Abstract
The molecular and functional changes in endothelial cells during disease progression such as cancer have been noted but the mechanism of their activation is still under-studied. Previously we discovered that tumor-derived Oncostatin M induced tumor-associated vascular phenotypes, and the activated endothelial cells in turn promoted tumor progression and metastasis of clear-cell renal cell carcinoma (ccRCC). However, the mechanism of Oncostatin M action remains unknown. Here, we reveal that Oncostatin M signaling triggers specific epigenetic reprogramming of endothelial cells through upregulation of lysine acetyltransferase 6B, leading to increased histone 3 lysine 14 acetylation (H3K14ac) in vitro and in vivo. H3K14ac-modified chromatins upregulate specific gene sets associated with hypoxic response, hyper-angiogenesis, inflammation, and mesenchymal transition. Targeting H3K14ac in endothelial cells by interfering with acetyltransferase 6B function or neutralizing Oncostatin M ameliorates the premalignant hyperplastic phenotypes in the autochthonous ccRCC mouse model and diminishes tumor growth and metastasis in the ccRCC xenograft model.
Similar content being viewed by others
Introduction
Tumor cells can profoundly influence the function and activity of surrounding stromal cells by altering their gene expression patterns, thereby creating a pro-tumorigenic microenvironment1,2. Although stromal cells exhibit few or no oncogenic mutations in their genome, they often display distinct gene expression profiles compared to their counterparts in normal tissues. This phenomenon is typically attributed to transient gene regulation at the transcriptional and post-transcriptional levels affected by growth factors and cytokine-induced signal transduction. Canonical signal transduction is usually transient, and the effective duration of a given signal is limited by the half-life of the ligand-receptor complex, the size of the genes (i.e., the time needed for transcription), and the half-life of mRNA or target protein. However, recent evidence has indicated that reprogramming of gene expression in the tumor microenvironment can also be induced by epigenetic modulation, which results in long-term changes in specific stromal cell populations3. Nonetheless, the mechanism that regulates epigenetic modulation in the stromal cells, and the extent of direct involvement of the tumor cells in such modulation, remains unclear.
Clear-cell renal cell carcinoma (ccRCC) is the most prevalent form of kidney cancer, the instances and death rates of which are projected to continue growing annually4. The initiation of ccRCC is closely associated with inactivation of the VHL tumor suppressor gene, which results in increased activity of hypoxia-inducible factors (HIFs), leading to pseudohypoxic responses5,6 such as heightened Warburg effect, tissue inflammation, and a disorganized vascular network that is tortuous, leaky, and increased in density7,8. Thus, the tumor microenvironment reconstituted by the VHL-deficient kidney tubule epithelial cells plays a critical role in the progression of ccRCC9.
Interestingly, cell type enrichment analysis indicates that endothelial cells (EC) is the most enriched cell type in ccRCC, and ccRCC is the only solid tumor type that is specifically enriched in the EC population among all other stromal cell types10, suggesting an indispensable and unique role of ECs in the initiation and progression of ccRCC. However, our understanding of the pro-tumorigenic role of ECs has largely been limited to receptor tyrosine kinase (RTK)-induced hyper-angiogenesis that supplies oxygen and nutrients to the growing tumor11. This may explain why anti-angiogenesis therapeutics based on RTK inhibitors often fail because of nearly inevitable development of drug resistance12,13. There is therefore an urgent need to gain a deeper understanding of the molecular and cellular mechanisms driving activation of ccRCC-associated ECs. Previously, we have generated conditional mouse Vhlh (the mouse VHL locus)14,15 knockout (KO) in a subpopulation of kidney epithelial cells by using Hoxb7 promoter-driven Cre (Hoxb7-Cre-GFP; VhlhloxP/loxP; henceforth VhlhKO)16. This mouse model recapitulated phenotypes of premalignant ccRCC, including immune cell infiltration, fibrosis, hyperplasia in cortical and medullary tubule segments, pseudohypoxic responses such as metabolic switch and hyper-angiogenesis, and appearance of clear-cell clusters. This mouse strain, therefore, represents an autochthonous early-stage ccRCC model16,17,18.
Using this ccRCC model, we previously discovered that Oncostatin M (OSM) secreted by the VHL-deficient kidney tubule cells could induce extensive transcriptomic changes in ECs, although these ECs did not harbor any exogenously induced genomic mutations. Attenuation of the Osm pathway by inactivation of the Osm receptor β (Osmrβ) gene in the Vhlh mutant background could rescue the premalignant ccRCC phenotypes, including reduced Ki67+ proliferating cells, decreased immune cell infiltration, attenuated angiogenesis, and diminished fibrosis18. In the current study, we analyze the oncogenic action of OSM and uncover an interesting interaction mechanism between tumor cells and ECs, in which OSM emanated from the tumor cells promotes epigenetic modification in ECs by inducing the activity of lysine acetyltransferase KAT6B. We used in vitro and in vivo experimental models to explore the underlying mechanism of epigenetic modification in ECs and tested the potential of targeting epigenetic changes in ECs for ccRCC treatment.
Results
OSM induces gene expression changes in ECs consistent with ccRCC-associated vascular phenotypes
It has been reported that ccRCC-associated ECs are activated by OSM derived from the VHL-deficient tumor cells18. The activated ECs exhibit angiogenic and endothelial-to-mesenchymal transition (EndoMT) phenotypes, and can promote immune cell infiltration and tumor metastasis. To gain deeper insights into the underlying mechanism of OSM-induced EC activation, we treated HMEC-1, a human microvasculature EC line, with recombinant human OSM (hOSM) in vitro. Total mRNAs from the hOSM-treated ECs and the untreated counterparts (CT) were then analyzed by next-generation mRNA sequencing (mRNA-Seq) (Fig. 1a). The complete RNA counts from the comparative transcriptomes are shown in Supplementary Data 1. Specific OSM-induced genes include known OSM responsive targets such as IL-6, OSM receptor (OSMR), and several genes typical of ccRCC-associated EC activation including CHI3L1, HIF-1α, ICAM-1, and SNAIL1 identified in VhlhKO mouse model18 (Fig. 1b). The transcriptome of hOSM-stimulated ECs promote biological processes that have been shown to be the molecular signature of VhlhKO-associated ECs18, including JAK-STAT signaling (the OSM signaling pathway), glycolysis and hypoxia (the HIF-1α pathway), inflammatory response, angiogenesis, and mesenchymal transition (Fig. 1c). Therefore the molecular signature of ECs stimulated by OSM in vitro recapitulates the underlying pathological phenotypes of VhlhKO-associated ECs in vivo.
a Left: HMEC-1 cells, a human microvascular EC cell line, were treated with recombinant human OSM (hOSM) at 10 ng/ml for 48 h. Total mRNAs of OSM-treated HMEC-1 and the untreated counterpart (CT) were harvested and analyzed by mRNA sequencing. Right: Heatmap of normalized gene-expression levels (TPM, transcripts per million; Supplementary Data 1) shows distinct gene expression patterns between untreated control (CT) and OSM-treated samples. Each column represents one individual sample. b Representative genes upregulated in OSM-treated ECs also identified previously as OSM-responsive genes in VhlhKO kidney18. c Biological process enrichment analysis shows enriched pathways in the OSM-treated EC transcriptome that underlie the phenotypes of ECs observed in vivo. NES: normalized enrichment score. d Gene set enrichment analysis (GSEA) shows Histone modification as the most significantly induced. e GSEA identifies “Histone modification” and “Histone 3 acetylation” as significantly enriched in OSM-treated vs CT groups. f Western blotting of different acetylation sites of histone 3 (H3K14ac, H3K9ac, H3K18ac, and H3K27ac) of OSM-treated ECs and their untreated counterparts was performed to validate the acetylation of histone H3 shown in (e). Statistical testing was performed by a two-tailed paired Student’s t-test. ns no significance; **P ≤ 0.01. Error bars show the standard deviation. Only acetylation at K14 was significantly induced by OSM. Uncropped blots are shown in Fig. S1. The source data points for quantification are provided in Supplementary Data 2.
Interestingly, gene set enrichment analysis (GSEA) identifies enrichment of histone modification, specifically histone 3 acetylation, in OSM-stimulated ECs (Fig. 1d, e). Remarkably, among the four common histone 3 acetylation sites examined, H3K14ac is found to be uniquely induced by OSM (Fig. 1f and Fig. S1). The result suggests an important role of H3K14ac in the activation of ECs induced by OSM.
H3K14ac is robustly induced in ECs of ccRCC and Vhlh KO tissues
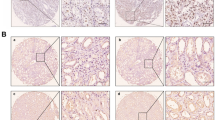
H3K14ac is not a “general-purpose” histone modification that regulates a wide range of genes throughout the genome. Instead, H3K14ac plays specific and important roles in embryonic development19,20, inflammation21, and adult stem cell maintenance22,23. As shown in Fig. 2a, H3K14ac is prominently expressed in ECs (CD31+ cells) of the peritumor (Peri) and the ccRCC tissues in human ccRCC biopsies, but not in normal adjacent tissue (NAT). In the mouse model (Fig. 2b), the expression of H3K14ac in ECs (CD34+) is strongly induced in VhlhKO tissue compared with WT or Osm receptor β (Osmrβ) KO tissue (Osmr−/−). Double KO of Osmrβ in VhlhKO tissue (Hoxb7-Cre-GFP; VhlhloxP/loxP; Osmrβ−/−; DBKO) reduces the expression of H3K14ac to the basal level. Note that in order to identify ECs, anti-CD34 and anti-CD31 antibodies were used for detecting mouse and human ECs, respectively. This is because of the limited availability of host-specific primary-secondary antibodies pair for double staining. Both markers have been widely used to identify ECs in tissues rich in vasculature24. The intensity of H3K14ac in the above genetic backgrounds are validated by Western blotting of whole-kidney tissue extracts (Fig. 2c and Fig. S2), confirming that H3K14ac is induced by OSM signaling.
Anti-CD34 and anti-CD31 antibodies were used for detecting mouse and human ECs, respectively, based on the available host-specific primary-secondary antibody pairs for double staining. a Double staining of H3K14ac (green) and CD31 (red) in human ccRCC tissues. Colocalization of H3K14ac and CD31 can be found in peritumor (Peri) and ccRCC tissues but not in normal adjacent tissue (NAT). Quantification of H3K14ac intensity in ECs (CD31+) is shown in the right panels. Each data point represents one clinical sample. NAT (n = 23), Peri (n = 9), ccRCC (n = 20). Scale bars are 20 μm. b Double staining of H3K14ac (green) and CD34 (red) in WT, Osmrβ KO (Osmr-/-), VhlhKO, and Hoxb7-Cre-GFP;VhlhloxP/loxP; Osmrβ−/− (DBKO) kidney tissues. The signal of H3K14ac is abundant and shows colocalization with CD34+ cells in VhlhKO tissue but not in WT, Osmr−/−, or DBKO. The quantification of H3K14ac intensity in ECs (CD34+) is shown in the right panels. Each data point represents one animal. n = 11. Scale bars are 20 μm. H3K14ac intensity was quantified by CellProfiler (version 4.2.5) per the supplier’s instructions. Statistical testing was performed by one-way ANOVA with Tukey’s post hoc test. ***P ≤ 0.001. Error bars show the standard deviation. c Western blotting for H3K14ac of whole-kidney tissue from WT, Osmrβ KO (Osmr−/−), VhlhKO, and DBKO mice. Strong expression of H3K14ac is observed in VhlhKO kidney tissue but not in WT or Osmr−/−. Statistical testing was performed by a two-tailed paired Student’s t-test. ***P ≤ 0.001. Error bars show the standard deviation. Knocking out Osmr in VhlhKO kidney tissue (DBKO) reversed the expression of H3K14ac to the basal level. The uncropped blots are shown in Fig. S2. The source data points for quantification are provided in Supplementary Data 2.
These results suggest that OSM-induced EC phenotypes in ccRCC and VhlhKO may be mediated via epigenetic changes of H3K14ac. We next explore this novel possibility.
Histone acetylation underlies OSM-induced EC activation
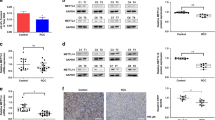
Histone acetylation is a dynamic process that is tightly regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs)25. Thus, we first examined the expression of HAT and HDAC genes in hOSM-modified EC (HMEC-1) transcriptome. Interestingly, among the cohort of 18 commonly known HAT/HDAC genes, 7/9 upregulated are HAT genes, while 8/9 downregulated are HDAC genes (Fig. 3a). Furthermore, OSM does not disrupt the balance of histone or DNA methylation/demethylation genes; that is, both methyl transferase and demethylase gene sets are upregulated by OSM treatment (Fig. S3). Therefore, OSM stimulation appears to exert a net effect of increased histone acetylation.
a Heatmap shows the expression of histone acetylation/deacetylation enzyme genes found in hOSM-treated (OSM) and untreated (CT) HMEC-1 transcriptomes. The order is arranged according to Log2fold change (FC). 7/9 upregulated genes encode acetyltransferases (in red), while 8/9 downregulated genes encode deacetylases (in blue). Each column represents one individual sample. b The volcano plot based on -Log10(P value) and Log2(FC) of HAT and HDAC genes shown in (a). Top three significantly upregulated genes are indicated. c Western blotting of selected HAT and HDAC genes to validate the mRNA results in (a). Uncropped blots are shown in Fig. S4. Statistical testing was performed by a two-tailed paired Student’s t-test. ns no significance; **P ≤ 0.01; *P < 0.05. Error bars show the standard deviation. d Double staining for KAT6B (green) and CD31 (red) in human ccRCC tissue. Colocalization of KAT6B and CD31 is found in peritumor (Peri) and ccRCC tissues but not in normal adjacent tissue (NAT). Quantification of KAT6B intensity is shown on the right. NAT (n = 17), Peri (n = 9), ccRCC (n = 14). Statistical testing was performed by one-way ANOVA with Tukey’s post hoc test. ***P ≤ 0.001. Error bars show the standard deviation. Scale bars are 20 μm. KAT6B intensity was quantified by CellProfiler (version 4.2.5) per the supplier’s instructions. e Kidney tissues from WT or VhlhKO mice were harvested, homogenized, and selected for primary ECs (ADD-7−CD45-CD31+). Primary ECs from WT (WT-EC) or VhlhKO (VhlhKO-EC) tissue were then cultured on a gelatin-coated culture dish. These primary ECs were propagated for three passages over 6 days. f Proteins of each passage of primary ECs from WT or VhlhKO were collected for Western blotting for detection of Kat6b, H3K14ac, Icam-1, and Snail1. Uncropped blots are shown in Fig. S5. Statistical testing was performed by a two-tailed paired Student’s t-test. ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05. Error bars show the standard deviation. Upregulation of OSM-induced markers are sustained for three passages over 6 days. g The primary ECs from WT and VhlhKO as shown in (e) were cultured for three passages over 6 days and observed under a light microscope. The mesenchymal morphology of VhlhKO-ECs is maintained. The source data points for quantification are provided in Supplementary Data 2.
Quantification of OSM-induced or suppressed HAT/HDAC genes based on -Log10(P value) and Log2(FC) shows that KAT6B, KAT6A, and EP300 (KAT3B) are the most significantly upregulated (Fig. 3b). We then aimed to validate the expression of representative HATs and HDACs by Western blotting (Fig. 3c and Fig. S4). Interestingly, at the protein levels, among the HATs, EP300 and KAT6B show the most striking response to OSM stimulation, while KAT7, KAT6A, and CLOCK levels are constitutive and do not respond significantly or respond only modestly to OSM. Among the HDACs, SIRT7 and SIRT6 show a moderate decrease in response to OSM.
Interestingly, EP300 (also known as p300 or KAT3B) is involved in the acetylation of almost all histone lysine residues, but KAT6B (also known as MYST4, MORF, or MOZ2) is mostly involved in the acetylation of the residue lysine 14 of histone 3 (H3K14ac) in ECs26. This is in agreement with the notion that EP300 is a component of the general transcription factor complex serving as a transcription activator27, while suggesting that KAT6B possesses a specific target range in ECs.
We next investigated the clinical relevance of KAT6B expression in human ccRCC. As shown in Fig. 3d, the expression of KAT6B is highly enriched in ECs (CD31+ cells) within peritumor (Peri) and ccRCC tissues compared with normal adjacent tissue (NAT). A characteristic feature of epigenetic modification is heritability without DNA sequence alterations, which allows for changes in gene expression to be transmitted across several generations. We isolated primary ECs from WT and VhlhKO tissues, propagated them in vitro, and assessed the expression of OSM target markers over multiple passages (Fig. 3e). Interestingly, upregulation of representative OSM-responsive markers Kat6b, H3K14ac, Icam-1, and Snail1 proteins in primary ECs from VhlhKO tissue compared with WT tissue, is maintained for three passages over 6 days in ex vivo culture (Fig. 3f and Fig. S5). This correlates well with the persistence of morphological changes of VhlhKO-associated primary ECs in culture (Fig. 3g). These findings indicate that the phenotypic feature of ECs in VhlhKO tissue may result from epigenetic modification, not from transient gene expression changes.
The gene expression profile induced by OSM in ECs is primarily controlled by H3K14ac
The above results suggest that the gene expression profile induced by OSM is controlled by KAT6B-induced H3K14ac. To verify this notion, we performed a variant of chromatin immunoprecipitation analysis, the CUT&Tag sequencing (CUT&Tag-Seq), on hOSM-treated and untreated ECs labeled with anti-H3K14ac antibody. Several OSM-responsive genes identified previously (Fig. 1b)18 indeed contain increased levels of H3K14ac upon OSM stimulation (Fig. 4a), supporting the notion that increased expression of these genes is the result of H3K14 acetylation in the corresponding genomic loci. OSM-induced H3K14ac peak changes are then compared with mRNA-Seq-based transcriptomic changes of OSM-treated and untreated ECs. 1595 genes with significant changes in H3K14ac contents also appear in OSM-altered mRNA-Seq transcriptome, constituting 62% of the altered transcriptome of OSM-treated ECs vs untreated (Fig. 4b). Since histone acetylation has mainly been associated with transcriptionally active genes28,29, we investigated genes that are commonly upregulated in CUT&Tag-Seq and mRNA-Seq. These commonly upregulated genes constitute enrichment in the biological processes related to vascular and endothelial functions consistent with those found in OSM-treated EC gene sets (compare Figs. 4c with 1c). Figure 4d shows representative peak plots of genes that are involved in EC phenotypes in ccRCC and VhlhKO mouse models and are known OSM targets18, including VEGFA (angiogenesis and VEGF pathway), IL-6 (inflammatory cytokine), ICAM-1 (cell-cell adhesion), SNAIL1 (mesenchymal transition), HIF-1α (hypoxia signaling pathway), OSMR (OSM receptor), and KAT6B. This is in agreement with the above finding that the increased expression of these markers can be maintained in ex vivo monoculture of primary ECs (Fig. 3f), and is likely the result of epigenetic modification.
a Volcano plot of genes with increased or decreased H3K14ac-containing chromatins in response to hOSM treatment determined by CUT&Tag-Seq of hOSM-treated compared with untreated HMEC-1 cells. Upregulation is defined by Log2(FC) ≥1 and P value ≤0.05. Downregulation is defined by Log2(FC) ≤−1 and P value ≤0.05. Non-significant genes are defined by P value ≥0.05. b Venn diagram shows the overlapped genes that are significantly altered in mRNA sequencing of hOSM-stimulated vs untreated datasets and CUT&Tag-Seq of H3K14ac-containing genes in hOSM-treated vs untreated datasets. c The Gene set enrichment analysis of commonly upregulated genes identified in (b). The identified enriched biological processes are the same as in Fig. 1C. d Peak plots of representative genes that are known OSM targets and are involved in angiogenesis (VEGFA), inflammatory response (IL-6), cell-cell adhesion (ICAM-1), regulation of mesenchymal cell shape (SNAIL1), and hypoxia/glycolysis (HIF-1α) signaling pathways from CUT&Tag-Seq of hOSM-stimulated (red) or untreated (blue) ECs. All show increased presence of H3K14ac upon hOSM stimulation.
OSM signaling is mediated by the JAK-STAT3 signaling downstream of the OSM receptor. Previously, we have shown that Tofacitinib (Tofa), a JAK1/2 inhibitor, can mitigate the phenotypes observed in VhlhKO tissue18. We therefore investigated whether Tofa can influence the OSM-induced epigenetic changes. As shown in Fig. 6a, the mesenchymal cell shape of OSM-stimulated ECs is reverted to the endothelial morphology upon treatment with Tofa in vitro. The expression of OSM downstream targets shown in Fig. 4d, including HIF-1α, p-STAT3, ICAM-1, and SNAIL1, is also reverted to the baseline levels following treatment with Tofa (Figs. S6b, S7). In accordance, KAT6B and H3K14ac levels in OSM-stimulated ECs are reduced with Tofa treatment (Figs. S6b, S7).
Taken all together, the results indicate that the gene expression profile of OSM-stimulated ECs that is relevant to the altered phenotypes is likely controlled by H3K14ac.
KAT6B is required for EC activation
Since both EP300 and KAT6B are induced by OSM at both transcript and protein levels (Fig. 3b, c), we sought to determine whether both of these two HATs play a role in EC activation. In the following assays, EP300-specific inhibitor CBP30030 and KAT6 inhibitor PF936331 were included for comparison. Note that because of the high molecular similarity between KAT6A and KAT6B, there are currently no inhibitors that can specifically inhibit KAT6A or KAT6B activity separately. However, since KAT6A protein expression is not induced by OSM (Fig. 3c), the effect of PF9363 on OSM-induced changes should be largely mediated by inhibition of KAT6B. For the histone acetylation markers, we also included H3K9ac, a known KAT6A target in ECs, for comparison26. As shown in Fig. 5a and Fig. S8, inhibiting EP300 activity has no effect or only modestly reduces H3K14ac and H3K9ac expression compared with untreated cells, while inhibiting KAT6 activity abolishes the expression of H3K14ac and only modestly reduces H3K9ac. Both inhibitors can reduce the level of HIF-1α induced by OSM (see Discussion).
a HMEC-1 cells were treated or untreated with recombinant human OSM and with histone acetyltransferase inhibitors (CBP300, a EP300 inhibitor; or PF9363, a KAT6 inhibitor). Total proteins from CT (0 ng hOSM), OSM (10 ng/ml hOSM), CBP300 (10 ng/ml hOSM + 10 μM CBP300), or PF9363 (10 ng/ml hOSM + 30 μM PF9363) were isolated after 48 h. The concentrations of the inhibitors followed the published protocols and were selected empirically. Western blotting was performed for the expression of H3K14ac, H3K9ac, and HIF-1α. Uncropped blots are shown in Fig. S8. b EC cord formation assay of CT, OSM, CBP300, and PF9363 experimental groups. Prior to seeding onto Matrigel (growth-factor-reduced), samples were labeled with CellTracker Green CMFDA (1 μM). Images were obtained after 6 h using fluorescence microscopy. Scale bars are 200 μm. c Quantification of the experiment in (b) by measuring total network length and number of nodes (branching points). OSM significantly induces cord formation compared with the CT group. Treating hOSM-stimulated ECs with KAT6 inhibitor PF9363 reduces the cord formation, while EP300 inhibitor CBP300 has no effect. Each data point represents an independent experiment. d Heatmap of HMEC-1 cells from CT, OSM, PF9363, and CBP300 groups by unsupervised hierarchical clustering based on normalized gene expression levels (TPM, transcripts per million; Supplementary Data 1). Heatmap shows that the OSM and CT groups are located in different branches of clusters. EP300 inhibitor (CBP300) has a limited effect on the overall gene expression profile altered by OSM. In contrast, CT and OSM + PF9363 show similarity in hierarchical grouping. In particular, two clusters (Clusters 1 and 5; bracketed) upregulated by hOSM are reverted to the CT levels by KAT6 inhibitor PF9363. Each column represents one individual sample. e Gene set enrichment analysis of HMEC-1 genes in Clusters 1 and 5 of the heatmap shown in (d). These clusters compare the PF9363-rescued gene sets (PF9363) with the corresponding clusters in the OSM treatment group and the control (untreated with OSM). These genes represent those reverted by treatment of PF9363 (OSM+inhibitor PF9362) versus the OSM treatment group, to a level similar to the control (CT, untreated with OSM or PF9362). They show gene set enrichment in biological processes similar to those identified as common pathways in hOSM-treated HMEC-1 cells (Fig. 1c). f Western blotting of proteins from HMEC-1 cells with (shKAT6B-51 and -63) or without (Scr) KAT6B knockdown, treated with (OSM) or without (CT) hOSM, for representative OSM downstream targets. hOSM can induce expression of KAT6B, and the knockdown efficiency is confirmed by the lack of KAT6B protein expression in shKAT6B lines. Correspondingly, H3K14ac expression is induced by hOSM, and the induction is ameliorated in KAT6B knockdown. Conversely, KAT6B knockdown has no effect on the expression of KAT6A and H3K9ac with or without hOSM treatment. Importantly, hOSM can induce the expression of ICAM-1 and SNAIL1, and loss of KAT6B reduces the extent of induction. Uncropped blots are shown in Fig. S9. This result also validates the specificity of anti-KAT6B and anti-H3K14ac antibodies, as no signal was detected in KAT6B knockdown samples. g The schematic showing how cancer cell invasion through the EC monolayer was evaluated (left panel). The invasive cancer cells (RFP positive cells) across the EC monolayer was observed using fluorescence microscopy. Quantification of invasive cell numbers is shown in the bar graph. Each data point represents one independent experiment. KAT6B knockdown reduces the extent of ccRCC cell invasion. Scale bars are 1 mm. h Cord formation assay to test the angiogenic ability of HMEC-1 treated with (OSM) or without (CT) hOSM, and with or without KAT6B knockdown. HMEC-1 were grown on Matrigel (growth-factor-reduced). Quantification of network length and number of nodes is shown on the right. Each data point represents one independent experiment. hOSM can induce cord formation, and KAT6B knockdown can ameliorate the induction. Scale bars are 200 μm. Statistical testing of Western blotting quantification was performed by a two-tailed paired Student’s t-test. ns no significance; *P ≤ 0.05; **P ≤ 0.01. Other statistical tests were performed by one-way ANOVA (Tukey post hoc test). ***P ≤ 0.001. Error bars show the standard deviation. The source data points for quantification are provided in Supplementary Data 2.
For testing the angiogenic capacity, the cord formation assay was performed. HMEC-1 were labeled with fluorescence dye, stimulated with OSM with or without HAT inhibitors, and seeded onto Matrigel (growth-factor-reduced). Interestingly, inhibitor PF9363 strongly suppresses EC cord formation in Matrigel induced by hOSM as measured by network length and number of nodes (branching points), whereas the inhibitor CBP300 has no significant effect (Fig. 5b, c). To further explore the role of KAT6 and EP300 in EC activation by OSM, we performed mRNA-Seq of samples from untreated (CT), hOSM-treated, and hOSM-treated HMEC-1 in the presence of PF9363 or CBP300. A heatmap is drawn with unsupervised hierarchical clustering (Fig. 5d) based on the transcriptomes of HMEC-1 RNA-seq (Supplementary Data 1). hOSM induces upregulation of genes in clusters 1, 4, and 5, and downregulation of genes in clusters 2 and 6. Interestingly, inhibiting KAT6 activity in the OSM-treated group by PF9363 reverses the expression of genes in clusters 1 and 5 to the levels comparable to those in CT (bracketed in Fig. 5d). We analyzed the genes in these two clusters and conducted GSEA to explore the functional changes reflected by these OSM-induced, KAT6-dependent gene sets. As shown in Fig. 5e, these gene sets are involved in JAK-STAT signaling, glycolysis, hypoxia, inflammatory response, and mesenchymal transition, as well as those related to vascular and endothelial functions. These are exactly the same pathways enriched in the overlapped genes in OSM-treated EC (HMEC-1) transcriptome and H3K14ac-marked genetic loci (compare with Fig. 4c). Interestingly, the EP300 inhibitor does not influence the overall gene-expression profile of OSM-stimulated HMEC-1 cells (Fig. 5d), indicating that the genes influenced by EP300 play no significant role in the OSM-induced EC phenotypic changes. Collectively, these results show that inhibiting KAT6 activity counteracts the activation of ECs induced by OSM signaling.
To verify that induced KAT6B is responsible for the EC phenotypes, we specifically attenuated the expression of KAT6B in ECs by using short-hairpin RNA (shRNA) in HMEC-1 cells. Two KAT6B knockdown constructs, shKAT6B-51 and shKAT6B-63, as well as control with scrambled sequence (Scr) were used. These HMEC-1 were treated with or without hOSM in the presence of shKAT6B-51, shKAT6B-63, or Scr. The cells were then collected and investigated for OSM downstream target expression. As shown in Fig. 5f and Fig. S9, the expression of KAT6B protein is strongly induced after OSM treatment in Scr and abolished in KAT6B knockdown cells (shKAT6B-51 and shKAT6B-63). Notably, knocking down KAT6B does not affect the expression of KAT6A protein, nor the levels of H3K9ac, while the basal and OSM-induced levels of H3K14ac are nearly completely inhibited by KAT6B knockdown. This also indicates that the modest reduction of H3K9ac by KAT6 inhibitor PF9363 (Fig. 5a) is likely the result of KAT6A inhibition by the inhibitor. Other representative OSM downstream targets, ICAM-1 and SNAIL1, are also reduced in hOSM-treated HMEC-1 when KAT6B is knocked down. This result also serves as validation of the specificity of anti-KAT6B and anti-H3K14ac antibodies, also used in immunofluorescence staining in Fig. 3d (human KAT6B) and Fig. 2a, b (mouse and human H3K14ac). The characteristics of tumor-associated ECs are increased angiogenesis and loosening of their cellular junctions that facilitates tumor cell metastasis. We therefore investigated these functions of OSM-activated ECs after knocking down KAT6B. HMEC-1 with or without KAT6B knockdown were seeded on the upper transwell insert with 8-μm pores, and allowed to grow into a confluent monolayer mimicking endothelium. The ECs were then treated with hOSM, before RFP-expressing ccRCC cells, 786-ORFP, were added over the EC monolayer. The number of 786-ORFP invaded through the endothelium and migrated to the underside of the insert was examined using fluorescence microscopy. As shown in Fig. 5g, knockdown of KAT6B significantly reduces hOSM-induced cancer cell invasion compared with Scr cells. In addition, the angiogenic capacity of the ECs was quantified by cord network length and number of nodes (branching points) in Matrigel. KAT6B knockdown significantly diminishes the HMEC-1 angiogenic capacity induced by hOSM (Fig. 5h). To exclude the potential off-target or cytotoxic effects that could confound the angiogenesis assay, we evaluated the impact of the inhibitors and shKAT6B-mediated knockdown on HMEC-1 viability and proliferation. As shown in Fig. S10, KAT6B inhibition did not significantly affect cell viability or proliferation in the absence of OSM compared with the control. In contrast, treatment with CBP300 markedly reduced both viability and proliferation, consistent with the notion that EP300 is part of a general transcription factor complex27. Taken together, these results support the use of KAT6B as a candidate for further experiments.
Inhibiting Kat6 activity reduces the phenotypes of conditional Vhlh KO mice
Having established that KAT6B, not KAT6A, is mainly responsible for mediating the ccRCC-associated EC phenotypes, we then tested whether a small-molecule Kat6 inhibitor could be used to ameliorate tumor progression in the autochthonous premalignant ccRCC mouse model VhlhKO that we have established. We chose to use the injection of small-molecule inhibitors instead of using genetic inactivation of Kat6b in the VhlhKO background, because double KO of Vhlh in tubule cells and Kat6b in ECs requires two different Cre lines for different tissues in the same animal, which often produces unexpected health problems, which will make results difficult to interpret. Constitutive KO of Kat6b in Vhlh conditional KO background can be generated, but this will result in unpredictable genetic interactions of the two KO genes during embryogenesis since Kat6b genes are involved in embryonic development19,20. The use of small molecules can also serve as a proof-of-principle test for therapeutic potential.
WM-1119 is a highly selective, potent Kat6 inhibitor that has been used for the treatment of ER+ breast cancer31 and acute myeloid leukemia32 in preclinical mouse models. In the subsequent treatment setup, 25 mg/kg WM-1119 (determined empirically) was injected into 2-month-old mice every day for 14 days and the mice were sacrificed on Day 15 (Fig. 6a). Four groups were included: WT mice with or without WM-1119 treatment and VhlhKO mice with or without WM-1119 treatment. WT mice treated with WM-1119 show no obvious deleterious health effects or changes in vasculature density. On the other hand, treatment with WM-1119 in VhlhKO significantly reduces H3K14ac intensity in ECs (Fig. 6b, c, upper panel). Consistently, WM-1119 reduces the vasculature density of VhlhKO mice to the WT level (Fig. 6b, c, lower panel).
a Two-month-old WT or VhlhKO mice were treated with WM-1119 (KAT6 inhibitor, 25 mg/kg) or vehicle [50% PEG400 in ddH2O (v/v)] by I.P. injection once per day for 14 days. The concentration of the inhibitor followed the published protocols and selected empirically. b Double staining of H3K14ac (green) and CD34 (red) in WT or VhlhKO kidney tissue treated with WM-1119 or vehicle. Scale bars are 20 μm. c Quantification of H3K14ac intensity in ECs (CD34+), and the percent area of CD34+ cells in WT or VhlhKO tissue treated with or without WM-1119. WM-1119 can reduce the levels of H3K14ac in CD34+ cells and vascular density compared with the untreated counterparts. The intensity of H3K14ac and percentage of CD34 positive cells were quantified by CellProfiler (version 4.2.5) per the supplier’s instructions. n = 10. Each data point represents one animal. d Triple staining for CD34 (blue), GFP (green), and Ki67 (red) in WT or VhlhKO tissue treated with WM-1119 or vehicle. Scale bars are 20 μm. e Quantification of Ki67+ cells in all cells or in GFP+ cells from (d). n = 10. Each data point represents one animal. Statistical testing was performed by one-way ANOVA (Tukey’s post hoc test). ns: no significance; ***P ≤ 0.001. Error bars show the standard deviation. The source data points for quantification are provided in Supplementary Data 2.
In addition, inhibiting Kat6 in ECs significantly reduces the number of Ki67+ proliferating cells among the total cell population and specifically in Vhlh-deficient (GFP+) tubule cells in VhlhKO tissue compared with those in untreated VhlhKO mice (Fig. 6d, e). This is consistent with the previous finding that activated ECs in VhlhKO kidney can promote Vhlh-deficient tubule cell proliferation18. Macrophage infiltration represents a prevalent characteristic of ccRCC tumors and VhlhKO kidney tissues, exerting a pivotal influence across all stages of cancer progression. As shown in Fig. S11, in untreated VhlhKO tissues, there is an abundance of F4/80+ macrophage infiltration, and this infiltration is significantly reduced after treatment with WM-1119. Importantly, these phenotypic rescue effects on VhlhKO by Kat6 inhibition recapitulate those observed previously in Osmrβ−/−-VhlhKO double knockout18. Taken together, the above results show that inhibiting Kat6 activity in ECs can ameliorate the tumorigenic phenotypes of conditional Vhlh KO mice.
KAT6B function in ECs is important for tumor growth and metastasis
We first validated the role of OSM in regulating H3K14ac in ECs using a malignant ccRCC xenograft model. Human ccRCC cells 786-OLUC-RFP were implanted into the subcapsular space of immunodeficient NOD/SCID mouse kidneys. Tumor-bearing mice were treated with either anti-human OSM neutralizing antibodies (αOSM) or isotype control IgG via intravenous injection once per week for 4 weeks (Fig. S12a). Given that human OSM shares only approximately 45–50% sequence homology with murine OSM33,34, we validated the activity of human OSM on murine endothelial cells MS1, a murine microvascular endothelial cell line. As shown in Figs. S13, S14, human OSM robustly activated the OSM-related signaling pathway in MS1 cells, including upregulation of pStat3, Hif-1α, Icam-1, Snail1, Kat6b, and H3K14ac. These results support the proper functioning of the implantation model. Thus, OSM neutralization significantly reduces H3K14ac intensity in ECs compared with those in the IgG control group (Fig. S12b). Notably, reduced H3K14ac levels in ECs within primary tumor tissue also correlate with lower levels of lung metastases (Fig. S12c).
To further clarify the role of H3K14ac in ECs, we next specifically target the expression of H3K14ac by interfering with the function of KAT6B in ECs in this xenograft model. It has been shown that exogenously added ECs can enhance neovascularization and promote tumor progression35, and the ccRCC xenograft possesses a unique property of maintaining the co-xenografted ECs36. We therefore reason that since xenografted cancer cells still require newly formed vasculature to grow, exogenously added ECs should further accelerate neovascularization and ccRCC progression. In such cases, interfering with the activity of added endothelial cells should impede cancer growth when compared with unaltered ECs. Malignant 786-OLUC-RFP cells were mixed with HMEC-1 with control (Scr) or KAT6B knockdown (shKAT6B), and the cell mixture was implanted in the sub-renal capsular space of the left kidney of NOD/SCID mice (Fig. 7a). Tumors in Scr xenograft grow rapidly after Week 4 post-implantation, resulting in significantly larger tumors compared with EC-shKAT6B xenograft (Fig. 7b). Further quantification of primary tumor area from whole tissue section staining shows larger tumors in the Scr xenograft compared with the EC-shKAT6B xenograft (Fig. 7c). Consistently, there is a higher number of proliferative cancer cells (human mitochondria+ and Ki67+) in primary tumor of the Scr xenograft compared with the EC-shKAT6B xenograft (Fig. S15).
a The schematic showing the experimental design. Human ccRCC cells 786-O (expressing luciferase and RFP; 786-OLUC-RFP) were mixed with HMEC-1 with (shKAT6B) or without (Scr) KAT6B knockdown, and the mixture was injected into the sub-renal capsular space of the left kidney of 2-month-old immunodeficient mice (NOD/SCID). Tumor-bearing mice were weighed weekly, and monitored for tumor growth by the IVIS system. b Left: Tumors of representative tumor-bearing mice visualized by the IVIS system at the end point of the experiment. Right: Tumor growth was monitored by the IVIS system weekly during the experiment. Tumors in the Scr group grow faster after Week 4 than those in the shKAT6B group. c Representative images of whole-kidney sections from Scr and shKAT6B. The primary tumor areas are marked by white lines. Quantification of primary tumor areas in whole-kidney sections is shown on the right. Each data point is one whole-kidney section containing the largest tumor area from one animal. n = 10. d Double staining for H3K14ac (green) and human CD31 (hCD31; red) of tumor-bearing kidney tissues from Scr and shKAT6B groups. Scale bars are 20 μm. Quantification of H3K14ac intensity in human ECs (hCD31+) cells, and the percentage of hCD31+ area, is performed by CellProfiler (version 4.2.5) and shown at the bottom. n = 10. Each data point represents one animal. e The tumor-bearing kidney (Primary site) and metastasis (Lung) tissues of Scr and shKAT6B groups were stained for 786-OLUC-RFP (RFP+ cells) to visualize cancer cells. White arrows show invasive cancer cells in the primary site of the Scr sample. N is normal mouse tissue. T is tumor tissue. Scale bars are 50 μm. Quantification of the metastatic area and the number of metastasis foci are shown at the bottom. n = 10. Each data point represents one animal. Statistical testing was performed by Student’s t-test. ns no significance; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. Error bars show the standard deviation. The source data points for quantification are provided in Supplementary Data 2.
In addition, the intensity of H3K14ac in the EC-shKAT6B xenograft is significantly reduced compared with that in the Scr xenograft (Fig. 7d). The reduction of H3K14ac in exogenous human ECs is also associated with decreased neovasculature formation in the EC-shKAT6B xenograft (Fig. 7d), confirming the heightened angiogenic capacity of the activated ECs and the dependence of this capacity on the KAT6B function. Importantly, cancer cells in the Scr xenograft show greater invasiveness (white arrows) than those in the EC-shKAT6B xenograft, correlating with larger and more numerous lung metastases (Fig. 7e). Collectively, the results indicate that EC-specific KAT6B function can promote tumor vasculature, tumor growth, and metastasis.
Discussion
The full spectrum of interactions between tumor cells and ECs in cancer progression is not well-studied. In this study we reveal a novel epigenetic mechanism by which OSM drives EC activation, vascular remodeling, and inflammatory response in progression of ccRCC through acetylation at lysine 14 residue of histone 3, mediated by KAT6B (summarized in Fig. 8). We show that H3K14ac is induced by OSM-JAK-STAT signaling and regulates the gene sets associated with OSM-induced EC activation, including angiogenesis, hypoxia, glycolysis, inflammatory response, and mesenchymal transition (Fig. 4c, d). In vitro, attenuation of H3K14ac by KAT6B inactivation in ECs can rescue EC phenotypes accompanied by reduced expression of the gene sets induced by OSM signaling pathway (Fig. 5). Remarkably, in vivo targeting of KAT6 activity in ECs ameliorates phenotypes observed in the autochthonous early-stage ccRCC model of conditional Vhlh KO mouse, including reduced angiogenesis, decreased macrophage infiltration, and inhibited proliferation of Vhlh-deficient cells (Fig. 6 and Fig. S11). In the xenograft model of malignant ccRCC cells mixed with exogenous ECs, silencing KAT6B expression specifically in ECs results in diminished tumor growth and metastatic dissemination, in step with reduced H3K14ac in ECs (Fig. 7).
Oncostatin M (OSM), secreted by VHL-deficient tubule cells, induces epigenetic changes in ccRCC-associated endothelial cells (ECs) specifically through histone 3 lysine 14 acetylation (K14ac) mediated by histone acetyltransferase 6B (KAT6B). The hyperacetylation facilitates chromatin opening, activating transcription of specific gene sets involved in hypoxic response, vascular remodeling, inflammatory response, and mesenchymal transition, thereby promoting tumor growth and metastasis. Inhibiting OSM or histone acetyltransferase activity could mitigate these pathological EC phenotypes and reduce tumor growth and metastasis.
ccRCC is characterized by its highly vascularized nature, usually attributed to the abundance of angiogenic factors secreted by VHL-deficient tumor cells within the tumor microenvironment, such as VEGF. As such, targeting angiogenesis through inhibition of the VEGF pathway has been a cornerstone in ccRCC treatment. Nonetheless, resistance to these therapies invariably emerges in advanced stages of the disease, which may be due to decreased sensitivity to the drugs, or the involvement of additional unidentified factors and cellular mechanisms governing the EC behavior. We have discovered that OSM is a major regulator of angiogenesis and inducer of tumor-associated vascular phenotypes18. In this study, we further explore the molecular and cellular mechanisms of the OSM action on tumor-associated endothelial cells. We show strong upregulation of hypoxia, glycolysis, inflammation, mesenchymal transition, and angiogenesis hallmarks in OSM-treated ECs (Fig. 1a–c). These features are consistent with the vascular phenotypes in ccRCC. Our results demonstrate that OSM induces the expression of HIF-1α (Figs. 4d, 5a and Fig. S6) in ECs, and this upregulation is at least in part associated with increased H3K14ac (Figs. 4d, 5a). Interestingly, OSM has been recognized as an inducer of metabolic switch in tumor cells37,38,39. Thus, the pseudohypoxic signaling pathway of OSM-HIF-1α in ECs may amplify the VHL-deficient kidney tubule cell-mediated pseudohypoxic condition in ccRCC tumor microenvironment and contribute to tumor angiogenesis. This also suggests a transmitted metabolic abnormality between cancer cells and stromal cells mediated by intercellular HIF-OSM-HIF pathways.
In addition, mesenchymal transition of the OSM-stimulated endothelium, probably promoted by SNAIL1 and adhesion molecule ICAM-1 (Figs. 1b, 3f, 4d, 5f and Fig. S6), can promote cancer cell invasiveness (Fig. 5g). We show that the vascular phenotypes associated with ccRCC tissues are regulated by KAT6B-mediated histone acetylation at H3K14, highlighting the critical role of epigenetic regulation in tumor-associated ECs (Figs. 6b, 7d). We should note that HATs are known to also acetylate and activate/stabilize transcription factors, including HIF-1α40; therefore, part of the pseudohypoxic response induced by OSM may be the result of acetylation of HIF-1α by HATs. However, our results show that at least at the transcription level, HIF-1α is induced by OSM (Fig. 1b) and the HIF-1α genomic locus is heavily modified by H3K14ac upon OSM induction (Fig. 4d).
In this report, we observe that EP300 is the second most significantly upregulated gene related to histone modification in OSM-stimulated ECs (Fig. 3b), but it has no specific role in the EC phenotypes associated with ccRCC development (Fig. 5a–d). Also surprising, in our system, inhibiting EP300 activity did not exert a pronounced effect on the H3K14ac level or on the transcriptomic profile induced by OSM (Fig. 5a, d). In contrast to EP300, KAT6B exhibits specific acetylation activity toward H3K14 in our system (Fig. 5a, f). Inhibition of KAT6B effectively counteracts the effects of OSM on ECs, notably abolishing their angiogenic, inflammatory, and prometastatic capacity (Figs. 5b, c, g, 6b, 7d, e). These findings suggest that KAT6B in ECs can be a potential target of anti-tumor therapy for ccRCC patients. Indeed, the Kat6 inhibitor WM-1119 can ameliorate the premalignant phenotypes of VhlhKO kidney (Fig. 6) and downregulate KAT6B expression in tumor-associated ECs can reduce ccRCC growth and metastasis (Fig. 7). On this point, we note that the KAT6 inhibitor WM-1119 has been tested in preclinical animal models and shows no obvious cytotoxicity 32.
It has been shown in other systems that histone acetylation indeed plays an important role in angiogenesis. For example, histone 3 lysine 56 acetylation (H3K56ac) is induced in ECs by fibroblast growth factor 2 (FGF2) and epidermal growth factor (EGF)41; and H3K14ac can promote sprouting during angiogenesis in retinal endothelium42. VEGF can upregulate HATs p300 and MRTF-A, leading to enhancement of histone 3 lysine 9 acetylation (H3K9ac) and H3K14ac43. On the other hand, it has been shown that HDACs are overexpressed in ccRCC tumor cells44,45, suggesting a tumor suppressor role of histone acetylation. HDAC inhibitors have therefore been suggested as potential therapeutics for ccRCC. However, in the ccRCC xenograft mouse model, we demonstrate that knockdown of KAT6B in exogenous ECs co-implanted with ccRCC cells can reduce tumor growth and metastasis compared with co-implantation with wild-type ECs (Fig. 7). Thus, the result highlights the prooncogenic role of histone acetylation in tumor-associated ECs, which may also explain why HDAC inhibitor therapy has not been successful in the preclinical settings for ccRCC.
OSM signaling-induced histone acetylation has not been observed widely. Rare examples include increased SMAD4 transcription in cardiac fibroblasts through H3K27 hyperacetylation46. Conversely, OSM can reduce H3K9ac on the promoter of the SLUG gene in lung adenocarcinoma cells in vitro47. Our finding places OSM-induced histone acetylation in the pathophysiological context, specifically in tumor-associated ECs, and provide a new insight into the unique connection of pseudohypoxic responses between ccRCC and the endothelial compartment.
It has been shown that epigenetic modification plays a critical role in organotypic EC diversity during murine development48. Our research presents a rare example of tumor cells reshaping the tumor microenvironment through alterations in the epigenetic profile of ECs. These findings expand our understanding of endothelial cell activation and suggest an alternative avenue for vascular-targeted therapies for ccRCC treatment.
Methods
Cell culture
Human renal cell adenocarcinoma cell line 786-O was obtained from Bioresource Collection and Research Center (BCRC; Taipei, Taiwan; RRID:CVCL_1051). Human microvascular endothelial cell line (HMEC-1) was purchased from American Type Culture Collection (ATCC; Gaithersburg, MD, USA; #CRL-3243; RRID:CVCL_0307). Mouse microvascular endothelial cells, MILE SVEN 1 (MS1), were purchased from Bioresource Collection and Research Center (BCRC; Taipei, Taiwan; RRID:CVCL_6502). All cells were used within ten passages to ensure no genetic variations accumulate and cell identities remain the same as the supplier’s source. The regrown cells were routinely stained with DAPI to ensure the lack of mycoplasma contamination. Primary mouse ECs were isolated from kidney tissues of 3-month-old wildtype or VhlhKO mice as described18. Details of media and supplements are shown in Table S1.
Animals
The Hoxb7-Cre-GFP; VhlhloxP/loxP (VhlhKO) mouse strain has been reported16. The nomenclature Vhlh denotes the mouse homolog of the human VHL gene, named VHL-homologous sequence 1, hence Vhlh15. The nomenclature was used for the original VhlhloxP/loxP strain generated by Volker Hasse and Rudy Jaenisch laboratories14. The original strain was used by us to generate VhlhKO in this and other studies16,17,18,49,50. Osm receptor β KO mouse (Osmr−/−; RRID:IMSR_RBRC02711) and Hoxb7-Cre-GFP; VhlhloxP/loxP; Osmr−/− double knockout mouse have also been reported18. Hoxb7-Cre-GFP was used as wild-type control. NOD/SCID mouse (NOD.CB17-Prkdcscid/NcrCrl; RRID:IMSR_CRL:394) was purchased from BioLASCO Taiwan Co., Ltd (Taipei, Taiwan). Using the samplesz.xls spreadsheet, we calculated that for a given genotype, e.g., vascular abnormality that is 100% penetrant with standard deviation ~5–10% and estimated error ~10% based on previous studies16,17,18,50, to have this be different from another genotype with a power of 80%, ~10 mice in each group are needed to have a 95% power of detecting this difference (P value of 0.05). No preset inclusion or exclusion criteria were used except that the male-to-female ratio remained constant in different experimental groups. No confounders such as cage location or weight differentials were noted during the experiments, and therefore, no adjustments were made. Group allocation was done using random.org software. Only the experimenter was aware of the group allocation. Outcome assessment and data analysis were performed with persons unaware of group allocation.
Mice were housed in pathogen-free facilities with climate and light-cycle control. Cages were provided with nest-building materials and an igloo for nesting. Anesthesia was used for all surgical procedures, and analgesia was administered after xenografting. We have complied with all relevant ethical regulations for animal use. All experiments were performed in accordance with a protocol prepared before the study and approved by the Institutional Animal Care and Use Committee of China Medical University (CMUIACUC-2024-034).
Human ccRCC tissue
The Institutional Review Board of the Triservice General Hospital approved the collection and use of archived human ccRCC tissues from patients at Triservice General Hospital in this study (IRB no. 2-106-05-079). Informed consent was obtained from all participants. All ethical regulations relevant to human research participants were followed. Tissue arrays containing human kidney cancer tissue paraffin sections were purchased from TissueArray.Com LLC (#KD244a; US Biomax Inc., Derwood, MD, USA).
CellProfiler image analysis
The percentage of stained area and expression intensity were quantified by CellProfiler Image Analysis Software (https://cellprofiler.org; RRID: SCR_007358) following the instructions of the supplier51. CellProfiler is an open-source software tool designed to quantitatively measure phenotypes from a large number of images automatically. Cell-specific marker staining is analyzed separately, and cell-specific expression of a given gene is identified. The images with multiple stained markers were merged to assess the intensity of a marker in a particular cell type, and the percentile of the particular cell type within the tissue of interest52.
Recombinant human OSM treatment
About 4 × 104 HMEC-1 cells were seeded in a 3.5-cm2 culture dish and grown overnight. The media were replaced with fresh complete media with or without 10 ng/mL of recombinant human OSM (hOSM) (PeproTech Proteins, ThermoFisher, #300-10; Waltham, MA, USA) and cells incubated for 48 h. Total proteins or total mRNAs of hOSM-treated or untreated HMEC-1 or MS1 cells were harvested for downstream analysis.
Inhibitor treatment
PF9363 (KAT6A/B inhibitor, 30 μg/mL; Selleckchem, #E0146; Houston, TX, USA), CBP300 (CREBBP/EP300 inhibitor, 10 μg/mL; Selleckchem, #S7256), or Tofacitinib (JAK1/2 inhibitor, 50 μM; Selleckchem, #S2789) was added into hOSM-treated HMEC-1 growth media. Total proteins or total mRNAs were harvested at the indicated time for downstream analysis.
In vitro cord formation
Matrigel Growth Factor Reduced (Corning #354230) was thawed at 4 °C overnight. About 120 μL of Matrigel were gently poured into each well of a 96-well flat clear-bottom black microplate (Corning #3603) and the culture plate was placed in 37 °C cell culture incubator for the Matrigel to congeal. ECs (HMEC-1) were incubated with CellTracker Green CMFDA (1 μM; Thermo Fisher, #C2925) at 37 °C for 30 min, and then incubated with OSM and with or without HAT inhibitors as described above. Labeled ECs were harvested and resuspended in growth media to a final concentration of 1.5 × 105 cells/ml in the presence of OSM with or without inhibitors. About 100 μL of resuspend ECs were added into each well of Matrigel-coated 96-well microplate and placed in 37 °C cell culture incubator for 6 hr before observation with fluorescence microscopy. Quantification of cord formation was performed using ImageJ (RRID:SCR_003070) with the Angiogenesis Analyzer plugin (https://biii.eu/angiogenesis-analyzer-imagej).
RNA sequencing
Total mRNAs were isolated using miRNeasy Mini Kit (Qiagen, #217084; Hilden, Germany) per the instructions of the manufacturer. mRNA sequencing was performed by Genomics BioSci&Tech Co. (New Taipei City, Taiwan) using the Illumina NovaSeq 6000 platform. Volcano plot and heatmap were drawn in the R Project for Statistical Computing (version 4.3.1; RRID:SCR_001905). Gene Ontology and Bioprocess enrichment analyses were performed by Gene Set Enrichment Analysis (GSEA; RRID:SCR_003199).
Cleavage under targets and tagmentation-sequencing (CUT&Tag-Seq)
CUT&Tag-Seq is a variant of the chromatin immunoprecipitation-coupled sequencing (ChIP-Seq), utilizing protein A-Tn5 fusion to target the antibody bound to a specific DNA-binding protein motif, thus providing highly specific Tn5 nuclease-mediated DNA cuts around target sites, compared with conventional ChIP-Seq that generates randomly sheared DNA fragments. CUT&Tag-Seq of HMEC-1 cells, with or without hOSM treatment, were performed by Topgen Biotechnology Co., Ltd., (Taipei, Taiwan). The experimental reaction system and library construction process followed the protocol of the Hyperactive Universal CUT&Tag Assay Kit for Illumina (Vazyme, TD903), in which permeabilized cells were incubated with the H3K14ac antibody (Abcam, #ab52946; RRID:AB_880442). The libraries were sequenced using the Illumina NovaSeq 6000 platform. Data quality was assessed with FastQC (v0.73). Raw sequencing reads were aligned to the human reference genome (hg38) using Bowtie2 (v2.4.5), and aligned reads were processed for peak calling using SEACR (v1.3.0). Peak annotation was performed with ChIPSeeker (v1.34.1), and differential peak analysis and normalization were conducted using DESeq2 (v1.38.3).
shRNA-mediated gene knockdown
Lentiviral vector pCMV-ΔR8.91 (RRID:Addgene_202687), pMD2.G (RRID:Addgene_12259), and short-hairpin RNAs (shRNAs) of scrambled sequence (Scr) or sequences specific for KAT6Bs (shKAT6Bs) were purchased from the National RNAi Core Facility (Academia Sinica, Taipei, Taiwan). The packaging of lentiviral particles of Scr or shKAT6Bs was performed per the instructions of the supplier. Transduced HMEC-1 cells were selected with 1 μg/mL puromycin (InvivoGen, #ant-pr-1; San Diego, CA, USA) for 7 days to establish KAT6B knockdown stable lines. The sequences of shRNAs are 5’-ATGGAAATGCCTCTAACTTTA-3’ (shKAT6B-51), 5’-GCTGTGAATAATGGGAGGTTA-3’ (shKAT6B-63), and 5’-CTACTCAGTATGCACGTCG-3’ (Scr, for negative control).
Viability and proliferation assay
Cells were pre-labeled with ViaFluor® SE Cell Proliferation Kit (Biotium, Cat# 30139-T) at a concentration of 1 µM, following the manufacturer’s instructions. Labeled cells were then seeded in complete medium containing either 10 µM CBP300 or 30 µM PF9363 (inhibitor groups). Scrambled control (Scr) and shKAT6B-transduced cells were seeded in complete medium without inhibitors. A portion of the labeled cells was analyzed immediately by flow cytometry to establish the starting point of fluorescence intensity (fluorescence intensity 106–107). After 48 h, cells were harvested and stained with 7-AAD (BioLegend, Cat# 420403) to assess cell viability. Dead cells were defined as 7-AAD–positive. Non-proliferative cells were defined by fluorescence intensity in the range of 106–107, low-proliferative cells as 105–106, and high-proliferative cells as 103–105.
Western blotting
Cellular proteins were harvested by RIPA Lysis and Extraction Buffer (Thermo Fisher, #89900) with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher, #78440), and quantified by Pierce BCA Protein Assay Kits (ThermoFisher, #23225). About 10 μg of total proteins were subjected to SDS-PAGE (acrylamide concentration 8–15%). Separated proteins were transferred to PVDF membranes (Millipore, #IPVH85R; Burlington, MA, USA) and then blocked with 5% non-fat milk for 1 h at room temperature. The membranes were then washed and hybridized with primary antibodies at 4 °C overnight, followed by washing and then hybridization with corresponding secondary antibodies for 1 h at room temperature. The membranes were then washed and imaged using Sapphire FL Biomolecular Imager (RRID: SCR_025635) in the chemiluminescence module with ECL (PerkinElmer, #NEL103E001EA; Waltham, MA).
In fluorescence Western blot, proteins separated in SDS-PAGE were transferred to low fluorescence Western membrane (Azure Biosystems, #AC2109; Dublin, CA, USA), and blocked with Azure Fluorescent Blot Blocking Buffer (Azure Biosystems, #AC2190) for 10 min at room temperature. Membranes were then hybridized with primary antibodies at 4 °C overnight, followed by washing and then hybridization with corresponding fluorescence secondary antibodies for 1 h at room temperature. The signals of detected proteins were recorded by Sapphire FL Biomolecular Imager in a fluorescent module. Quantification of protein expression levels was performed by AzureSpot software (version 2.2.167). Details of primary and secondary antibodies are listed in Table S2.
Three-month-old WT, Osmr−/−, VhlhKO, and DBKO mice were perfused with 30 mL of HBSS buffer to remove residual blood. Kidneys were collected and minced into ~1 mm pieces. The tissues were then homogenized in 1 mL of RIPA Lysis and Extraction Buffer (Thermo Fisher, #89900) supplemented with Halt™ Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher, #78440) using a Dounce homogenizer. Total protein concentrations were measured using the Pierce™ BCA Protein Assay Kit (Thermo Fisher, #23225). Western blotting was performed as described above.
Fluorescence immunohistochemistry
Tissues were collected, washed with PBS, and immediately fixed in 10% neutral PBS-buffered formalin (Leica Microsystems, #3800600; Deerfield, IL, USA) at 4 °C overnight. Tissues were then embedded in paraffin blocks. The paraffin sections were de-paraffinized, rehydrated, and retrieved with retrieval buffer (Table S3). The sections were then blocked with 5% normal serum and hybridized with primary antibodies at 4 °C overnight. Hybridized sections were washed and hybridized with corresponding secondary antibodies for 1 h at room temperature. To reduce auto-fluorescence, sections were covered with the reagents in Vector TrueVIEW Autofluorescence Quenching Kit (Vector Labs, #SP-8400-15; Newark, CA) for 5 min at room temperature. Sections were mounted with VECTASHIELD Antifade Mounting Medium (Vectorlabs, #H-1000-10) and imaged with an Olympus fluorescence microscope IX81. The staining conditions were listed in Table S3. The specificity of the primary antibodies used is controlled by the lack of any fluorescence signals when only fluorescence-labeled secondary antibodies (Table S3) were used (Fig. S16).
Inhibitor treatment in vivo
WM-1119 (25 mg/kg; Selleckchem, #S8776) or vehicle (50% PEG400 in ddH2O; ThermoFisher, #B21992.30) was injected into the peritoneum (I.P. injection) once per day for two weeks. In the neutralizing OSM experiment, tumor-bearing mice were treated with 100 µg anti-human OSM antibodies (R&D System, MAB295-500) or IgG control (BioXCell, BE0085) once a week (for a total of 4 weeks) by I.V. injection.
Cancer cell implantation
The implantation of ccRCC cell line 786-OLUC-RFP and human microvascular endothelial cells HMEC-1 was based on the method of orthotopic xenograft described previously, with modification17,18. Briefly, 4.5 × 105 786-OLUC-RFP cells (expressing luciferase and red fluorescent protein) were mixed with 0.5 × 105 HMEC-1 cells with or without KAT6B knockdown in 10 μL serum-free RPMI medium with 50% Matrigel Growth Factor Reduced (Corning, Glendale, Arizona, USA; #356238). The mixture was then injected into the sub-renal capsular space of the left kidney of immunodeficient mice (NOD.CB17-Prkdcscid/NcrCrl). The growth and metastasis of cancer cells were monitored weekly by luminescence signal using the IVIS Lumina LT Series III system (Revvity; Waltham, MA, USA). At the end of the experiment, tissues were harvested and embedded in paraffin blocks for analysis. The hematoxylin and eosin (H&E) staining of whole-kidney sections was analyzed by TissueFAXS SPECTRA Multispectral Tissue Cytometer (TissueGenostics; Vienna, Austria). The tumor area in the primary site was quantified by ImageJ (https://imagej.net/ij/).
For the OSM neutralization experiment, 786-OLUC-RFP cells (5 × 105) were injected as described above. One week after implantation, mice were treated weekly for 4 weeks via intravenous (I.V.) injection with 100 μg of IgG control (BioXCell, BE0085) or anti-human OSM antibody (R&D Systems, MAB295-500).
The humane endpoints for the animal experiments were defined as an average tumor diameter not exceeding 20 mm or 10% of the animal’s original body weight. None of the experiments exceeded these limits.
Statistics and reproducibility
The unpaired two-tailed Student’s t-test was used for two-group comparison. One-way ANOVA with Tukey’s post hoc test was used for multiple-group comparison for statistical analysis. The statistic is significant when its two-sided P value is ≤0.05. Sample sizes and number of replicates are denoted in each relevant figure legend.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mRNA sequence and CUT&Tag sequence in this study have been deposited to Gene Expression Omnibus (GEO) (RRID:SCR_005012) with accessible numbers: GSE271554 for OSM-treated HMEC-1 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE271554) and GSE271723 for CUT&Taq-Seq (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE271723). All other data were available from the corresponding author (or other sources, as applicable) on reasonable request.
References
Bejarano, L., Jordao, M. J. C. & Joyce, J. A. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 11, 933–959 (2021).
Aran, D. et al. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 8, 1077 (2017).
Pidsley, R. et al. Enduring epigenetic landmarks define the cancer microenvironment. Genome Res. 28, 625–638 (2018).
Hsieh, J. J. et al. Renal cell carcinoma. Nat. Rev. Dis. Prim. 3, 17009 (2017).
Linehan, W. M. et al. The metabolic basis of kidney cancer. Cancer Discov. 9, 1006–1021 (2019).
Rathmell, W. K., Rathmell, J. C. & Linehan, W. M. Metabolic pathways in kidney cancer: current therapies and future directions. J. Clin. Oncol. https://doi.org/10.1200/JCO.2018.79.2309 (2018).
Kaelin, W. G. Jr. Cancer and altered metabolism: potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb. Symp. Quant. Biol. 76, 335–345 (2011).
Semenza, G. L. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin. Cancer Biol. 19, 12–16 (2009).
Vuong, L., Kotecha, R. R., Voss, M. H. & Hakimi, A. A. Tumor Microenvironment dynamics in clear-cell renal cell carcinoma. Cancer Discov. 9, 1349–1357 (2019).
Aran, D., Hu, Z. & Butte, A. J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, 220 (2017).
Sjoberg, E. Molecular mechanisms and clinical relevance of endothelial cell cross-talk in clear cell renal cell carcinoma. Ups J. Med. Sci. https://doi.org/10.48101/ujms.v129.10632 (2024).
Bergers, G. & Hanahan, D. Modes of resistance to anti-angiogenic therapy. Nat. Rev. Cancer 8, 592–603 (2008).
Liu, Z. L., Chen, H. H., Zheng, L. L., Sun, L. P. & Shi, L. Angiogenic signaling pathways and anti-angiogenic therapy for cancer. Signal Transduct. Target Ther. 8, 198 (2023).
Biju, M. P. et al. Vhlh gene deletion induces Hif-1-mediated cell death in thymocytes. Mol. Cell Biol. 24, 9038–9047 (2004).
Gao, J. et al. Cloning and characterization of a mouse gene with homology to the human von Hippel-Lindau disease tumor suppressor gene: implications for the potential organization of the human von Hippel-Lindau disease gene. Cancer Res. 55, 743–747 (1995).
Pritchett, T. L., Bader, H. L., Henderson, J. & Hsu, T. Conditional inactivation of the mouse von Hippel-Lindau tumor suppressor gene results in wide-spread hyperplastic, inflammatory and fibrotic lesions in the kidney. Oncogene 34, 2631–2639 (2015).
Nguyen, T. N., Nguyen-Tran, H. H., Chen, C. Y. & Hsu, T. IL-6 and CCL18 mediate crosstalk between VHL-deficient kidney cells and macrophages during development of renal cell carcinoma. Cancer Res. 82, 2716–2733 (2022).
Nguyen-Tran, H. H., Nguyen, T. N., Chen, C. Y. & Hsu, T. Endothelial reprogramming stimulated by oncostatin M promotes inflammation and tumorigenesis in VHL-deficient kidney tissue. Cancer Res. 81, 5060–5073 (2021).
Karmodiya, K., Krebs, A. R., Oulad-Abdelghani, M., Kimura, H. & Tora, L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13, 424 (2012).
Regadas, I. et al. A unique histone 3 lysine 14 chromatin signature underlies tissue-specific gene regulation. Mol. Cell 81, 1766–1780.e1710 (2021).
Lu, Y. et al. Increased acetylation of H3K14 in the genomic regions that encode trained immunity enzymes in lysophosphatidylcholine-activated human aortic endothelial cells - Novel qualification markers for chronic disease risk factors and conditional DAMPs. Redox Biol. 24, 101221 (2019).
Kueh, A. J. et al. Stem cell plasticity, acetylation of H3K14, and de novo gene activation rely on KAT7. Cell Rep. 42, 111980 (2023).
Yang, Y. et al. The histone lysine acetyltransferase HBO1 (KAT7) regulates hematopoietic stem cell quiescence and self-renewal. Blood 139, 845–858 (2022).
Pusztaszeri, M. P., Seelentag, W. & Bosman, F. T. Immunohistochemical expression of endothelial markers CD31, CD34, von Willebrand factor, and Fli-1 in normal human tissues. J. Histochem. Cytochem. 54, 385–395 (2006).
Shvedunova, M. & Akhtar, A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 23, 329–349 (2022).
Fang, Z., Wang, X., Sun, X., Hu, W. & Miao, Q. R. The role of histone protein acetylation in regulating endothelial function. Front. Cell Dev. Biol. 9, 672447 (2021).
Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H. & Nakatani, Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959 (1996).
Johnsson, A. et al. HAT-HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Rep. 10, 1009–1014 (2009).
Pokholok, D. K. et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527 (2005).
Ibrahim, Z. et al. Structural insights into p300 regulation and acetylation-dependent genome organisation. Nat. Commun. 13, 7759 (2022).
Sharma, S. et al. Discovery of a highly potent, selective, orally bioavailable inhibitor of KAT6A/B histone acetyltransferases with efficacy against KAT6A-high ER+ breast cancer. Cell Chem. Biol. 30, 1191–1210.e1120 (2023).
Baell, J. B. et al. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature 560, 253–257 (2018).
Hermanns, H. M. Oncostatin M and interleukin-31: Cytokines, receptors, signal transduction and physiology. Cytokine Growth Factor Rev. 26, 545–558 (2015).
Adrian-Segarra, J. M. et al. The AB loop and D-helix in binding site III of human Oncostatin M (OSM) are required for OSM receptor activation. J. Biol. Chem. 293, 7017–7029 (2018).
Warner, K. A. et al. Endothelial cells enhance tumor cell invasion through a crosstalk mediated by CXC chemokine signaling. Neoplasia 10, 131–139 (2008).
Sanz, L. et al. Differential transplantability of human endothelial cells in colorectal cancer and renal cell carcinoma primary xenografts. Lab. Invest. 89, 91–97 (2009).
Battello, N. et al. The role of HIF-1 in oncostatin M-dependent metabolic reprogramming of hepatic cells. Cancer Metab. 4, 3 (2016).
Komori, T. & Morikawa, Y. Oncostatin M in the development of metabolic syndrome and its potential as a novel therapeutic target. Anat. Sci. Int. 93, 169–176 (2018).
Stephens, J. M. & Elks, C. M. Oncostatin M: potential implications for malignancy and metabolism. Curr. Pharm. Des. 23, 3645–3657 (2017).
Geng, H. et al. HIF1alpha protein stability is increased by acetylation at lysine 709. J. Biol. Chem. 287, 35496–35505 (2012).
Dutta, D. et al. Regulation of angiogenesis by histone chaperone HIRA-mediated incorporation of lysine 56-acetylated histone H3.3 at chromatin domains of endothelial genes. J. Biol. Chem. 285, 41567–41577 (2010).
Grant, Z. L. et al. The histone acetyltransferase HBO1 promotes efficient tip cell sprouting during angiogenesis. Development 148, dev199581 (2021).
Shu, X. Z. et al. Histone acetyltransferase p300 promotes MRTF-A-mediates transactivation of VE-cadherin gene in human umbilical vein endothelial cells. Gene 563, 17–23 (2015).
Cheng, F. et al. Comprehensive analysis of a new prognosis signature based on histone deacetylases in clear cell renal cell carcinoma. Cancer Med. 10, 6503–6514 (2021).
Pili, R. et al. Combination of the histone deacetylase inhibitor vorinostat with bevacizumab in patients with clear-cell renal cell carcinoma: a multicentre, single-arm phase I/II clinical trial. Br. J. Cancer 116, 874–883 (2017).
Xu, P. et al. Oncostatin M/oncostatin M receptor signal induces radiation-induced heart fibrosis by regulating SMAD4 in fibroblast. Int. J. Radiat. Oncol. Biol. Phys. 118, 203–217 (2024).
Pan, C. M., Wang, M. L., Chiou, S. H., Chen, H. Y. & Wu, C. W. Oncostatin M suppresses metastasis of lung adenocarcinoma by inhibiting SLUG expression through coordination of STATs and PIASs signalings. Oncotarget 7, 60395–60406 (2016).
Cantu Gutierrez, M. E. et al. Mapping the transcriptional and epigenetic landscape of organotypic endothelial diversity in the developing and adult mouse. Nat. Cardiovasc. Res. 4, 473–495 (2025).
Bader, H. L. & Hsu, T. Inactivation of the tumor suppressor gene von Hippel-Lindau (VHL) in granulocytes contributes to development of liver hemangiomas in a mouse model. BMC Cancer 16, 797 (2016).
Kuo, C. Y., Lin, C. H. & Hsu, T. VHL Inactivation in Precancerous Kidney Cells Induces an Inflammatory Response via ER Stress-activated IRE1alpha signaling. Cancer Res. 77, 3406–3416 (2017).
Stirling, D. R. et al. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinformatics 22, 433 (2021).
Carpenter, A. E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Acknowledgements
This work is supported by a grant to T.H. from the National Science and Technology Council-Taiwan (#NSTC 112-2320-B-039-019) with supplemental funding from the China Medical University-Taiwan (#CMU113-MF-01), and a grant to T.H. from the National Health Research Institute-Taiwan (#NHRI-EX111-11101BI). H.-H.N.-T. is supported by a postdoctoral fellowship from the National Science and Technology Council-Taiwan (#NSTC 112-2811-B-039-029).
Author information
Authors and Affiliations
Contributions
H.-H.N.-T performed all the experiments and analysis of the data, and wrote the initial draft of the manuscript. T.-N. N. helped perform animal experiments in Figs. 6, 7, and assisted in experiments relating to macrophage function. T.H. provided overall scientific guidance, analyzed data, and finalized the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Ahmed Hegazy and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editors: Derrick Ong and Johannes Stortz.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nguyen-Tran, HH., Nguyen, TN. & Hsu, T. Oncostatin M induces epigenetic reprogramming in renal cell carcinoma-associated endothelial cells. Commun Biol 8, 1541 (2025). https://doi.org/10.1038/s42003-025-08907-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08907-x