig. 5G). TSCV cells displayed consistent clustering in the stem state and following STB and EVT cell lineage differentiation (Fig. 5G). Differential gene expression analysis of RNA-sequencing datasets from each TSCV cell line showed individual gene differences (Supplementary Data 6–13). Results from correlation analyses performed to compare cell expression profiles are indicative of comparable transcriptomic changes across TSCV and TSCT cell lines (Fig. 5H and Supplementary Fig. 5). Transcriptomic changes in TSCVK09 cells differentiated to STB and EVT showed the highest correlation to TSCT (TSCT27 and TSCT29) cells (STB R = 0.79; EVT R = 0.75; Supplementary Fig. 5B, F). TSCVK24 showed the lowest correlation in both STB and EVT differentiation states (STB R = 0.65; EVT R = 0.60; Supplementary Fig. 5D, H). Correlation values for TSCVK01 (STB R = 0.76; EVT R = 0.67; Supplementary Fig. 5A, E) and TSCVK23 (STB R = 0.72; EVT R = 0.67; Supplementary Fig. 5C, G) were in a similar range. Overall, these results indicate that TSCV cells are capable of self-renewal and effective differentiation into both STB and EVT cell lineages and can be considered Bonafide TS cells.
Discussion
Our understanding of placenta development and function has benefited from the availability of in vitro model systems. In the human, these model systems have included primary cell and explant cultures, choriocarcinoma-derived cell lines, and immortalized cell lines36,37. Each in vitro approach has had merits but also limitations3,35. Over two decades ago, Rossant and colleagues reported a procedure for culturing TS cells from the mouse38. These cells could be maintained in a proliferative stem state or induced to differentiate. Furthermore, TS cells could be reintroduced into blastocysts and shown to possess the capacity to contribute to mouse placentas38. Mouse TS cells became an effective model system to elucidate gene regulatory networks controlling trophoblast cell differentiation and placental development39,40,41,42. Efforts ensued to establish TS cells in other species with some success43,44, but human TS cells represented an enigma37,45. Culture protocols for sustaining mouse TS cells were ineffective in the human45. The discovery of culture conditions for propagating and differentiating human TS cells represented a major advancement10. Utilizing these human TS cell culture tools, we have demonstrated the feasibility of capturing and expanding authentic TS cells from human chorionic villus specimens. Unique to these newly derived stem cells is the ability to obtain clinical outcomes that can be used to study placental development, leading to healthy outcomes and disease states.
The initial human TS cell lines were derived from either blastocysts or cytotrophoblast from first-trimester pregnancy terminations10. These human TS cell lines represent the benchmark for all TS cell lines subsequently derived. Chorionic villus biopsies are an alternative tissue source for deriving TS cells. They are retrieved during the first trimester of pregnancy as part of standard medical care30,31,46,47,48,49,50,51. Thus, chorionic villus-derived TS cell lines can be connected to robust pregnancy outcome information. Human TS cell lines have also been derived from miscarriages18, term human placenta tissue52, and reprogrammed from pluripotent stem cells23,24,25,26,27,28,29,53,54,55. These alternative TS cell models are potentially useful tools for investigating trophoblast cell development, but each offers caveats for consideration. TS cells derived from trophoblast tissue obtained from miscarriages may best contribute to understanding trophoblast cell-related mechanisms linked to pregnancy failure and the impact of a failed pregnancy on TS cells. TS cells recovered from term placental tissue reflect the culmination of events transpiring throughout the duration of pregnancy, as epigenomic differences are observed in the placenta during gestation32,33. Pluripotency is established through extensive genomic reprogramming56,57, which minimizes the impact of the epigenetic landscape established during pregnancy on the TS cell phenotype. Additionally, unlike trophoblast tissue-derived TS cells, derivation of TS cells from pluripotent stem cells undermines genomic imprinting58,59, which is fundamental to development of the trophoblast lineage and placentation59,60,61. It is reasonable to assume that genetic background and source of trophoblast tissue for TS derivation will influence TS cell behavior. Culturing TS cells under optimized conditions may normalize some features attributed to an adverse pregnancy and maternal environment, whereas in other cases, the aberrant behavior may persist. Advantages of using chorionic villus-derived TS cells for investigating trophoblast cell-gene regulatory networks contributing to placental development are evident.
Chorionic villus-derived TS cells could be interrogated in the stem state and following differentiation into either STB or EVT cell lineages. The phenotypic and functional parameters evaluated revealed similarities between cytotrophoblast and chorionic villus-derived TS cells when cultured for stem state maintenance or following STB and EVT cell differentiation. Some differences in the capacity for EVT cell differentiation among chorionic villus-derived TS cells were noted. Variability in the capacity for human TS cell differentiation into EVT cells has been previously reported10,19,24,62. Numerous factors, such as unreported clinical characteristics, undetected genomic differences, or inherent sample variability, could be contributing to the differences observed. Thus, chorionic villus-derived TS cells represent a unique in vitro model to investigate functional variability in TS cells isolated from a temporally relevant tissue source.
TS cell lines were successfully derived from chorionic villus biopsies possessing both normal and abnormal karyotypes. TS cells with a triploid karyotype have also been established from human blastocysts63. Most recently, trophoblast organoids with abnormal karyotypes have been derived from chorionic villus biopsies64. The true impact of the chromosomal abnormalities on TS cells and their differentiation into STB or EVT cells will require successful cultivation and characterization of multiple cell lines possessing the same abnormal karyotype.
Mosaicism is a characteristic feature of the human placenta65,66,67. Trophoblast cells possess a tolerance for karyotypic abnormalities not evident in the embryo or fetus65,67,68. Each cotyledon of the placenta exhibits elements of trophoblast cell clonality65. Placental mosaicism is manifested in genetic and functional differences among cotyledons within a human placenta65,69,70,71. Among the TS cell lines derived from chorionic villus biopsies, some exhibited a karyotype consistent with the karyotype of chorionic villus tissue used for the clinical genetic analysis, whereas others differed. Chorionic villus biopsies contain a mixture of trophoblast and extraembryonic mesoderm6. Thus, differences in TS cells versus chorionic villus tissue could be attributed to confined placental mosaicism or alternatively, linked to an unappreciated consequence of culture conditions required to establish the TS cell lines.
In summary, the generation of chorionic villus biopsy-derived human TS cells expands the genetic diversity of existing TS cell models available for basic research and provides an opportunity to associate pregnancy outcomes with trophoblast cell biology. Importantly, this tissue source for capturing TS cells is available during early stages of human placental development, is part of standard medical care, is not associated with the political and ethical concerns that accompany deriving TS cells from human embryos or elective pregnancy terminations, and does not require modifications of the epigenome. Finally, investigating chorionic villus-derived TS cells offers the potential for obtaining insights into the significance of genetic anomalies and mosaicism in trophoblast cell development and introduces a novel precision medicine approach to the study of placentation.
Materials and methods
Chorionic villus tissue collections, karyotypic analysis, and clinical phenotyping
Chorionic villus tissue was obtained by highly experienced perinatologists as part of standard medical care between 10 and 14 weeks of gestation for clinical genetic diagnosis at Cedars-Sinai Medical Center31,69. Clinical cytogenetic analysis was performed on tissue specimens by direct and long-term culture and reviewed by a team of cytogeneticists69. Residual trophoblast tissue fragments not required for clinical cytogenetic analysis were recovered, suspended in Complete TS Cell Medium [DMEM/F12 (11320033, Thermo Fisher, Waltham, MA), 100 μM 2-mercaptoethanol, 0.2% (vol/vol) fetal bovine serum (FBS), 50 μM penicillin, 50 U/mL streptomycin, 0.3% bovine serum albumin (BSA, BP9704100, Thermo Fisher), 1% Insulin-Transferrin-Selenium-Ethanolamine (ITS-X) solution (vol/vol, 51500056, Thermo Fisher)], 8.5 μM L-ascorbic acid (A8960, Sigma-Aldrich, St. Louis, MO), 50 ng/mL epidermal growth factor (EGF, E9644, Sigma-Aldrich), 2 μM CHIR99021 (04-0004, Reprocell, Beltsville, MD), 0.5 μM A83-01 (04-0014, Reprocell), 1 μM SB431542 (04-0010, Reprocell), 800 μM valproic acid (P4543, Sigma-Aldrich), and 5 μM Y27632 (04-0012-02, Reprocell)10, shipped overnight to the University of Kansas Medical Center, and used for TS cell derivation. Demographic data were collected from patients and included parental ages, races and ethnicities, and ancestry (Supplementary Data 1).
Derivation of TS cells from chorionic villus tissue specimens
Chorionic villus biopsy tissue fragments were dissected and transferred to complete human TS cell medium. Briefly, individual villus fragments were minced and transferred to a 1.7 mL tube, washed with PBS, and centrifuged at 500×g for 3 min. Tissue pellets were resuspended in HBSS (with Ca2+ and Mg2+) supplemented with 1.25U/mL dispase II, 0.4 mg/mL collagenase IV and 80 U/mL DNase I. Samples were then agitated for 15 min at 37 °C. After incubation, tissue suspensions were centrifuged at 500×g for 3 min. and washed with basal TS cell medium. Finally, cells and tissue suspensions were centrifuged at 500×g for 3 min, resuspended in complete human TS cell medium and plated in 5 mg/mL Corning® mouse type IV collagen (35623, Discovery Labware Inc., Billerica, MA) coated dishes containing complete human TS cell medium. Cells and remaining tissue fragments attached within 2–5 days. Medium was replaced with fresh TS cell culture medium after initial attachment and every 2 days thereafter. Time to first passage was unique to each sample and determined by the extent of the outgrowth, but commonly occurred around 21 days post-plating. Cells and attached tissue fragments were washed with PBS and detached with TrypLE Express (12604021, Thermo Fisher). Cell and tissue fragments were replated in human TS cell culture conditions in a 24-well plate format. Colonies emerged after the first passage. Cells were maintained in a 24-well plate format for 3–5 passages and then expanded into a 6-well plate format.
TS cell culture
Following TS cell derivation, TS cells were cultured in dishes pre-coated with iMatrix511 (1:2000 dilution; NP892-01, Reprocell). TS cells were maintained in Modified Complete TS Cell Medium [DMEM/F12 (11320033, Thermo Fisher), 50 U/mL penicillin, 50 μg/mL streptomycin, 0.15% BSA (BP9704100, Thermo Fisher), 1% ITS-X solution (vol/vol; 51500056, Thermo Fisher)], 200 μM L-ascorbic acid (A8960, Sigma-Aldrich), 1% KnockOut Serum Replacement (KSR, 10828028, Thermo Fisher), 25 ng/mL EGF (E9644, Sigma-Aldrich), 2 μM CHIR99021 (04-0004, Reprocell), 5 μM A83-01 (04-0014, Reprocell), 800 μM valproic acid (P4543, Sigma-Aldrich), and 2.5 μM Y27632 (04-0012-02, Reprocell)21 medium was replaced every 2 days of culture. TSCT27 (XX) and TSCT29 (XY)10 were used as reference lines.
For single-cell cloning, cells were seeded at a density of no more than one cell/well in a 96-well plate with TS Cell Medium supplemented with 0.5 µg/mL iMatrix-511 (NP892-01, Reprocell) and 10 ng/mL BMP4 (314-BP, R&D Systems, Minneapolis, MN). Medium was replaced after 3 days, and colony inspection was performed on day 6 of culture.
STB differentiation
To induce STB cell differentiation, we utilized ST2D and ST3D protocols.
ST2D differentiation. TS cells were plated in a 6-well plate pre-coated with 2.5 μg/mL collagen IV at a density of 100,000 cells per well and cultured in ST2D Medium [DMEM/F12 (11320033, Thermo Fisher), 50 U/mL penicillin, 50 μg/mL streptomycin, 0.15% BSA (BP9704100, Thermo Fisher), 1% ITS-X solution (vol/vol; 51500056, Thermo Fisher)], 200 μM L-ascorbic acid (A8960, Sigma-Aldrich), 5% KSR (10828028, Thermo Fisher), 2.5 μM Y27632 (04-0012, Reprocell), 2 μM forskolin (F6886, Sigma-Aldrich)10. Medium was replaced on day 3, and the cells were analyzed on day 6 of STB cell differentiation.
ST3D differentiation. TS cells were plated into 6 cm petri dishes at a density of 300,000 cells per dish and cultured in ST3D Medium [DMEM/F12 (11320033, Thermo Fisher), 50 U/mL penicillin, 50 μg/mL streptomycin, 0.15% BSA (BP9704100, Thermo Fisher), 1% ITS-X solution (vol/vol; 51500056, Thermo Fisher)], 200 μM L-ascorbic acid (A8960, Sigma-Aldrich), 5% KSR (10828028, Thermo Fisher), 2.5 μM Y27632 (04-0012, Reprocell), 2 μM forskolin (F6886, Sigma-Aldrich), and 50 ng/mL of EGF (E9644, Sigma-Aldrich)10. On day 3 of cell differentiation, 3 mL of fresh ST3D medium was added to the culture dishes. Cells were analyzed on day 6 of STB cell differentiation.
EVT cell differentiation
EVT cell differentiation was induced by plating human TS cells onto 6-well plates pre-coated with 1 μg/mL of mouse type IV collagen at a density of 80,000 cells per well. Cells were cultured in EVT Differentiation Medium [DMEM/F12 (11320033, Thermo Fisher), 100 μm 2-mercaptoethanol, 50 U/mL penicillin, 50 μg/mL streptomycin, 0.3% bovine serum albumin (BP9704100, Thermo Fisher), 1% Insulin-Transferrin-Selenium-Ethanolamine solution (vol/vol; 51500056, Thermo Fisher)], 100 ng/mL of neuregulin 1 (NRG1, 5218SC, Cell Signaling, Danvers, MA), 7.5 μM A83-01 (04-0014, Reprocell), 2.5 μM Y27632 (04-0012, Reprocell), 4% KSR (10828028, Thermo Fisher), and 2% Matrigel® (CB-40234, Thermo Fisher). On day 3 of EVT cell differentiation, the medium was replaced with EVT Differentiation Medium, excluding NRG1 and with a reduced Matrigel® concentration of 0.5%. On day 6 of EVT cell differentiation, the medium was replaced with EVT Differentiation Medium with a Matrigel® concentration of 0.5% and excluding NRG1 and KSR. Cells were analyzed on day 8 of EVT cell differentiation.
Cell line karyotyping
Chromosome analysis of TSCV cells was performed using standard cytogenetic methods31,69. GTG banded chromosomes were analyzed at 450–550 band levels. Cytogenetic and fluorescence in situ hybridization results were described according to the current International Standing Committee on Human Cytogenetic Nomenclature (ISCN, 2009).
Immunocytochemical analysis
Cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) for 15 min at room temperature. Fixed cells were incubated with primary antibody against HLA-G (ab52455, Abcam) or E-cadherin (4A2, Cell Signaling Technology), followed by Alexa488-conjugated goat-anti-mouse immunoglobulin G (IgG; A32723, Thermo Fisher Scientific) secondary antibody and 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes, Eugene, OR). Fluorescence images were captured on a Nikon 80i upright microscope (Nikon) with a Photometrics CoolSNAP-ES monochrome camera (Roper Technologies, Inc., Sarasota, FL).
Western blotting
Cell lysates were prepared using RIPA buffer (sc-24948A, Santa Cruz Biotechnology, Dallas, TX). Proteins were separated via SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes (10600023, GE Healthcare, Chicago, IL). Membranes were blocked with 5% Nonfat dry milk in TBS containing 0.1% Tween-20 and then incubated with primary antibodies against HLA-G (1:5000, ab52455, Abcam), CCR1 (1:1000, PB9991, Boster Bio, Pleasanton, CA), and/or GAPDH (1:5000, AM4300, Thermo Fisher). The following HRP-conjugated antibodies were used: horse anti-mouse IgG (1:5000, 7076S, Cell Signaling Technology) and goat anti-rabbit IgG (1:5000, 7074S, Cell Signaling Technology). Immunoreactive proteins were visualized using chemiluminescence detection (Immobilon Crescendo, WBLUR0500, EMD Millipore).
iPS cell culture
Human iPS cells were propagated in tissue culture plates pre-coated with Matrigel® (1:100 dilution; 356231, Corning Life Sciences, Tewksbury, MA). iPS cells were maintained in complete iPS Cell Medium [mTeSR1 Basal Medium + mTeSR1 5X Supplement (85850, STEMCELL Technologies, Inc., Vancouver, CA) and 10 μM Y27632 (04-0012-02, Reprocell)] and incubated at 37 °C and 5% CO2. After the first day of culture, cells were cultured in complete iPS cell medium without Y27632. Medium was replaced every other day of culture. Cells were passaged or harvested at 80% confluency.
DNA methylation analysis
Genomic DNA was isolated using the DNeasy Blood and Tissue Kit (69504, Qiagen, Germantown, MD). Bisulfite convertion of 500 ng of DNA was performed using the EZ DNA Methylation-Gold Kit (D5005, Zymo Research, Irvine, CA) according to instructions. Following bisulfite conversion, the ELF5 promoter region was amplified using a nested PCR approach with previously reported primers (Primer Set A: forward: 5’-GGAAATGATGGATATTGAATTTGA-3’; reverse: 5’-CAATAAAAATAAAAACACCTATAACC-3’ Primer Set B: forward: 5’-GAGGTTTTAATATTGGGTTTATAATG-3’; reverse: 5’-ATAAATAACACCTACAAACAAATCC-3’; Supplementary Data 1535,54). PCR was performed with a hot start DNA polymerase, ZymoTaq (E2001, Zymo Research). After the second PCR, Taq polymerase-amplified PCR products were gel-purified with QIAquick Gel Extraction Kit (28706×4, Qiagen), using manufacturer protocols. The purified DNA was inserted directly into a plasmid vector using TOPO® TA Cloning® Kits for Sequencing (450030, Thermo Fisher) according to the manufacturer’s instructions. One microliter of purified PCR product was cloned into the plasmid vector (pCR™4-TOPO®) for 5 min at room temperature. Competent E. coli cells were transformed with the pCR4-TOPO construct, cultured, and minipreps were prepared using the QIAprep Spin Miniprep Kit (27106×4, Qiagen). Purified DNA was sequenced (GENEWIZ, South Plainfield, NJ).
CG enzyme-linked immunosorbent assay (ELISA)
Conditioned medium was collected following 6 days of STB culture for each cell line. CG levels were measured using an ELISA kit (HC251F, Calbiotech, El Cajon, CA), following the manufacturer’s protocol. The measurements were normalized to total cell protein content.
miRNA/mRNA isolation, cDNA preparation, and quantitative real-time PCR
For miRNA, total RNA was isolated using mirVana kit (AM1560, Thermo Fisher), and RNA concentration was measured with the Qubit™ RNA BR Assay Kit (Thermo Fisher). cDNA synthesis was performed with TaqMan® Advanced miRNA cDNA Synthesis kit (A28007, Thermo Fisher). RT-qPCR was performed using TaqMan™Fast Advanced Master Mix (4444556, Thermo Fisher) and targeted miRNAs MIR517a-3p, MIR517-5p, MIR-526b-3p, and housekeeping miRNA MIR103a-3p (479485_mir, 478996_mir, and 478253_mir; TaqMan™ Advanced miRNA Assays, Thermo Fisher; Supplementary Data 14). Relative expression of each transcript was calculated using the ΔΔCT method and normalized to hsa-miR-103a-3p.
For mRNA, total RNA was isolated using TRIzol®/chloroform precipitation (15596018, Thermo Fisher) as previously reported22. cDNA was synthesized from 1 μg of total RNA using the High-Capacity cDNA Reverse Transcription Kit (4368813, Thermo Fisher) and diluted 10 times with ultra-pure distilled water. qPCR was performed using PowerSYBR® Green PCR Master Mix (4367659, Thermo Fisher) and primers (250 nM each). RT-qPCR primer sequences are presented in Supplementary Data 16. Amplification and fluorescence detection were measured with a QuantStudio 5 Flex Real-Time PCR System (Thermo Fisher). An initial step (95 °C, 10 min) preceded 40 cycles of a two-step PCR (92 °C, 15 s; 60 °C, 1 min) and was followed by a dissociation step (95 °C, 15 s; 60 °C, 15 s; 95 °C 15 s). The comparative cycle threshold method was used for relative quantification of the amount of mRNA for each sample normalized to the housekeeping genes B2M or POLR2A.
RNA library preparation and RNA-Seq
Stranded mRNA-sequencing was performed on the Illumina NovaSeq 6000 Sequencing System in the Genomics Core at the University of Kansas Medical Center. Quality control was completed with the RNA Screen Tape Assay kit (5067-5576, Agilent Technologies, Santa Clara, CA) on the Agilent TapeStation 4200. Total RNA (1 μg) was processed in the following steps: (1) oligo dT bead capture of mRNA, (2) fragmentation, (3) reverse transcription, (4) cDNA end repair, (5) Unique Dual Index (UDI) adapter ligation, (6) strand selection, and (7) library amplification using the Universal Plus mRNA-Seq with NuQuant library preparation kit (0520-A01, Tecan Genomics, Männedorf, Switzerland). Library validation was performed with the D1000 Screen Tape Assay kit (5067-5582, Agilent Technologies) on the Agilent Tape Station 4200. Library concentrations were determined with the NuQuant module using a Qubit 4 Fluorometer (Thermo Fisher). Libraries were pooled based on equal molar amounts and the multiplexed pool was quantified, in triplicate, using the Roche Lightcycler96 with FastStart Essential DNA Green Master (06402712001, Roche, Indianapolis, IN) and KAPA Library Quant (Illumina, Inc., San Diego, CA) DNA Standards 1–6 (KK4903, KAPA Biosystems, Wilmington, MA). Using the qPCR results, the RNA-Seq library pool was adjusted to 2.125 nM for multiplexed sequencing. Pooled libraries were denatured with 0.2 N NaOH (0.04 N final concentration), neutralized with 400 mM Tris-HCl, pH 8.0, and diluted to 425 pM. Onboard clonal clustering of the patterned flow cell was performed using the NovaSeq 6000 S1 Reagent Kit (200 cycle, 20012864, Illumina). A 2 × 101 cycle sequencing profile with dual index reads was completed using the following sequence profile: Read 1 – 101 cycles x Index Read 1–8 cycles x Index Read 2–8 cycles x Read 2 – 101 cycles. Sequence data were converted from .bcl to FASTQ file format using bcl2fastq software and de-multiplexed. Raw FASTQ files were trimmed using default parameters (-r 0.1 -d 0.03) in Skewer (Version 0.2.2), and reads shorter than 18 bp were discarded. Transcripts were quantified using Kallisto (Version 0.46.2). Differentially expressed genes (FDR of 0.05) were discovered using the Bioconductor package DESeq2 in R (Version 1.32.0).
Live cell imaging
Cells were placed into an EVOS Onstage Incubator attached to an EVOS FL Automated Imaging System (Thermo Fisher). The live cell chamber was maintained at constant temperature (37 °C), humidity, and 5% CO2. For stem culture, TS cells were maintained in stem state culture conditions described above, and images were acquired 1–2 days after passage and immediately following culture medium change. EVT cell differentiation was induced as described above. On the fourth day of the EVT cell differentiation protocol, cells were placed into the live cell chamber. Phase contrast images were acquired every 10 min continuously from days 2–4 of stem cell growth or days 4–6 of EVT cell differentiation.
Statistical analysis
Statistical analysis was completed with the GraphPad Prism 9 software. Welch’s t tests, Brown-Forsythe and Welch ANOVA tests were applied when appropriate. The figures depict the data represented as mean ± standard deviation with a statistical significance level of p < 0.05.
Study approval
All human tissue specimens used for research purposes were collected following informed written consent, deidentified, and approved by institutional review boards at both Cedars-Sinai Medical Center and the University of Kansas Medical Center.
Data availability
All numerical source data and uncropped blots are provided in this manuscript (Supplementary Data 17 and Supplementary Fig. 6). All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/; GSE221617).
Materials availability
Materials will be made available upon reasonable request to the investigators.
Code availability
Only publicly available tools were used for data analysis and are described where relevant in the methods.
References
Amoroso, E. C. The evolution of viviparity. Proc. R. Soc. Med. 61, 1188–1200 (1968).
Knöfler, M. et al. Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell Mol. Life Sci. 76, 3479–3496 (2019).
Soares, M. J., Varberg, K. M. & Iqbal, K. Hemochorial placentation: development, function, and adaptations. Biol. Reprod. 99, 196–211 (2018).
Turco, M. Y. & Moffett, A. Development of the human placenta. Development 146, dev163428 (2019).
Rossant, J. & Tam, P. P. L. New insights into early human development: lessons for stem cell derivation and differentiation. Cell Stem Cell 20, 18–28 (2017).
Aplin, J. D. & Jones, C. J. P. Cell dynamics in human villous trophoblast. Hum. Reprod. Update 27, 904–922 (2021).
Sun, T. et al. Sexually dimorphic crosstalk at the maternal-fetal interface. J. Clin. Endocrinol. Metab. 105, e4831–e4847 (2020).
Brosens, I., Puttemans, P. & Benagiano, G. Placental bed research: I. The placental bed: from spiral arteries remodeling to the great obstetrical syndromes. Am. J. Obstet. Gynecol. 221, 437–456 (2019).
Burton, G. J., Fowden, A. L. & Thornburg, K. L. Placental origins of chronic disease. Physiol. Rev. 96, 1509–1565 (2016).
Okae, H. et al. Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50–63.e6 (2018).
Bhattacharya, B. et al. Atypical protein kinase C iota (PKCλ/ι) ensures mammalian development by establishing the maternal-fetal exchange interface. Proc. Natl. Acad. Sci. USA 117, 14280–14291 (2020).
Hornbachner, R. et al. MSX2 safeguards syncytiotrophoblast fate of human trophoblast stem cells. Proc. Natl. Acad. Sci. USA 118, e2105130118 (2021).
Ishiuchi, T. et al. Zfp281 shapes the transcriptome of trophoblast stem cells and is essential for placental development. Cell Rep. 27, 1742–1754.e6 (2019).
Jaju Bhattad, G. et al. Histone deacetylase 1 and 2 drive differentiation and fusion of progenitor cells in human placental trophoblasts. Cell Death Dis. 11, 311 (2020).
Muto, M. et al. Intersection of regulatory pathways controlling hemostasis and hemochorial placentation. Proc. Natl. Acad. Sci. USA 118, e2111267118 (2021).
Perez-Garcia, V. et al. BAP1/ASXL complex modulation regulates epithelial-mesenchymal transition during trophoblast differentiation and invasion. Elife 10, e63254 (2021).
Ruane, P. T. et al. Trophectoderm differentiation to invasive syncytiotrophoblast is promoted by endometrial epithelial cells during human embryo implantation. Hum. Reprod. 37, 777–792 (2022).
Saha, B. et al. TEAD4 ensures postimplantation development by promoting trophoblast self-renewal: an implication in early human pregnancy loss. Proc. Natl. Acad. Sci. USA 117, 17864–17875 (2020).
Shannon, M. J. et al. Cell trajectory modeling identifies a primitive trophoblast state defined by BCAM enrichment. Development 149, dev199840 (2022).
Sheridan, M. A. et al. Characterization of primary models of human trophoblast. Development 148, dev199749 (2021).
Takahashi, S. et al. Loss of p57KIP2 expression confers resistance to contact inhibition in human androgenetic trophoblast stem cells. Proc. Natl. Acad. Sci. USA 116, 26606–26613 (2019).
Varberg, K. M. et al. ASCL2 reciprocally controls key trophoblast lineage decisions during hemochorial placenta development. Proc. Natl. Acad. Sci. USA 118, e2016517118 (2021).
Castel, G. et al. Induction of human trophoblast stem cells from somatic cells and pluripotent stem cells. Cell Rep. 33, 108419 (2020).
Cinkornpumin, J. K. et al. Naive human embryonic stem cells can give rise to cells with a trophoblast-like transcriptome and methylome. Stem Cell Rep. 15, 198–213 (2020).
Dong, C. et al. Derivation of trophoblast stem cells from naïve human pluripotent stem cells. Elife 9, e52504 (2020).
Guo, J. et al. Single-cell analysis of the developing human testis reveals somatic niche cell specification and fetal germline stem cell establishment. Cell Stem Cell 28, 764–778.e4 (2021).
Liu, X. et al. Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature 586, 101–107 (2020).
Wei, Y. et al. Efficient derivation of human trophoblast stem cells from primed pluripotent stem cells. Sci. Adv. 7, eabf4416 (2021).
Yanagida, A. et al. Naive stem cell blastocyst model captures human embryo lineage segregation. Cell Stem Cell 28, 1016–1022.e4 (2021).
Stranc, L. C., Evans, J. A. & Hamerton, J. L. Chorionic villus sampling and amniocentesis for prenatal diagnosis. Lancet 349, 711–714 (1997).
Pisarska, M. D. et al. Optimization of techniques for multiple platform testing in small, precious samples such as human chorionic villus sampling. Prenat. Diagn. 36, 1061–1070 (2016).
Flowers, A. E. et al. Sex differences in microRNA expression in first and third trimester human placenta. Biol. Reprod. 106, 551–567 (2022).
Gonzalez, T. L. et al. High-throughput miRNA sequencing of the human placenta: expression throughout gestation. Epigenomics 13, 995–1012 (2021).
Yan, J. & Huangfu, D. Epigenome rewiring in human pluripotent stem cells. Trends Cell Biol. 32, 259–271 (2022).
Lee, C. Q. E. et al. What is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Rep. 6, 257–272 (2016).
Ringler, G. E. & Strauss, J. F. In vitro systems for the study of human placental endocrine function. Endocr. Rev. 11, 105–123 (1990).
Shibata, S. et al. Unique features and emerging in vitro models of human placental development. Reprod. Med. Biol. 19, 301–313 (2020).
Tanaka, S., Kunath, T., Hadjantonakis, A. K., Nagy, A. & Rossant, J. Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075 (1998).
Hada, M. et al. Highly rigid H3.1/H3.2-H3K9me3 domains set a barrier for cell fate reprogramming in trophoblast stem cells. Genes Dev. 36, 84–102 (2022).
Hemberger, M., Hanna, C. W. & Dean, W. Mechanisms of early placental development in mouse and humans. Nat. Rev. Genet. 21, 27–43 (2020).
Latos, P. A. & Hemberger, M. From the stem of the placental tree: trophoblast stem cells and their progeny. Development 143, 3650–3660 (2016).
Lee, B.-K. et al. Super-enhancer-guided mapping of regulatory networks controlling mouse trophoblast stem cells. Nat. Commun. 10, 4749 (2019).
Asanoma, K. et al. FGF4-dependent stem cells derived from rat blastocysts differentiate along the trophoblast lineage. Dev. Biol. 351, 110–119 (2011).
Grigor’eva, E. V. et al. FGF4 independent derivation of trophoblast stem cells from the common vole. PLoS ONE 4, e7161 (2009).
Kunath, T. et al. Developmental differences in the expression of FGF receptors between human and mouse embryos. Placenta 35, 1079–1088 (2014).
Adusumalli, J., Han, C. S., Beckham, S., Bartholomew, M. L. & Williams, J. Chorionic villus sampling and risk for hypertensive disorders of pregnancy. Am. J. Obstet. Gynecol. 196, 591.e1–7 (2007).
Dong, L., Falk, R. E., Williams, J., Kohan, M. & Schreck, R. R. Tetrasomy 12p–unusual presentation in CVS. Prenat. Diagn. 23, 101–103 (2003).
McIntosh, N., Rubin, C., Wang, B. & Williams, J. Transcervical CVS sample size: correlation with placental location, cytogenetic findings, and pregnancy outcome. Prenat. Diagn. 13, 1031–1036 (1993).
Wang, B. T. et al. Chorionic villi sampling: laboratory experience with 4,000 consecutive cases. Am. J. Med. Genet. 53, 307–316 (1994).
Williams, J., Medearis, A. L., Chu, W. H., Kovacs, G. D. & Kaback, M. M. Maternal cell contamination in cultured chorionic villi: comparison of chromosome Q-polymorphisms derived from villi, fetal skin, and maternal lymphocytes. Prenat. Diagn. 7, 315–322 (1987).
Williams, J., Wang, B. B., Rubin, C. H. & Aiken-Hunting, D. Chorionic villus sampling: experience with 3016 cases performed by a single operator. Obstet. Gynecol. 80, 1023–1029 (1992).
Wang, L.-J. et al. Functional antagonism between ΔNp63α and GCM1 regulates human trophoblast stemness and differentiation. Nat. Commun. 13, 1626 (2022).
Jang, Y. J., Kim, M., Lee, B.-K. & Kim, J. Induction of human trophoblast stem-like cells from primed pluripotent stem cells. Proc. Natl. Acad. Sci. USA 119, e2115709119 (2022).
Soncin, F. et al. Derivation of functional trophoblast stem cells from primed human pluripotent stem cells. Stem Cell Rep. 17, 1303–1317 (2022).
Viukov, S. et al. Human primed and naïve PSCs are both able to differentiate into trophoblast stem cells. Stem Cell Rep. 17, 2484–2500 (2022).
Hanna, J. H., Saha, K. & Jaenisch, R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell 143, 508–525 (2010).
Papp, B. & Plath, K. Epigenetics of reprogramming to induced pluripotency. Cell 152, 1324–1343 (2013).
Fischer, L. A. et al. Tracking and mitigating imprint erasure during induction of naive human pluripotency at single-cell resolution. Stem Cell Rep. 20, 102419 (2025).
Kobayashi, E. H. et al. Genomic imprinting in human placentation. Reprod. Med. Biol. 21, e12490 (2022).
Monk, D. Genomic imprinting in the human placenta. Am. J. Obstet. Gynecol. 213, S152–S162 (2015).
Hanna, C. W. Placental imprinting: emerging mechanisms and functions. PLoS Genet. 16, e1008709 (2020).
Haider, S. et al. Transforming growth factor-β signaling governs the differentiation program of extravillous trophoblasts in the developing human placenta. Proc. Natl. Acad. Sci. USA 119, e2120667119 (2022).
Kong, X., Chen, X., Ou, S., Wang, W. & Li, R. Derivation of human triploid trophoblast stem cells. J. Assist. Reprod. Genet. 39, 1183–1193 (2022).
Schäffers, O. J. M. et al. Single-cell atlas of patient-derived trophoblast organoids in ongoing pregnancies. Organoids 1, 106–115 (2022).
Coorens, T. H. H. et al. Inherent mosaicism and extensive mutation of human placentas. Nature 592, 80–85 (2021).
Robinson, W. P. & Del Gobbo, G. F. Mistakes are common; should we worry about them? Trends Mol. Med. 27, 721–722 (2021).
Yuen, R. K. C. & Robinson, W. P. A high capacity of the human placenta for genetic and epigenetic variation: implications for assessing pregnancy outcome. Placenta 32, S136–S141 (2011).
Shahbazi, M. N. et al. Developmental potential of aneuploid human embryos cultured beyond implantation. Nat. Commun. 11, 3987 (2020).
Huang, A. et al. Prevalence of chromosomal mosaicism in pregnancies from couples with infertility. Fertil. Steril. 91, 2355–2360 (2009).
Rubin, C. H., Williams, J. & Wang, B. B. Discrepancy in mosaic findings between chorionic villi and amniocytes: a diagnostic dilemma involving 45,X, 46,XY, and 47,XYY cell lines. Am. J. Med. Genet. 46, 457–459 (1993).
Wang, B. B., Rubin, C. H. & Williams, J. Mosaicism in chorionic villus sampling: an analysis of incidence and chromosomes involved in 2612 consecutive cases. Prenat. Diagn. 13, 179–190 (1993).
Acknowledgements
We thank Stacy Oxley and Brandi Miller for their administrative assistance and Clark Bloomer and Rosanne Skinner in the KUMC Genome Sequencing Facility for their work in library preparation and DNA sequencing. The research was supported by a National Institutes of Health (NIH) National Research Service Award postdoctoral fellowship (F32HD096809) to K.M.V. and pathway to independence awards (K99HD107262) to K.M.V. and (K99HD115834) to A.M, NIH grants (M.D.P.: AI154535; M.J.S.: HD020676, HD105734, HD112559), and the Sosland Foundation.
Author information
Authors and Affiliations
Contributions
K.M.V., A.M., M.D.P., and M.J.S. conceived and designed the research; A.N., J.W., and M.D.P. collected and obtained tissue specimens. J.G., I.S., H.O., and T.A. provided reagents, protocols, or equipment; K.M.V., A.M., B.M., and K.I. performed experiments and/or analyzed data; K.M.V., A.M., J.G., I.S., M.D.P., and M.J.S. interpreted results of experiments; K.M.V., A.M., J.M.V., and M.J.S. prepared figures and drafted the manuscript; K.M.V., A.M., M.D.P., and M.J.S. edited and revised the manuscript. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary handling editor: Dario Ummarino.
Additional information
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Varberg, K.M., Moreno-Irusta, A., Novoa, A. et al. Leveraging chorionic villus biopsies for the derivation of patient-specific trophoblast stem cells. Commun Biol 8, 964 (2025). https://doi.org/10.1038/s42003-025-08393-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08393-1