Abstract
The membrane (M) protein of coronaviruses is essential for maintaining structural integrity during membrane virion budding and viral pathogenesis. Given its high conservation in lineages within the betacoronavirus genus, such as sarbecoviruses, the M protein presents as an attractive therapeutic target; however, developing broad-spectrum antivirals targeting coronaviruses such as MERS-CoV is challenging due to lower sequence conservation and limited structural information available beyond that of the SARS-CoV-2 M protein. In this study, we report 3-3.2 Å resolution structures of MERS-CoV M protein, engineered with a SARS-CoV-2-like antibody interface, representing the first human merbecovirus M protein structure, and SARS-CoV M protein structures, with and without a previously identified SARS-CoV-2 M protein inhibitor, JNJ-9676. We highlight the structural differences between the MERS-CoV, SARS-CoV and SARS-CoV-2 M proteins, and present insights into the conservation of the JNJ-9676 binding pocket as well as key differences that could be targeted to accelerate the design of specific MERS-CoV and broad-spectrum antivirals targeting coronavirus M proteins.
Similar content being viewed by others
Introduction
The membrane (M) protein of coronaviruses is the most abundant of the four structural proteins in the Coronaviridae family, which encompasses a wide range of enveloped positive stranded RNA viruses1. The M protein is a small multi-pass membrane protein in the virion that forms a 50 kDa homodimer; each monomer consisting of a short N-glycosylated ectodomain, three transmembrane domain helices and a cytosolic intravirion beta-sheet domain2,3. The primary role of the M protein is to maintain viral structural integrity through interactions with the other structural proteins: interactions with envelope (E) and spike (S) proteins facilitate membrane budding, while interactions with the nucleocapsid (N) protein direct encapsidation for viral biogenesis3,4,5,6,7. Viral assembly is regulated by two distinct conformations of the M protein which exist in dynamic equilibrium: a long and short form3,8,9,10. Elongated M protein is linked with membrane rigidity through interactions with S proteins resulting in limited membrane curvature, whereas short M protein is associated with flexibility and reduced S protein interactions.
Among the four subgenera within the Coronaviridae family, alpha and betacoronaviruses are able to infect humans, causing mild to severe upper respiratory tract illnesses11. Notably, betacoronaviruses such as severe acute respiratory syndrome coronavirus (SARS-CoV)12,13, Middle East respiratory syndrome coronavirus (MERS-CoV)14,15 and SARS-CoV-216 have caused significant outbreaks and pandemics over the past two decades with profound impacts on public health, economies and healthcare systems. Betacoronaviruses are comprised of four subgroups, sarbecoviruses, nobecoviruses, embecoviruses and merbecoviruses. To date, structural studies have primarily concentrated on sarbecoviruses, SARS-CoV and SARS-CoV-2, revealing through immunofluorescence, electron microscopy and tomography imaging studies that the host cellular membranes undergo remodeling during viral infection17,18. Additionally, structures have been reported for both apo and inhibitor bound SARS-CoV-2 M protein3,8,9,19,20, showing the apo M protein can form a homodimer in either a short or long conformation, while inhibitor-bound structures are locked in either a short or altered conformation between the long and short form. The development of M protein inhibitors targeting SARS-CoV-2 is relatively new and remains an active area of research. Currently, two first-in-class, orally efficacious inhibitors are reported to target the M protein of SARS-CoV-2, JNJ-9676 and CIM-83419,20. Both inhibitors have overlapping binding sites and block the conformational switch of the M protein from short to long form, thus preventing the assembly of infectious virions during the late stages of the replication cycle. In vitro studies have shown that JNJ-9676 exhibits activity against SARS-CoV-2 and SARS-CoV with an EC50 of approximately 20 nM and modest activity against MERS-CoV with an EC50 of 600 nM19. In contrast, CIM-834 has an EC50 of 112 nM against SARS-CoV-2 and 640 nM for SARS-CoV but lacks activity against MERS-CoV20. Notably, the induced conformation of the M protein when bound to JNJ-9676 is significantly different from the short conformation observed with CIM-834, attributed to additional lipophilic interactions of JNJ-9676 with the induced pocket in the dimeric transmembrane domain. Both inhibitors serve as promising starting points for further optimization to broaden the antiviral spectrum to other coronaviruses. To gain a clearer understanding of the structural variations among the M proteins of betacoronaviruses, we concentrated our study on human coronaviruses that have caused significant outbreaks in recent years: SARS-CoV-2, SARS-CoV and MERS-CoV. SARS-CoV and SARS-CoV-2 share approximately 80% identity at the genome level, while MERS-CoV exhibits about 50% similarity21. SARS-CoV and SARS-CoV-2 are closely related and belong to the sarbecovirus subgenus of betacoronaviruses22, whereas MERS-CoV has a distinctly divergent evolutionary trajectory from SARS-CoV and SARS-CoV-2, and belongs to the merbecovirus subgenus11.
Here, we build on previous work on SARS-CoV-2, and present cryo-electron microscopy (cryo-EM) structures of apo MERS-CoV and SARS-CoV M protein. We also generated an inhibitor-bound structure of the SARS-CoV M protein to specifically interrogate the binding pocket that has been recently described for JNJ-967619. With this work, we highlight key structural differences in the MERS-CoV and SARS-CoV M proteins as observed from comparisons with their SARS-CoV-2 homolog. This work establishes a foundation for future efforts in structure-based drug discovery for the development of specific MERS-CoV and broad-spectrum antivirals targeting betacoronavirus M proteins.
Results
Design and evaluation of SARS-CoV and MERS-CoV FAb complexes for structural studies
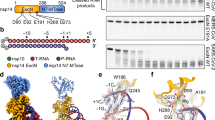
The M protein mirrors the overall genomic similarity within betacoronaviruses; the M protein of SARS-CoV-2 shares 90% sequence identity with that of SARS-CoV, whereas MERS-CoV exhibits 40% and 43% sequence identity with SARS-CoV-2 and SARS-CoV, respectively (Fig. 1a, b, Supplementary Fig. 1). To expand the structural understanding of M protein beyond SARS-CoV-2, we aimed to determine the structures of SARS-CoV and MERS-CoV M protein.
a Sequence alignment of M protein. b Sequence identity matrix of M protein. c Domain architecture of M proteins, highlighting designed MERS mutations as space-filling models (pink) at the M protein (protomer A—blue, protomer B—orange) and FAb B (gray) interface; the viral envelope is indicated with dashed lined (PDB: 8W2E).
Given that the M protein can adopt both short and long conformations2,7,8 and based on our previous cryogenic electron microscopy (cryo-EM) work with the SARS-CoV-2 M protein, where we successfully used a short form conformation-specific antibody fragment (FAb B) as a fiducial marker to improve particle alignment19, we applied a similar approach for SARS-CoV and MERS-CoV M proteins. The residues at the binding interface of M protein-FAb B exhibit a sequence identity that parallels the overall M protein sequence identity, with SARS-CoV and MERS-CoV showing 87% and 39% sequence identity, respectively (Supplementary Fig. 2a). We engineered and purified recombinant wild-type SARS-CoV and MERS-CoV M proteins (Fig. 1c, Supplementary Fig. 3a) and assessed their binding to FAb B. As anticipated based on the sequence identity at the M protein-FAb B interface, SARS-CoV effectively formed a complex with FAb B, whereas MERS-CoV did not (Supplementary Fig. 3b).
To achieve a MERS-CoV M protein-Fab B complex, the importance of SARS-CoV-2 M protein residues for FAb B interaction were assessed by estimating the change in affinity resulting from single point mutations to alanine using the Molecular Mechanics Generalized Born Surface Area (MM-GBSA) approach23,24. Estimated affinity changes greater than 1 kcal/mol were used to prioritize interface residues for mutation design (Fig. 1c, Supplementary Table 1). Two MERS-CoV mutant M protein constructs were designed (Supplementary Table 2), wherein MERS-CoV M protein residues were replaced with corresponding SARS-CoV-2 M protein residues and subsequently screened for FAb B binding. Of the two mutant MERS-CoV M protein constructs, one, featuring seven mutations (K199R, N145R, T127I, N165K, H196S, A200I, A177Y), successfully formed a complex with FAb B, and will be addressed as MERSmut-CoV M protein going forward (Fig. 1c, Supplementary Fig. 2, Supplementary Fig. 3a, b).
Cryo-EM structure of SARS-CoV M protein is similar to SARS-CoV-2
We acquired a cryo-EM structure of apo SARS-CoV M protein-FAb B at 3.23 Å resolution (protein data bank [PDB]: 9NZ4; Fig. 2a, Supplementary Fig. 4). Like apo SARS-CoV-2 M protein (PDB: 7VGS), the apo SARS-CoV M protein forms a dimer that adopts a short form conformation, with a root-mean-square deviation (RMSD) value of 1.73 Å. The transmembrane domain (TM), TM1 in one protomer is domain-swapped to form a three-helix bundle with TM2 and TM3 of the second protomer3. Although the three-helix bundle is largely consistent, minor variations in the side chain orientations of Phe36, and Trp57 were observed, while Met32 represents a mutation from the Cys33 residue found in SARS-CoV-2 M protein (Fig. 2a).
a Superimposed structural comparison of apo SARS-CoV-2 M protein (gray, PDB:7VGS) versus apo SARS-CoV M protein (orange, PDB: 9NZ4) in cartoon ribbon representation, highlighting JNJ-9676 binding pocket residues as sticks and lines. Key residue differences are highlighted with (*). b Superimposed structural comparison of apo SARS-CoV-2 (protomer A gray, protomer B blue—gray, PDB: 7VGS), apo SARS-CoV (protomer A orange, protomer B green, PDB: 9ZN4) and apo MERSmut-CoV (protomer A cyan, protomer B, blue, PDB: 9NZ5) in cartoon ribbon representation. SARS-CoV and MERSmut-CoV M proteins were aligned to SARS-CoV-2 (PDB: 7VGS). The N-terminal regions of the TMs are highlighted. c Superimposed JNJ-9676 binding pocket formed by protomer B TM1, protomer A TM2 and TM3 shown by cartoon ribbon representation (coloring as shown in (b)), highlighting key differences in the binding pocket residues shown as sticks and lines. d, Two-dimensional (2D) interaction pattern of JNJ-9676 and SARS-CoV-2 M protein (protomer A: gray, protomer B: blue-gray), and SARS-CoV M protein (protomer A: orange, protomer B: green), highlighting key residue differences as shown in (b), with MERS-CoV M protein (protomer A: cyan, protomer B: blue) differences shown with (*), and interactions are indicated with dashed lines (π-π stacking: purple).
MERSmut-CoV Cryo-EM structure reveals differences in the transmembrane portion of JNJ-9676 binding pocket
Single-particle cryo-EM analysis of apo MERSmut-CoV M protein-FAb B complex resulted in a three-dimensional (3D) reconstruction with a global resolution of 3.15 Å (Fig. 2b–d, Supplementary Fig. 5), displaying C1 symmetry and a short form conformation of the M protein dimers.
Apo MERSmut-CoV M protein has significant movement of the cytoplasmic termini of transmembrane (TM) domains in both protomers compared with the apo SARS-CoV-2 (PDB: 7VGS, RMSD 3.86 Å, Fig. 2b) and apo SARS-CoV M proteins (PDB: 9NZ5, RMSD 4.10 Å, Supplementary Fig. 6). Specifically, protomer A of MERSmut-CoV shows conformational changes in transmembrane domain 1 (TM1) with shifts of 3.3 and 3.5 Å, TM2 with shifts of 5.4 and 6.3 Å, and TM3 with shifts of 7.4 and 8.1 Å compared to SARS-CoV-2 and SARS-CoV, respectively, when structures are aligned to SARS-CoV-2 (PDB:7VGS) (Fig. 2b). Notably, protomer B of MERSmut-CoV exhibits less pronounced conformational shifts for TM2 (1.9 and 3.2 Å shift) than protomer A, while TM1 (6.4 and 6.2 Å shift) and TM3 (3.2 and 4.2 Å shift) of protomer B show significant movement from SARS-CoV-2 and SARS-CoV, respectively.
MERSmut-CoV M protein, in its short form conformation, protomer B TM1 forms a three-helix bundle with protomer A TM2 and TM3, as seen with SARS-CoV-2 and SARS-CoV; however, there are key differences in the northern lipophilic pocket between the proteins (Fig. 2c, Supplementary Fig. 7). For instance, in TM1, MERSmut-CoV has a polar uncharged Thr31 residue which differs from SARS-CoV-2 Ile32 and SARS-CoV Ile31, that participate in hydrophobic interactions when bound to JNJ-9676 (Fig. 2d). Other notable variations in the side chains of TM1 MERSmut-CoV are Ile32, Gln35, and Phe36, for which the corresponding residues in SARS-CoV-2 and SARS-CoV form hydrophobic aromatic-aromatic interactions with JNJ-9676 (Fig. 2d). Furthermore, due to the conformational shifts of the TMs for MERSmut-CoV, TM2 residue Trp57 is shifted deeper in the lipophilic pocket, while Ser60 side chain faces away compared to SARS-CoV-2 and SARS-CoV (Fig. 2c, d). Finally, MERSmut-CoV TM3 residues V86 and S87 show minor shifts compared to SARS-CoV-2 and SARS-CoV (Fig. 2c).
In the short form conformation of SARS-CoV-2 M protein (PDB: 7VGS), Glu115, a hinge region residue important in the structural transition between long and short conformation, forms a hydrogen bond with Tyr47 in the TM1-TM2 loop, while in the long form (PDB: 7VGR), Glu115 forms a hydrogen bond with Ala403. SARS-CoV maintains the corresponding hinge region Glu114-Tyr46 hydrogen bond in both protomers, whereas MERSmut-CoV M protein exhibits variations. In MERSmut-CoV protomer A, Glu114 could potentially form a weak hydrogen bond interaction with Tyr46, while in protomer B, both residues flip outwardly, disrupting this interaction. (Supplementary Fig. 6).
To study the effect of FAb B on the structure of the MERS-CoV M protein dimer, and to assess the impact of the mutations introduced in MERSmut-CoV on the stability of the dimer short form conformation, we performed molecular dynamics (MD) simulations of the wild-type MERS-CoV M protein dimer model initiated from the cryo-EM structure (PDB: 9NZ5). The analysis of the MD trajectories shows that the short form conformation is locally stable for wild-type MERS-CoV in absence of FAb B (Supplementary Fig. 2b), with an average RMSD over C-alpha atoms of 3 Å from the MERSmut-CoV Cryo-EM structure.
Investigation of the JNJ-9676 binding pocket in SARS-CoV
We previously reported on JNJ-9676 (Fig. 3a), a small molecule inhibitor that targets SARS-CoV-2 and SARS-CoV M protein as well as other sarbecoviruses; compound binding stabilized the M protein in an altered conformational state, preventing the release of infectious virus19. JNJ-9676 demonstrated in vitro activity against SARS-CoV-2 and SARS-CoV in infected cells with an EC50 of 14–26 nM and 20 nM, respectively, and modest activity against MERS-CoV with an EC50 of 600 nM19.
a Chemical structure of JNJ-967619. b nanoDSF melting temperatures (Tm) of M protein dimethyl sulfoxide (DMSO) versus JNJ-9676; table indicates average Tm, ΔTm and standard deviations from three technical replicates. Mean differences between groups are calculated using paired t-test Holm-Sidak method where ** P ≤ 0.01, * P ≤ 0.05, ns P > 0.05. c Superimposed structural comparison of JNJ-9676 (orange) bound SARS-CoV-2 M protein (gray, PDB:8W2E) versus JNJ-9676 (magenta) bound SARS-CoV M protein (cyan, PDB: 9NZ3) in cartoon ribbon representation, highlighting JNJ-9676 binding pocket residues as sticks and lines. Key residue differences are highlighted with (*), and interactions are indicated with dashed lines (H-bonding: yellow; π-π stacking: green).
We used a nano differential scanning fluorimetry (nanoDSF) method to confirm JNJ-9676 target engagement with the purified recombinant proteins. SARS-CoV-2 and SARS-CoV M proteins were stabilized in the presence of JNJ-9676 (ΔTm = 2.59 ± 0.18 °C, ΔTm = 1.87 ± 0.21 °C, respectively), but no stabilization was observed for the MERS-CoV and MERSmut-CoV M proteins, both of which exhibited similar melting patterns (Fig. 3b, Supplementary Fig. 3c).
In addition, similar to findings with JNJ-9676 and SARS-CoV-219, selection of compound-resistant SARS-CoV mutants by serial passaging in the presence of increasing doses of JNJ-9676 led to the accumulation of mutations in the SARS-CoV M protein, which were absent when viruses were passaged without drug pressure (Supplementary Table 3). Introduction of the mutation that was most frequently found in independent compound-resistant lineages, P131S, resulted in an increase of the EC50 value of JNJ-9676 of up to 60-fold, relative to the parental SARS-CoV cDNA clone-derived virus (Supplementary Table 4, Supplementary Fig. 8).
We acquired a 3.07 Å cryo-EM structure of SARS-CoV M protein-FAb B complexed with JNJ-9676 (PDB: 9NZ3; Fig. 3c, Supplementary Fig. 9), processed with C1 symmetry. In the samples of MERSmut-CoV M protein incubated with JNJ-9676, no electron density for the compound was observed.
Upon binding with JNJ-9676, the overall structures of the SARS-CoV and SARS-CoV-2 M proteins remain largely comparable (RMSD 1.82 Å). JNJ-9676 interacts with an induced pocket within both proteins, leading to a conformational shift from the apo state that stabilized the M protein in an inhibited configuration19 (Fig. 3c). The JNJ-9676 binding pocket consists of a southern polar pocket (Asn116/117, Ser98/99), and a northern lipophilic pocket between the helices of the two protomers in the transmembrane domain, which interacts with the difluorophenylmethyl group (Supplementary Fig. 7). Gln35 and Tyr94 flip out of the pocket to accommodate compound binding in SARS-CoV M protein, suggesting a similar mode of binding for both M proteins19. JNJ-9676 establishes similar interactions with residues in SARS-CoV M protein, as seen in the SARS-CoV-2 protein (PDB: 8W2E), though there are discernible differences in the orientation of residues such as Met32 and Met83 in SARS-CoV compared to their counterparts Cys33 and Met84 in SARS-CoV-2. In SARS-CoV, Met32 shifts closer to the CF2 linker of JNJ-9676 with respect to the position of the Cys33 side chain in SARS-CoV-2. Met83 in SARS-CoV remains positioned away from the binding pocket, similar to the apo structure (8.4 Å from JNJ-9676), whereas in SARS-CoV-2, Met84 rotates towards JNJ-9676 to make hydrophobic interactions (3.9 Å from JNJ-9676). Notably, a water electron density was identified in chain A of JNJ-9676-bound SARS-CoV M protein which was not previously observed in SARS-CoV-2, that facilitates hydrogen bonds between N116, S98, N111and the oxygen atom of the JNJ-9676 exocyclic amide.
Discussion
Over the last two decades there have been several outbreaks caused by coronaviruses, the most recent of which, COVID-19, caused by SARS-CoV-2, resulted in a global pandemic with more than 700 million confirmed cases and just over 7 million confirmed deaths based on the World Health Organization. The M protein of coronaviruses is an attractive target for antiviral therapeutics, as M proteins are well-conserved in lineages within the betacoronavirus subgenus and plays a critical role in viral replication and assembly. The lower sequence conservation for the M protein between betacoronavirus subgenus’ suggests that developing a broad-spectrum antiviral targeting the M protein of both sarbecoviruses such as SARS-CoV-2 and SARS-CoV as well as more distantly related merbecoviruses such as MERS-CoV is challenging. Our work reveals structures of SARS-CoV and MERSmut-CoV M proteins that are structurally similar to previously determined SARS-CoV-2 M protein structures; however, we have identified differences in the relative arrangement of the TM helices, that are forming lipophilic interactions with JNJ-9676 both for SARS-CoV-2 and SARS-CoV.
As with SARS-CoV-2, each subunit of SARS-CoV M protein forms a dimer, with three transmembrane helices and a C-terminal beta-sheet domain. Apo SARS-CoV M protein adopts a short conformation, further supporting evidence that M proteins have a conserved structural fold. In the compound-bound SARS-CoV M protein structure, subtle differences at Met32 and Met83 were observed in the lipophilic transmembrane pocket compared to SARS-CoV-2. Together, these observations suggest that targeting the northern lipophilic region of the pocket placed between TM1 and TM2 could be key for broad-spectrum activity against SARS-CoV-2, SARS-CoV and MERS-CoV M proteins.
JNJ-9676 has approximately 30-fold weaker antiviral activity for MERS-CoV M protein than SARS-CoV-2 and SARS-CoV19 and our structural analysis showed no compound density for MERSmut-CoV M protein samples incubated with JNJ-9676. This is also consistent with nanoDSF results in which no stabilization of MERS-CoV or MERSmut-CoV M protein is observed in the presence of JNJ-9676, pointing to weak binding. Significant conformational shifts of the TMs in the apo MERSmut-CoV M protein structure were observed compared to SARS-CoV-2 and SARS-CoV. MERSmut-CoV M protein differs from SARS-CoV-2 and SARS-CoV in 6 of 22 binding pocket residues19, primarily at TM1 residues (Thr32, Ile32, Tyr37). There are minor variations in secondary binding pocket residues at TM2 (Ser60) and TM3 (Val86, Ser87); however, these residues do not directly interact with JNJ-9676 in the SARS-CoV-2 and SARS-CoV M proteins. Conversely, the TM2 (residues 55-59), TM3 (residues 91-99) and hinge region between TM3 and the C-terminal beta-sheet domain (residues 107-118) in MERSmut-CoV are conserved in SARS-CoV-2 and SARS-CoV M proteins. In the M protein structures of SARS-CoV-2 and SARS-CoV bound to JNJ-9676 (PDB: 8W2E, 9NZ3), the TM2 residues engage with the benzonitrile group while the TM3 residues and the hinge region interact with the bicyclic core of JNJ-9676, respectively. These structural and sequence differences may represent a challenge for the design of broad-spectrum inhibitors, that could potentially be addressed by targeting interactions with transmembrane lipophilic residues or conserved polar residues such as Ser60/61. The MERSmut-CoV structure also reveals MERS-specific polar residues in the northern pocket that could be targeted (Thr31/Ser87). Overall, this data highlights the structural distinctions between the MERS-CoV, SARS-CoV-2 and SARS-CoV M proteins, which could aid in the development of effective antivirals that target a broader range of viruses beyond sarbecoviruses.
The TM1-TM2 loop and the hinge region of TM3 have been implicated in playing an important role in the structural transition between the short and long form conformations of the SARS-CoV-2 M protein3. The re-arrangement of the helical bundles and C-terminal beta-sheet domain, in these conformations is specifically attributed to Glu115 in the hinge region. Structures of apo SARS-CoV and MERSmut-CoV M protein were captured in a short form conformation, providing further structural evidence that this conformation of M protein is conserved across betacoronaviruses3,8,19. Interestingly, the SARS-CoV M protein maintains the hydrogen bond between the corresponding residues of Glu114 and Tyr46, as observed in the short form conformation of SARS-CoV-2 M protein, with Glu115 and Tyr47 (PDB: 7VGS), whereas the MERSmut-CoV M protein exhibits variations in this interaction. In MERSmut-CoV protomer A, Glu114 potentially forms a weak hydrogen bond interaction with Tyr46, while in protomer B, both residues flip outwards, disrupting this interaction. (Supplementary Fig. 6). This suggests that Glu114 could be more dynamic for MERSmut-CoV M protein than SARS-CoV and SARS-CoV-2 and possibly reflects the equilibrium between the known long and short conformations. The mutations in the C-terminal beta-sheet domain of the MERSmut-CoV M protein are solvent-exposed and distant from Glu114, suggesting the mutations are likely not responsible for the differences observed between the MERSmut-CoV protomer transmembrane domains. MD simulations of MERS-CoV M protein, in which MERSmut-CoV (PDB: 9NZ5) mutations were reversed to WT residues, showed that its short form fold is stable in absence of the FAb. It should also be noted that while SARS-CoV-2 M protein datasets (PDB: 7VGS, 7VGR, 8CTK, 9EXA) were processed using C2 symmetry8,20,25, SARS-CoV-2, SARS-CoV and MERSmut-CoV datasets (PDB: 8W2E, 9NZ3, 9ZN3, 9ZN5) were processed with C1 symmetry. In our hands, C2 symmetry led to poor density maps for previous JNJ-9676 bound structures and for the MERSmut-CoV structure. C1 processing allowed us to detect subtle differences in the asymmetry of the MERSmut-CoV M protein dimer. Although this asymmetry was not observed in the JNJ-9676 bound and apo SARS-CoV datasets processed with C1 symmetry, processing the data with C2 symmetry did not notably enhance map quality or resolution.
In conclusion, our work provides the framework for understanding the structural conservation of the M protein across betacoronaviruses. The overall structural fold between SARS-CoV-2, SARS-CoV, and MERSmut-CoV is conserved, suggesting the function of the M protein between lineages within the betacoronavirus genus are similar. The conformation induced by binding of JNJ-9676 to SARS-CoV-2 is conserved in SARS-CoV and the key polar interactions with Asn117 and Ser99 and the stacking interaction with Tyr95 are maintained. While the lipophilic interactions with TM1, TM2 and TM3 transmembrane helices are also conserved, we highlight differences in the arrangement of side chains for SARS-CoV. The structure of the apo MERSmut-CoV M protein is the first human merbecovirus M protein structure and showed several significant conformational shifts of the transmembrane domains and differences in binding pocket residues and side chain orientations. The southern polar pocket is mostly conserved across coronaviruses and can be used for the design of broad-spectrum antiviral, while the northern lipophilic pocket is less conserved for MERS-CoV M protein and could be leveraged for the design of both specific and broad acting MERS-CoV antivirals. Taken together, the structural insights presented in this work provide a rationale for structure-guided antiviral design against MERS-CoV as well as broad-spectrum M protein inhibitors as potential therapeutics for emerging coronaviruses in the future.
Methods
Ethics and inclusion statement
The research process was a collaboration between Europe-based and United States of America researchers. Roles and responsibilities were defined between the authors based on their specific area of expertise to ensure the highest quality of standards. All experiments using pathogens were conducted in the appropriate biosafety containment level labs.
Compounds
The synthesis of JNJ-9676 is described in patent WO-2024/00890919. For in vitro experiments, JNJ-9676 was dissolved in 100% dimethyl sulfoxide (DMSO) as a 5–50 mM stock.
In vitro resistance selection assay and site-directed mutant virus generation
SARS-CoV virus Frankfurt strain FFM1; (GenBank Accession Number: AY291315) was used for in vitro resistance selection experiments19. Three independent IVRS runs have been performed in A549-hACE2 cells, where cells were seeded at 5000 cells/well in a 96-well plate and inoculated with SARS-CoV at MOI of 0.01 or 0.02 in the presence of compound or DMSO. Compound resistant viral samples were collected for RNA extraction followed by next generation sequencing. Only samples with an average coverage exceeding 1000 reads per position were included for further bioinformatic analysis. A read frequency threshold of 15% was established for variant calling. Mutations from compound-resistant viruses were filtered by comparing them to the virus control samples to exclude any potential mutations related to cell adaptation.
To confirm the mutations identified from the resistance selection are correlated to the compound resistance mutations of interest were introduced in the SARS-CoV full genome via circular polymerase extension reaction (CPER) reverse engineering platform19. CPER-primers with an overlapping region of more than 20 bp were designed using Geneious software (Supplementary Table 4). For site-directed mutagenesis, the desired mutation was first introduced in the corresponding fragment via overlap extension PCR followed up by full-genome assembly by CPER. For recovery of the mutant virus as well as the wild-type viruses, the above generated full-length circular DNA (up to 2.5 µg) was transfected directly in BHK-21 followed by re-infection of Vero E6 using the virus-containing BHK-21 supernatants. Recombinant viruses were harvested upon appearance of clear cytopathic effects (CPE). The impact of these mutations on the antiviral activity of JNJ-9676 was assessed by an HCI-based antiviral assay, in which cells were fixated with formaldehyde at room temperature for 20 min, washed, permeabilized, blocked and stained for spike protein, nucleus, whole cell, and lysosomes26. Following staining, plates were analyzed by HCI on the Cell Voyager 8000 (Yokogawa) confocal microscope. The analysis was performed with Phaedra HCI analysis software (version 1.0.10.202309011029). The fold-change in EC50 for a mutant virus compared to recombinant WT virus was determined.
Sequence alignment phylogenetic tree
The amino acid sequences for the M proteins were downloaded from Uniprot (P0DTC5, P59596, R9UNX5) and aligned followed by consensus tree generation using unweighted pair group method with arithmetic mean (UPGMA) using Biomatters Geneious Prime v2024.03 software.
Alanine scanning mutation analysis
The original PDB structure for the SARS-CoV-2 M protein dimer (PDB: 7VGS) was downloaded and prepared using Preparation Wizard in Maestro (Schrodinger Release 2022-4; Schrodinger). The ionization states of the residues were predicted using Propka at pH 7.0 and the resulting structures were minimized with restraints on the heavy atom using the OPLS4 force field, with a convergence cutoff on the heavy-atom root-mean-square deviation of 0.3 Å.
The importance of the M protein residue for Fabs interactions in the short and long conformations was assessed estimating the change in the binding free energy ΔΔGpred of the M protein/Fabs complex resulting from single point mutations to Alanine, using the Residue Scanning24 module in BioLuminate (Schrodinger Release 2022-4). A threshold on the estimated affinity change greater than 1 kcal/mol was used to prioritize the interface residues that are most important for the binding of the M protein and FAb, which guided subsequent mutation design of MERSmut M protein targeting binding to Fab B. Residue analysis is reported in Supplementary Table 1.
Cloning, protein expression and purification of SARS-CoV-2 M, SARS-CoV M, MERS-CoV M, Fab B
The gene encoding Fab B, SARS-CoV-2 M protein (1-222, Uniprot: P0DTC5), SARS-CoV M protein (1–221, Uniprot: P59596), MERS-CoV M protein (1-219, Uniprot: R9UNX5) and MERSmut-CoV M (1-219, K199R, N145R, T127I, N165K, H196S, A200I, A177Y, Uniprot: R9UNX5) and FAb B3 were cloned, transfected and purified19.
M proteins were transfected into Expi293F cells (Invitrogen) and incubated at 37 °C with 8% CO2 for 72 h, collected by centrifugation and frozen at −80 °C. Cells pellets were thawed, re-suspended in Buffer 1 (20 mM HEPES pH 7.5, 250 mM NaCl, 5% glycerol [v/v], protease inhibitor [Roche], 50 unit/mL nuclease), homogenized using a glass dounce homogenizer and lysed using a M110Ymicrofluidizer (Microfluidics). Cell lysates were centrifuged at 167,900×g for 1 h, and collected membranes were re-suspended in Buffer 1 and solubilized for 2 h at 4 °C with 1% (w/v) lauryl maltose neopentyl glycol (LMNG, Anatrace), and 0.1% (w/v) cholesteryl hemisuccinate (CHS, Anatrace). The supernatant was collected by centrifugation at 167,900×g and incubated with C-tag resin (Thermo Scientific) for 2 h at 4 °C. The resin was washed with 10 CV of Buffer 2 (20 mM HEPES pH 7.5, 250 mM NaCl, 1.25% glycerol [v/v], 1 mM EDTA, 0.0025% LMNG [w/v], 0.00025% CHS [w/v]), and eluted with 3 CV of Buffer 3 (as Buffer 2, 3 mM C-tag peptide). Proteins were further purified on a Superose 6 Increase 10/300 GL column (Cytiva) in Buffer 4 (20 mM HEPES pH 7.5, 150 mM NaCl, 0.001% LMNG [w/v], 0.0001% CHS [w/v], 0.00033% glycol diosgenin [GDN; w/v].
FAb B heavy and light chains were co-transfected into Expi293F cells (Invitrogen) and incubated for 96 h at 37 °C with 8% CO2. Conditioned media was loaded onto a 10 mL HisTrap excel column (Cytiva), column was washed with 6 CV of buffer (20 mM sodium phosphate pH 6.5, 150 mM NaCl, 20 mM imidazole) and eluted over 5CV using an imidazole gradient with buffer (20 mM sodium phosphate pH 6.5, 150 mM NaCl, 500 mM imidazole). FAb B was further purified on a HiLoad 16/600 Superdex 75 pg column (Cytiva) in buffer (20 mM HEPES sodium phosphate pH 6.5, 150 mM NaCl).
Purification and formation of SARS-CoV M/MERS-CoV M-FAb B complex
SARS-CoV M or MERSmut-CoV M and Fab B were mixed in a 1:2.5 ratio and incubated on ice for 1 h. The complexes were loaded into a Superose® 6 Increase 10/300 GL column (Cytvia) with buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.001% LMNG [w/v], 0.0001% CHS [w/v], 0.00033% GDN [w/v]). Peak fractions containing the complex assessed by SDS-PAGE was pooled. Samples were prepared ±100 µM JNJ-9676 and incubated for 1 h on ice. The sample was concentrated and prepared at 0.5–4 mg/mL with size exclusion chromatography (SEC) buffer ±100 µM JNJ-9676 for cryo-EM.
Nano differential scanning fluorimetry
Experiments were performed in a total volume of 10 µL. A Prometheus PANTA instrument (NanoTemper Technologies) was used to measure the melting temperatures. Samples were prepared in 0.5 mL microcentrifuge tubes with 0.5 mg/mL of each purified recombinant SARS-CoV-2, SARS-CoV, MERS-CoV, MERSmut-CoV, with 100 µM of JNJ-9676 in 20 mM HEPES pH 7.5, 150 mM NaCl, 0.001% LMNG (w/v), 0.0001% CHS (w/v), 0.00033% GDN (w/v), and 1% DMSO (v/v). The samples were loaded into standard-grade glass capillaries were measured in temperature range 20–95 °C with a temperature gradient of 1 °C/min and the intrinsic protein fluorescence at 330 nm and 350 nm was recorded using PR.PANTA.control (3 technical replicates). The data was analyzed with PR.PANTA.analysis (NanoTemper Technologies) and GraphPad Prism v8.4.2
Cryogenic electron microscopy (cryo-EM)
Quantifoil Au 1.2/1.3 300 mesh grids were subjected to glow discharge for 15 s at 15 mA using PELCO easiGlow Discharge Cleaning System. Three micrlitres of SARS-COV complex (1, 1.5 mg/mL) ± 100 µM JNJ-9676 and MERSmut-CoV complex (3 mg/mL), prepared as described above, were applied to the EM grids, and vitrified with a Vitrobot (Thermo Fisher Mark IV) using the settings: blot time 3 s, blot force 2, wait time 0 s, inner chamber temperature 4 °C, and 100% relative humidity. Flash-freezing in liquid ethane cooled by liquid nitrogen was performed.
Cryo-EM data collection for SARS-CoV was automated on a 200 kV Thermo Scientific™ Glacios™ microscope controlled by EPU software. Micrographs were taken at 105,000× magnification using a Falcon4 detector (Gatan) in counting mode. Each 6 s exposure recorded 40 frames with a total dose of 40 electrons/Å2. The calibrated physical pixel size for all digital micrographs was 0.910 Å. Cryo-EM data collection for MERSmut-CoV was automated on a 300 kV Thermo Scientific Titan Krios microscope with a post-column Gatan Image Filter (GIF), equipped with a Gatan K3 Summit direct electron detector and an energy filter slit width of 20 eV. Images were recorded with Leginon in counting mode with a pixel size of 0.831 Å and a nominal defocus range of −1.4 to −2.3 µm. The images were acquired with a 1.5 s exposure and 40 ms subframes (38 total frames), corresponding to a total dose of 51.96 electrons/Å2. All details corresponding to individual datasets are summarized in Table 1.
Cryo-EM data collection and image quality were monitored using cryoSPARC Live v4.2.1 for SARS-CoV datasets and cryoSPARC Live v4.6.0 for MERSmut-CoV dataset. Image pre-processing steps, including patch motion correction, patch contrast transfer function (CTF) estimation, blob particle picking (100–200 Å diameter), and extraction, were performed simultaneously. A total of 7573 and 8950 raw micrographs were recorded for apo and JNJ-9676 SARS-CoV, respectively, with the Glacios microscope and 19, 995 raw micrographs were recorded for MERSmut-CoV with the Titan Krios microscope. Acceptable 2-dimensional (2D) classes served as templates for particle repicking. One round of live 2D image classification yielded approximately 1,589789 and 1,294,716 good particle images for apo and JNJ-9676 SARS-CoV, respectively and 2,739, 392 particle images for the MERSmut-CoV dataset. These particles were used for three-dimensional (3D) reconstruction for each dataset using C1 symmetry. Four, five and five starting 3D models were calculated, for apo, JNJ-9676 SARS-CoV and MERSmut-CoV datasets, respectively, resulting in one major 3D class. One major class underwent non-uniform 3D refinement and local refinement using 1,210,771, 674,469, and 827,876 particles and was further refined to a 3D EM map with an average resolution of 3.23 Å, 3.07 Å, 3.15 Å for apo, JNJ-9676 SARS-CoV and MERSmut-COV, respectively.
Resolutions were estimated by applying a soft mask around the protein complex density using the gold standard (two halves of data refined independently) Fourier shell correlation (FSC) = 0.143 criterion. Prior to visualization, all density maps were sharpened by applying different negative temperature factors along with the half maps and used for model building. Local resolution was determined using ResMap. Detailed statistics about the cryo-EM data processing can be found in Supplementary Figs. 4, 5, and 9.
Cryo-EM modeling, refinement, and validation
Human SARS-CoV-2 M protein dimer (short form) in complex with Fab B (PDB: 7VGS) was used as the initial model for atomic model building of the apo EM maps of SARS-CoV and MERSmut-COV, while PDB: 8W2E was used as the initial model for JNJ-9676 bound SARS-CoV map. The M protein and Fab B were fitted into the 3D map using DiffModeler27 (apo SARS-CoV, MERSmut-CoV) and Chimera28 (JNJ-9676 SARS-CoV) and then residues were mutated (JNJ-9676 SARS-CoV) and further refined manually with COOT followed by real-space refinement in Phenix29 (all datasets). Detailed data collection and structural refinement statistics are provided in Table 1. Structure representations were generated using Pymol v4.6.030 and Chimera28.
MERS-CoV molecular dynamics simulations and trajectory analysis
The starting structure for the MERS M protein dimer was extracted from the experimental Cryo-EM model after removing Fabs. The terminal residues were capped. The dimer was positioned in a square 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC) membrane using the addMembrane OpenMM method and the trimBox.py scripts31. The system was solvated with TIP3P water and 0.15 M KCl. The amberff14SB32 and lipid1733 forcefields were respectively used for the protein and the lipids. All simulations were performed with OpenMM v8.1.234. The solvated protein/membrane system was minimized for 1000 steps, and temperature was gradually taken to 310 K before equilibrating the system for 5 ns in the NVT ensemble, with restraints placed on non-solvent heavy atoms. The restraints on lipids atoms were gradually relaxed during 15 ns of equilibration in the NPT ensemble, and the restraints on protein atoms were relaxed during additional 20 ns of simulation. The system was then simulated for 500 ns in the NPT ensemble. Hydrogen mass repartitioning and a 4 fs time step were used with the OpenMM Middle Langevin integrator35 and a coupling constant of 1.0 ps−1. All bonds were constrained. Structural snapshots were saved every 100 ps. This simulation protocol, including the initial 40 ns equilibration step, was repeated for 10 independent replicates using a different seed for the random number generator, for an accumulated simulation time of 5 µs. Conformational clusters were calculated from the merged trajectories using TTClust36 with default parameters. The optimal number of clusters was determined by inspection of the dendrogram of the hierarchical distance between frames. This analysis identified three main clusters, shown in Supplementary Fig. 2b.
Statistics and reproducibility
NanoDSF statistical analyses were performed in GraphPad v10.3.1 by a paired t-test using the Holm-Sidak method with a significance level of 0.05. Nonlinear regression analysis was used to derive SDM potency and dose response values by fitting inhibition data to a four-parameter logistic equation. Descriptive statistics were used to summarize the data, including mean and standard deviation, as indicated in relevant figure legends and tables. Refer to the legends for the number of independent or technical replicates performed. Technical replicates were performed on different days or at different times of the day.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank (accession codes: EMD-49949, EMD-49950, EMD-49951), while the atomic coordinates have been deposited in the Protein Data Bank (accession codes: 9NZ3, 9NZ4, 9NZ5). Numerical source data is provided in the Supplementary Data. Other data are available from the corresponding author on reasonable request. The uncropped images of the SDS-PAGE shown in Supplementary Fig. 3a are presented in Supplementary Fig. 10.
Code availability
In this paper, no custom code or mathematical algorithms were used.
References
Bracquemond, D. & Muriaux, D. Betacoronavirus Assembly: Clues and Perspectives for Elucidating SARS-CoV-2 Particle Formation and Egress. mBio 12, e0237121 (2021).
Rottier, P. J. M. in The Coronaviridae (ed Stuart G. Siddell) 115–139 (Springer US, 1995).
Zhang, Z. et al. Structure of SARS-CoV-2 membrane protein essential for virus assembly. Nat. Commun. 13, 4399 (2022).
Minigulov, N., Boranbayev, K., Bekbossynova, A., Gadilgereyeva, B. & Filchakova, O. Structural proteins of human coronaviruses: what makes them different?. Front. Cell Infect. Microbiol 14, 1458383 (2024).
Katiyar, H., Arduini, A., Li, Y. & Liang, C. SARS-CoV-2 assembly: gaining infectivity and beyond. Viruses 16, 1648 (2024).
Boson, B. et al. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 296, 100111 (2021).
Hardenbrook, N. J. & Zhang, P. A structural view of the SARS-CoV-2 virus and its assembly. Curr. Opin. Virol. 52, 123–134 (2022).
Dolan, K. A. et al. Structure of SARS-CoV-2 M protein in lipid nanodiscs. eLife 11, e81702 (2022).
Wang, X., Yang, Y., Sun, Z. & Zhou, X. Crystal structure of the membrane (M) protein from a bat betacoronavirus. PNAS Nexus 2, pgad021 (2023).
Neuman, B. W. et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 174, 11–22 (2011).
Cui, J., Li, F. & Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 17, 181–192 (2019).
Marra, M. A. et al. The Genome Sequence of the SARS-associated coronavirus. Science 300, 1399–1404 (2003).
Rota, P. A. et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300, 1394–1399 (2003).
Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D. & Fouchier, R. A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012).
van Boheemen, S. et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3, e00473–00412 (2012).
Zhu, N. et al. A novel coronavirus from patients with Pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020).
Cortese, M. et al. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe 28, 853–866 e855 (2020).
Snijder, E. J. et al. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80, 5927–5940 (2006).
Van Damme, E. et al. A small-molecule SARS-CoV-2 inhibitor targeting the membrane protein. Nature 640, 506–513 (2025).
Laporte, M. et al. A coronavirus assembly inhibitor that targets the viral membrane protein. Nature 640, 514–523 (2025).
Goyal, R. et al. Comparative highlights on MERS-CoV, SARS-CoV-1, SARS-CoV-2, and NEO-CoV. Excli J. 21, 1245–1272 (2022).
Helmy, Y. A. et al. The COVID-19 Pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med 9, 1225 (2020).
Massova, I. & Kollman, P. A. Computational alanine scanning to probe protein−protein interactions: a novel approach to evaluate binding free energies. J. Am. Chem. Soc. 121, 8133–8143 (1999).
Beard, H., Cholleti, A., Pearlman, D., Sherman, W. & Loving, K. A. Applying physics-based scoring to calculate free energies of binding for single amino acid mutations in protein-protein complexes. PLoS ONE 8, e82849 (2013).
Wu, A. et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27, 325–328 (2020).
Doijen, J. et al. A flexible, image-based, high-throughput platform encompassing in-depth cell profiling to identify broad-spectrum coronavirus antivirals with limited off-target effects. Antivir. Res. 222, 105789 (2024).
Wang, X., Zhu, H., Terashi, G., Taluja, M. & Kihara, D. DiffModeler: Large macromolecular structure modeling in low-resolution cryo-EM maps using diffusion model. bioRxiv https://doi.org/10.1101/2024.01.20.576370 (2024).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Adams, P. D. et al. The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 (2011).
Rosignoli, S. & Paiardini, A. Boosting the full potential of PyMOL with structural biology plugins. Biomolecules 12, 1764 (2022).
Rodrigues, J. P. G. L. M. openmm_scripts: Scripts to setup and run OpenMM simulations. (2019).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Dickson, C. J., Walker, R. C. & Gould, I. R. Lipid21: complex lipid membrane simulations with AMBER. J. Chem. Theory Comput. 18, 1726–1736 (2022).
Eastman, P. et al. OpenMM 8: molecular dynamics simulation with machine learning potentials. J. Phys. Chem. B 128, 109–116 (2024).
Zhang, Z., Liu, X., Yan, K., Tuckerman, M. E. & Liu, J. Unified efficient thermostat scheme for the canonical ensemble with holonomic or isokinetic constraints via molecular dynamics. J. Phys. Chem. A 123, 6056–6079 (2019).
Tubiana, T., Carvaillo, J.-C., Boulard, Y. & Bressanelli, S. TTClust: a versatile molecular simulation trajectory clustering program with graphical summaries. J. Chem. Inf. Model. 58, 2178–2182 (2018).
Acknowledgements
We thank Anne Henze (Akkodis Belgium) for coordination support on behalf of Janssen Pharmaceutica NV. This work was supported by Janssen Research & Development, LLC. This project has also been funded in part with federal funds from the Biomedical Advanced Research and Development Authority (BARDA), under Other Transaction Agreement (OTA) number HHSO100201700018C.
Author information
Authors and Affiliations
Contributions
M.K.M. and P.A. contributed to the conceptualization and overall study design. M.K.M. designed constructs, coordinated activities for expression, completed protein purification, target engagement by NanoDSF and structure determination of apo SARS-CoV and MERS-CoV by Cryo-EM including structure modeling and analysis, figures preparation, paper writing and editing. P.A. contributed to directing the study including experimental designs, data analysis, and paper writing and editing. Y.Y. contributed to the conceptualization of Cryo-EM experiments, completed the structure determination and modeling of JNJ-9676 bound SARS-CoV and analysis, and provided supervision for Cryo-EM experiments. R.M. performed expression of recombinant proteins. M.P. contributed to the purification of the recombinant FAb B. S.M. completed alanine scanning mutation analysis, MD simulations, figure preparation, paper writing and editing. J.X. and N.V.D.B. contributed to experiment design, data analysis, figure preparation and paper writing for the in vitro resistance selection, site-directed mutagenesis and SDM potency shift assay sections. J.D. contributed to paper writing and editing. J.D., N.V.D.B., C.K.K., and K.K. performed and coordinated IVRS and SDM studies. A.A.L., Hd.G., E.J.S., and Mv.H. contributed to IVRS studies. E.V.D. contributed to the critical revision of results, paper writing, editing and supervision. X.Y., M.V.G., M.V.L., A.K. and S.S. provided guidance and supervision throughout this study.
Corresponding author
Ethics declarations
Competing interests
E.V.D., M.V.L., and M.V.G. have been named an inventor in a pending patent application claiming inhibitors of coronavirus (WO 2024008909), which was filed by the Applicant Janssen Pharmaceutica NV. M.K.M., Y.Y., P.A., R.M., M.P., M.V.L., M.V.G., E.V.D., S.M., J.D., C.K., J.X., X.Y., and S.S. were/are employees of J&J Innovative Medicine and may possess stocks of Johnson & Johnson. N.V.D.B. and K.K. are employees of Charles River Laboratories, a contract research organization and may possess stocks of Johnson & Johnson. A.A.L., Hd.G., E.J.S., and Mv.H. received funding from Janssen Pharmaceutica to perform contract research. The remaining authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Manidipa Banerjee and Tobias Goris.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mann, M.K., Yin, Y., Marsili, S. et al. Structural insights into MERS and SARS coronavirus membrane proteins. Commun Biol 8, 1651 (2025). https://doi.org/10.1038/s42003-025-09042-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09042-3