Abstract
Hirschsprung’s disease (HSCR) is a congenital malformation of the enteric nervous system (ENS) caused by deficient enteric neural crest cell (ENCC) colonization. However, the mechanisms inducing impaired ENCC colonization remain unclear. This study investigates the involvement of extracellular vesicles (EVs), pivotal mediators of intercellular communication, in ENS development and HSCR pathogenesis. We find that treatment with plasma-derived EVs from children with HSCR (HSCR-EV) significantly inhibits ENCC proliferation and migration. MIR22HG is identified as the key mediator of the HSCR-EV-induced suppression of ENCC proliferation and migration. Schwann cells within the aganglionic colon of HSCR children derive EVs containing upregulated MIR22HG. In vitro, Schwann cell-derived EV-MIR22HG inhibits ENCC proliferation and migration. In vivo, it blocks ENCC colonization in the distal colon and disrupts ENS formation. MPP3 is identified as a potential downstream target of MIR22HG, with MIR22HG downregulating its expression via the m6A-dependent ALKBH5/IGF2BP3 axis. Collectively, Schwann cell-derived EVs transfer MIR22HG to ENCCs, downregulating MPP3 through the m6A/ALKBH5/IGF2BP3 pathway, and thereby disrupting ENCC colonization and impairing ENS formation.

Similar content being viewed by others
Introduction
The enteric nervous system (ENS) is a complex neural network composed of intermuscular and submucosal nerve plexuses and is equipped with intrinsic sensory neurons, excitatory and inhibitory interneurons, motor neurons, and a large number of enteric glial cells (EGCs). The ENS regulates the secretion, digestion, motility, and immune defense of the intestine independently of the central nervous system (CNS)1. ENS development is strongly dependent on the colonization of enteric neural crest cells (ENCCs) in the colon. This process involves the internal factors of ENCCs, as well as the extracellular microenvironment, which are important for maintaining a precise balance between the migration, proliferation, and differentiation of ENCCs2,3. The colonization of ENCCs may be disrupted by any abnormality occurs in this process, which may lead to ENS malformations such as Hirschsprung’s disease (HSCR).
HSCR is a rare congenital malformation of the ENS, with an incidence rate of approximately 1/5000, and is caused by the incomplete colonization of ENCCs into the distal colon during embryonic development4. Multiple factors, mainly genetic factors, regulate the colonization of ENCCs5,6. RET is a tyrosine kinase transmembrane receptor expressed on the surface of ENCCs. Mutations in the RET gene have been identified in approximately half of familial cases and nearly 1/5 of sporadic cases, establishing it as a major susceptibility gene for HSCR7. Endothelin Receptor B (EDNRB) represents the second most critical signaling pathway associated with HSCR pathogenesis. Through interaction with its ligand Endothelin-3 (END3), EDNRB maintains ENCC proliferation and undifferentiated status. Deficiency in EDNRB function can lead to the premature differentiation of ENCCs and disruption of normal migration, thereby impeding their colonization in the distal colon8. Additional studies suggest that signaling pathways including Sonic hedgehog9, Notch10, and Semaphorin11 have been reported to be involved in HSCR development, though their exact mechanisms of action remain unclear. Nevertheless, abnormalities in these key genes and pathways can only explain the pathogenesis of a subset of HSCR cases. While epigenetic modifications and small endogenous peptides have been increasingly implicated12,13, the pathogenesis of HSCR has not been fully elucidated.
Extracellular vesicles (EVs) are critical components of the extracellular microenvironment. They can be secreted and released by almost all cell types and are widely present in body fluids such as peripheral blood, cerebrospinal fluid, urine, and saliva14. As major carriers mediating intercellular communication, EVs can be transported to target cells, thereby delivering informational molecules such as nucleic acids, proteins, and lipids from parent cells to recipient cells while further regulating the gene expression levels and biological functions in recipient cells15,16. EVs are enclosed by a phospholipid bilayer, which enables them to stably exist in the circulatory system without being disrupted, thereby protecting their contents from degradation and facilitating their efficient delivery to recipient cells. Increasing evidence has shown that the components carried by plasma EVs in patients and healthy individuals are not exactly the same17,18, indicating that EVs in the blood may reflect the state of a disease, act as biomarkers, and regulate the occurrence and development of diseases. Fang et al.19 found that plasma-derived EVs from patients with non-small cell lung cancer exhibited higher expression levels of circARHGAP10, which can accelerate tumor progression after delivery to lung cancer cells. Gao et al. 20 demonstrated that after myocardial infarction, miR-1956 expression in plasma EVs from model mice was abnormally increased, and transferring EVs to adipose-derived bone marrow mesenchymal stem cells could enhance the angiogenic effects mediated by the stem cells. In the CNS, EVs have been confirmed to be closely associated with processes such as neurogenesis, neuroprotection, and neural repair, and are deeply involved in the pathogenesis and progression of various CNS diseases, including neurodegenerative diseases, neuroinflammation, and nervous system tumors21,22. However, research on EVs in ENS formation is lacking, especially regarding whether EVs can mediate information exchange between neurons, between neurons and glial cells, or between neurons and the external microenvironment within the ENS. The effects of EVs on impaired ENS formation and the occurrence of HSCR remain poorly understood.
In the present study, we investigated whether EVs are involved in the defective ENS formation in HSCR by delivering cargo to ENCCs, thus disrupting their colonization. We extracted plasma EVs from children with HSCR (HSCR-EVs) and controls (Ctrl-EVs), and co-cultured them with ENCCs to explore the influence of HSCR-EVs on ENCC proliferation and migration. The critical component, MIR22HG, in HSCR-EVs was further identified. Our study provides insights into exploring the pathogenesis of HSCR by focusing on EV-mediated intercellular crosstalk within the ENS, and elucidates the inhibitory role of EV-encapsulated MIR22HG during ENCC colonization.
Results
HSCR plasma-derived EVs inhibit ENCC migration and proliferation
EVs extracted from the plasma of children with HSCR and controls showed a typical cup-shaped morphology and bilayer membrane structure (Fig. 1A, B) under the TEM. NTA analysis indicated that the diameters of the plasma-derived EVs in both groups were around 100 nm (Supplementary Fig. 1), characteristic of small EVs. Figure 1C and D showed representative images of NTA. EV marker proteins, including ALIX, CD63, and TSG101, were detected in the HSCR-EVs and Ctrl-EVs, whereas the negative protein calnexin was not detected (Fig. 1E, F). ENCCs were cultured, and the extraction process and characterization are shown in Supplementary Fig. 2. After treatment with PKH67-labeled EVs, the IF assay showed the uptake of Ctrl-EVs and HSCR-EVs by ENCCs (Fig. 1G). Transwell and EdU assays illustrated that HSCR-EVs significantly inhibited the migration and proliferation of ENCCs (Fig. 1H, I). Cell migration is mainly driven by the dynamic formation of the cytoskeleton, in which actin filaments are crucial components23,24. We found that the formation of actin filaments was attenuated by treatment with HSCR-EVs (Fig. 1J).
A, B The typical cup-shaped morphology of EVs isolated from the plasma of children with HSCR (HSCR-EV) and controls (Ctrl-EV) (scale bar=100 nm). C, D NTA showing that the diameters of HSCR-EVs and Ctrl-EVs were mainly distributed between 100 and 200 nm. E, F Detection of EV positive marker proteins (ALIX, TSG101, and CD63) and negative markers (Calnexin). G Uptake of PKH67-labeled Ctrl-EVs or HSCR-EVs by ENCCs (scale bar=20 μm). HSCR-EVs significantly inhibit the migration (n = 5, scale bar=100 μm) (H), proliferation (n = 5, scale bar=50 μm) (I), and cytoskeleton formation (n = 3, scale bar=50 μm) (J) of ENCCs. Data are mean ± SD. ns not significant, *P < 0.05, **P < 0.01, ***P < 0.001.
MIR22HG mediates the function of HSCR-EVs
To elucidate the critical components mediating the function of HSCR-EVs, the RNA expression profiles of the HSCR-EVs and Ctrl-EVs were determined. The significantly differentially expressed genes (DEGs) in HSCR-EV are shown in Supplementary Table 1. Since EVs primarily exert their functions through cargo delivery, we focused on the significantly upregulated DEGs in HSCR-EVs. Functional enrichment analysis was further performed using GO and KEGG analyses (Supplementary Fig. 3). Given the inhibitory effects of HSCR-EVs on the colonization capacity of ENCCs, we further analyzed these genes based on their functional relevance to the suppression of cell proliferation and migration. We identified MIR22HG, which was significantly upregulated in HSCR-EVs and has previously been reported to exert inhibitory functions25. RT-qPCR validated the elevated expression of MIR22HG in HSCR-EVs (Fig. 2A). Treatment with RNase A did not affect the expression of MIR22HG in HSCR-EVs, whereas Triton X-100 treatment significantly reduced its expression (Supplementary Fig. 4A), confirming that MIR22HG is encapsulated within EVs. Tissue-level analysis in HSCR specimens showed that MIR22HG expression was markedly higher in the stenotic segment of HSCR (HSCR-S) compared to that in dilated segments (HSCR-D) and control colon tissues (Fig. 2B). Overexpression of MIR22HG inhibited ENCC migration, proliferation, and cytoskeletal formation (Fig. 2C-E). These findings suggested that upregulated MIR22HG in HSCR-EVs might critically contribute to the impairment of ENCC colonization. The diagnostic potential of plasma EV-derived MIR22HG for HSCR was also evaluated using an ROC analysis, with an area under the curve value of 0.78 (P = 0.02) (Supplementary Fig. 4B).
A MIR22HG was significantly upregulated in HSCR-EVs compared to that in Ctrl-EVs (n = 12). B MIR22HG exhibited higher expression in the stegnotic segments in children with HSCR (HSCR-S) compared to that in control tissues or the dilated segments from children with HSCR (HSCR-D) (n = 21). Overexpression of MIR22HG inhibited ENCC migration (n = 5, scale bar=100 μm) (C), proliferation (n = 5, scale bar=50 μm) (D), and cytoskeleton formation (n = 3, scale bar=50 μm) (E). Data are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Schwann cell-derived EV-MIR22HG inhibits ENCC migration and proliferation
In the aganglionic colon segments from children with HSCR, enteric neurons are lacking, whereas Schwann cells (SCs) are located within the hypertrophic external nerve plexus. The presence of SCs in the aganglionic segments was detected (Fig. 3A). We isolated SCs from the stenotic and dilated colon tissues of Ednrb-/- mice and normal colon tissues of Ednrb+/- mice. No significant differences in morphology were observed among the Schwann cells from the stenotic segment (S-SCs), dilated segment (D-SCs), and normal segment (N-SCs) (Fig. 3B). The expression of MIR22HG in EVs derived from S-SCs was higher than that in EVs derived from the D-SCs and N-SCs (Fig. 3C), indicating that Schwann cells within the aganglionic colon tissues may have been an essential source of the upregulated EV-MIR22HG in plasma of children with HSCR. EVs obtained from SCs were characterized (Fig. 3D–F). After co-culture with ENCCs, the uptake of SCs derived EVs by ENCCs was observed (Fig. 3G). MIR22HG-enriched EVs were isolated from SCs with MIR22HG overexpression, and the enrichment of MIR22HG was validated (Supplementary Fig. 5A). To determine the optimal working concentration of EV-MIR22HG, different concentration gradients were established to treat recipient cells. The CCK-8 assay showed that EV-MIR22HG inhibited cell proliferation in a dose-dependent manner (Supplementary Fig. 5B), and we selected 10 μg/ml as the working concentration for subsequent experiments to ensure robust biological effects while minimizing potential non-specific effects. Treatment with EV-MIR22HG attenuated ENCC migration and proliferation, as well as cytoskeletal formation (Fig. 3H–J). To validate that MIR22HG function is EV-mediated, we compared the effects of control EVs, MIR22HG-containing EVs and free MIR22HG on cell function. The EV-MIR22HG treatment group exhibited a significant inhibition of cell proliferation and cytoskeletal reorganization, whereas the free MIR22HG group failed to induce comparable effects (Supplementary Fig. 6A, B). Moreover, we pre-incubated cells with siRNA against MIR22HG prior to MIR22HG-containing EV uptake. Functional recovery was observed with the siRNA+EV-MIR22HG treatment but not the siRNA+EV-NC treatment (Supplementary Fig. 6C, D), indicating that the function is mainly due to MIR22HG and not to companion components. Theses findings suggest that EV-MIR22HG derived from Schwann cells may play an inhibitory role during the colonization of ENCCs by delivering MIR22HG.
A Expression of NCAM and Tuj1 in control, dilated (HSCR-D), and stenotic (HSCR-S) colon segments. The dashed boxes indicate the myenteric plexuses, which are relatively hypertrophic in the aganglionic segment (HSCR-S) (scale bar=100 μm). B Identification of Schwann cells isolated from the stenotic (S-SC), dilated (D-SC), and normal (N-SC) segments, as indicated by NCAM and GFAP staining (scale bar=100 μm). C MIR22HG was upregulated in S-SCs but not in D-SCs nor N-SCs (n = 6). D Cup-shaped morphology of EVs isolated from Schwann cells (SC-EV) under a TEM (scale bar=100 nm). E NTA showing that the main peak particle size of SC-EVs was around 100 nm. F Detection of EV positive markers (ALIX, TSG101, and CD63), and negative marker (Calnexin) in Schwann cells and SC-EVs. G Uptake of PKH67-labeled SC-EVs by ENCCs (Scale bar=20 μm). EV-MIR22HG inhibited the migration (n = 5, scale bar=100 μm) (H), proliferation (n = 5, scale bar=50 μm) (I), and cytoskeleton formation (n = 3, scale bar=50 μm) (J) of ENCCs. Data are mean ± SD. *P < 0.05, ***P < 0.001.
MIR22HG inhibits ENCC migration and proliferation by downregulating MPP3
RNA-seq analysis was employed to elucidate the potential downstream genes of MIR22HG. The top 20 downregulated DEGs by MIR22HG in ENCCs are shown in Supplementary Table 2. Among them, MAGUK P55 Scaffold Protein 3 (MPP3) with significant difference has been reported to be essential for the neuronal migration26. RT-qPCR and western blotting assays further verified the downregulation of MPP3 by MIR22HG (Fig. 4A, B). In the clinical aganglionic colon tissue samples, MPP3 exhibited lower mRNA and protein levels (Fig. 4C, D). Ectopic expression of MPP3 partly reversed the inhibition of ENCC migration and proliferation by MIR22HG, as well as that of cytoskeleton formation (Fig. 4E-G). Theses results indicated that MPP3 mediated the effects of MIR22HG on ENCC migration and proliferation.
A, B Overexpression of MIR22HG decreased the mRNA (n = 6) and protein (n = 3) levels of MPP3 in ENCCs. C, D MPP3 mRNA (n = 21) and protein (n = 3) levels were lower in HSCR-S segments than in control and HSCR-D segments. Introduction of MPP3 partially reversed the inhibitory effects of MIR22HG on ENCC migration (n = 5, scale bar=100 μm) (E), proliferation (n = 5, scale bar=50 μm) (F), and cytoskeleton formation (n = 3, scale bar=50 μm) (G). Data are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
MIR22HG reduces MPP3 expression in an m6A-dependent manner through the ALKBH5/IGF2BP3 pathway
Following treatment with MIR22HG, MPP3 mRNA stability was markedly impaired (Fig. 5A). Dot blot and m6A enrichment assays showed that MIR22HG significantly reduced the m6A modification level in ENCCs (Fig. 5B, C). MeRIP-qPCR assay determined that MIR22HG could decrease the m6A modification level of MPP3 mRNA (Fig. 5D). Subsequent to MIR22HG overexpression, a significant upregulation in the demethylase ALKBH5 expression level was observed, whereas no significant alterations were detected in either the demethylase FTO or critical methyltransferases METTL3 and METTL14 (Fig. 5E). RNA pull-down and RIP assays revealed an interaction between MIR22HG and the ALKBH5 protein (Fig. 5F, G). The SRAMP prediction analysis identified potential m6A motifs (GGACU) in the 3’UTR region of MPP3 mRNA (Supplementary Fig. 7A), and the dual-luciferase reporters were synthetized accordingly. Compared to that in the MUT reporter group, ALKBH5 markedly reduced luciferase activity in the WT reporter group (Fig. 5H), suggesting that ALKBH5 could recognize and bind to the MPP3 mRNA m6A motif. MeRIP-qPCR revealed that ALKBH5 was able to remove the m6A modification of MPP3 mRNA (Fig. 5I). Upon ALKBH5 overexpression, MPP3 was significantly downregulated (Fig. 5J, K). Knockdown of ALKBH5, however, reversed the inhibitory effects of MIR22HG on MPP3 expression (Fig. 5L, M) and mRNA stability (Fig. 5N). The fate of mRNA modified by m6A is primarily determined by recognition proteins, among which IGF2BP proteins, including IGF2BP1, IGF2BP2, and IGF2BP3, are mainly responsible for maintaining target mRNA stability27. The RIP assay indicated that IGF2BP3 enriched MPP3 mRNA more abundantly than IGF2BP1 and IGF2BP2 did (Supplementary Fig. 7B), whereas the upregulation of IGF2BP3, instead of IGF2BP1/2, enhanced MPP3 expression (Supplementary Fig. 7C). Furthermore, the overexpression of IGF2BP3 blocked MPP3 mRNA degradation (Fig. 5O). Based on these findings, MIR22HG was determined to reduce the abundance of m6A modifications in MPP3 mRNA by enhancing the expression of the demethylase ALKBH5, thus hindering the recognition and binding of IGF2BP3, which impaired the stability of MPP3 mRNA and decreased MPP3 expression.
A Overexpressed MIR22HG impaired the mRNA stability of MPP3, as shown by an ActD assay. B Dot blot (n = 3) and (C) m6A enrichment detection (n = 6) demonstrating that MIR22HG reduced the overall m6A modification level in ENCCs. D MeRIP-qPCR showing that the overexpression of MIR22HG reduced the m6A enrichment on the mRNA of MPP3 (n = 4). E MIR22HG enhanced ALKBH5 protein expression instead of that of METTL3, METTL14, and FTO (n = 3). F RNA pull-down and (G) RIP (n = 4) assays confirming the interaction between MIR22HG and ALKBH5. H Dual-luciferase reporter assay confirming that ALKBH5 could bind to the m6A site of the 3’UTR region of MPP3 mRNA (n = 6). I MeRIP-qPCR showing that ALKBH5 could reduce the m6A enrichment on the mRNA of MPP3 (n = 4). J, K Overexpression of ALKBH5 decreased the mRNA (n = 4) and protein (n = 3) levels of MPP3. Knockdown of ALKBH5 reversed the inhibitory effects of MIR22HG on MPP3 mRNA (n = 4) (L) and protein (n = 3) expression (M) and mRNA stability (N) in ENCCs. O The mRNA stability of MPP3 was enhanced by the overexpression of IGF2BP3. Data are mean ± SD. ns: not significant, *P < 0.05, **P < 0.01, ***P < 0.001.
EV-MIR22HG inhibits the formation of ENS in vivo
To further explore the influence of EV-MIR22HG on ENS formation in vivo, we first determined whether EVs could enter embryos through the placental barrier. After injections of DiR-labeled EVs, EVs could be absorbed by the pregnant mice (Supplementary Fig. 8A). Twelve hours later, the embryos were harvested, and ex vivo imaging revealed that EVs could pass through the placental barrier into the embryos (Supplementary Fig. 8B). The whole-mount staining assay showed that EV-MIR22HG attenuated the colonization of ENCCs into the distal colon (Fig. 6A). When additional mice were maintained for 2 weeks, immunohistochemistry experiments suggested a smaller number of ganglions in the mouse colons from the EV-MIR22HG group than in the EV-NC group (Fig. 6B). According to the time at which the first Evans blue-stained fecal pellet was expulsed, the gastrointestinal transit was slower in the mice treated with EV-MIR22HG than in the mice with EV-NC (Fig. 6C). In addition, colonic transit was weaker in the EV-MIR22HG group than in the EV-NC group, based on the time required for a glass bead to be expelled from the colon (Fig. 6D). These results indicated that EV-MIR22HG from Schwann cells blocked the formation of the ENS in vivo and suppressed the motility of the mouse colon. Furthermore, the ALKBH5 protein levels increased in the intestines of the EV-MIR22HG treatment group (Fig. 6E), whereas the mRNA level of MPP3 was significantly downregulated (Fig. 6F).
A Whole-mount staining of embryonic mouse colon, showing that EV-MIR22HG can impair the colonization of ENCCs to the distal colon. The white arrows indicate the migration end of ENCCs (n = 3, scale bar=1000 μm). B Immunohistochemistry assay showing fewer ganglions in the colon of EV-MIR22HG mice than in the EV-NC group. The black arrows indicate the ganglions in the myenteric plexus (n = 3, scale bar=50 μm). Treatment with EV-MIR22HG prolonged the time for the expulsion of the first blue pellet (n = 6) (C) and the time for the expulsion of the glass bead (n = 6) (D) in mice. The protein level of ALKBH5 was increased (n = 3) (E), whereas the mRNA level of MPP3 was decreased (n = 3) (F) in the colon of EV-MIR22HG mice (N1-3: EV-NC, M1-3: EV-MIR22HG). Data are mean ± SD. *P < 0.05, **P < 0.01.
Discussion
EVs have been shown to play a critical role in the development of the CNS and pathogenesis of CNS-related diseases by mediating the communication between glial cells and neurons; however, their involvement in the development and formation of ENS remains unclear. HSCR is a congenital developmental disorder of the ENS. To determine whether EVs are responsible for impaired ENS formation in children with HSCR, we isolated EVs from the plasma, and found that HSCR-EVs significantly blocked the migration of ENCCs, indicating that EVs may play a potent role during the occurrence of HSCR by inhibiting the colonization of ENCCs. Since HSCR is a rare congenital disease, obtaining appropriate clinical blood samples is highly challenging, and EV recovery rate of the PEG method is higher than that of ultracentrifugation, we thus employed the PEG precipitation method to isolate EVs from plasma to balance sample preciousness and yield requirements. Supplementary Fig. 9 demonstrated that the plasma-EVs obtained by our optimized method exhibited acceptable purity, whereas the gradient ultracentrifugation yielded EVs with higher purity. To identify the key components in HSCR-EV, we conducted a transcriptome analysis and validated our findings using clinical samples. Our results suggest that MIR22HG, which is upregulated in HSCR-EVs, may be an important factor. MIR22HG has been widely studied and exerts inhibitory effects on cell proliferation, migration, and invasion28. It has been found that MIR22HG relies on the production of miR-22-3p and targets histone deacetylase 4, thereby promoting the differentiation of skeletal muscle cells and the repair and regeneration of damaged skeletal muscles29. Ge et al.30 revealed that MIR22HG exacerbates oxygen-glucose deprivation and reoxygenation-induced myocardial cell damage through the miR-9-3p/SH2B3 axis. However, there have been no reports on the role of MIR22HG in the formation of ENS, and it has been unclear whether MIR22HG is involved in ENS-related developmental disorders, such as HSCR, by interfering with the migration and colonization of ENCCs. In the present study, we found that overexpression of MIR22HG significantly attenuated ENCC proliferation and migration, indicating that the transfer of MIR22HG might be responsible for the impairment of ENS formation in children with HSCR. Given the potential of plasma EVs as diagnostic biomarkers, we also investigated the diagnostic value of plasma EV-MIR22HG for HSCR in this study. However, because HSCR is a rare congenital disease and sample collection is challenging, the current study remains a small-sample exploratory investigation, which have limited the stability of the ROC curve analysis. We will further expand the sample size or conduct multicenter studies to validate the diagnostic value of plasma EV-MIR22HG for HSCR.
The lack of ganglion cells in the distal colon is a hallmark feature of HSCR. Therefore, within the microenvironment of aganglionic segments in children with HSCR, EVs that inhibit ENCC colonization are unlikely to originate from enteric neurons. Distinct from neurons, EGCs represent another crucial cell population in the ENS. Initially regarded as a structural support for neurons without specialized functions, EGCs have been increasingly recognized as key regulators of gastrointestinal functions, including motility, secretion, and immune responses. Although children with HSCR lack intrinsic enteric neurons in the aganglionic segments, they still exhibit hypertrophic and hyperplastic external nerve plexuses, which contain a high density of Schwann cells31,32. A recent study using single-cell sequencing to profile EGCs in the intestinal tissues of children with HSCR has revealed that the aganglionic segments exhibited an exclusive enrichment of the Schwann cell subpopulation33. Moreover, Schwann cells have been shown to deliver RNA to neurons via EVs, thereby influencing neurogenesis and axonal regeneration in the peripheral nervous system34,35. Indeed, murine models of HSCR have demonstrated that Schwann cells can dedifferentiate and remodel into functional enteric neurons, highlighting their neuroregenerative potential36,37. However, the intrinsic role of SCs within the aganglionic microenvironment remains poorly understood, particularly regarding whether these cells contribute to the impaired colonization of ENCCs under pathological conditions. Our study, however, investigated SC-ENCC crosstalk in the context of HSCR pathogenesis. We demonstrated that SCs isolated from the stenotic segment secrete EVs enriched in MIR22HG, which are internalized by ENCCs to suppress their migratory and colonizing capacities, uncovering a previously unrecognized inhibitory role of Schwann cell-derived EVs in ENCC biology. This dual function (beneficial or detrimental) highlights the complexity of Schwann cell role in HSCR and provides valuable insights into investigating HSCR pathogenesis and therapeutic intervention. Although our findings robustly demonstrated that EV-encapsulated MIR22HG exerts dose-dependent effects on recipient cell function, the MIR22HG copy number per EV was not precisely quantified. To overcome this limitation, future work will focus on optimizing single-vesicle analysis techniques to quantify MIR22HG loading efficiency at the individual EV level, thereby facilitating the determination of the precise copy number required for its functional effects.
MPP3 belongs to the MAGUK protein family, which is a class of multidomain scaffold proteins located near cell junctions that play important roles in cell adhesion, polarity, migration, synaptic development, and signal transduction38. In this study, we demonstrated that delivery of MIR22HG by EVs repressed MPP3 expression in ENCCs, whereas the overexpression of MPP3 rescued MIR22HG-induced impairments in ENCC proliferation and migration, suggesting MPP3 as an essential regulator of ENCC colonization. Consequently, reduced MPP3 expression may contribute to the development of HSCR by inhibiting the colonization of ENCCs. Targeting MPP3 in vivo may potentially simulate the HSCR pathophysiology; however, considering the important role of MPP3 in the CNS, global knockout or knockdown could induce severe neurological defects and subsequent mortality. The Cre-LoxP system would enable precise targeting of MPP3 in migratory ENCCs. Based on the above, we will further utilize SOX10-Cre mice to achieve specific knockout of MPP3 in ENCCs during the embryonic stage, thereby avoiding embryonic lethality while recapitulating the congenital HSCR disease process. Successful establishment of this model will provide mechanistic insights into how MPP3 deficiency impairs cell-cell adhesion and migration signaling pathways during ENS development, potentially identifying promising therapeutic targets for HSCR.
The m6A modification is closely associated with various processes, such as mRNA splicing, transport, translation, and stability, and plays a critical role in multiple physiological processes, including embryonic development39. Using the SRAMP tool, we identified potential methylation sites in MPP3 mRNA. Additionally, we observed that MIR22HG could regulate the methylation level of MPP3, indicating that MIR22HG might modulate the expression of MPP3 by influencing m6A methylation. Furthermore, the demethylase ALKBH5, the expression of which was enhanced by MIR22HG, was found to mediate the regulation of MPP3 m6A modifications by MIR22HG. This may have been related to the ability of MIR22HG to act as a “scaffold” that recruits the ALKBH5 protein. After m6A modifications occur on mRNA, its fate is primarily determined by m6A recognition proteins. Known m6A recognition proteins include YTHs, HNRNPs, eIF3, and IGF2BPs. Given that MIR22HG could impair the stability of MPP3 mRNA and that IGF2BPs are responsible for recognizing and maintaining the stability of m6A-modified target mRNAs, we systematically profiled IGF2BP family members for their ability to bind MPP3 mRNA. Among them, IGF2BP3 exhibited the highest enrichment and was able to enhance the stability of MPP3 mRNA. Therefore, MIR22HG was determined to recruit the m6A demethylase ALKBH5 to remove m6A modifications on MPP3 mRNA. This epitranscriptomic remodeling ablated IGF2BP3 binding, thereby diminishing MPP3 mRNA stability and suppressing its expression.
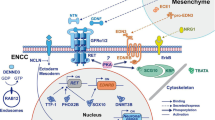
In summary, our findings demonstrated that EVs from Schwann cells within the aganglionic colon can transfer MIR22HG to ENCCs. Subsequently, in an m6A-dependent manner, MIR22HG downregulats MPP3 expression through the ALKBH5/IGF2BP3 axis, thereby inhibiting the migration and colonization of ENCCs in the distal colon and ultimately leading to the abnormal development of the ENS. This study provides notable insights into the pathogenesis of disorders associated with abnormal ENS development, particularly HSCR, by highlighting Schwann cell-to-ENCC communication within the colon microenvironment. However, there remain some limitations. Future studies should investigate whether MIR22HG regulates MPP3 expression via other pathways. The specific mechanisms by which Schwann cells selectively sort high levels of MIR22HG into EVs remain to be explored.
Materials and methods
Clinical samples
Twenty-one samples from children with HSCR and twenty-one control samples without ENS malformations were analyzed in this study. All the children with HSCR underwent surgery at the Children’s Hospital of Nanjing Medical University. Plasma samples were obtained from the children before surgery. Tissue samples were harvested during surgery and immediately stored in liquid nitrogen. This study was approved by the Institutional Ethics Committee of Children’s Hospital of Nanjing Medical University (approval number is 202205050-1), and obtained informed consent from the guardians of the pediatric patients. All ethical regulations relevant to human research participants were followed.
Isolation and characterization of EVs
To extract plasma EVs, plasma samples were centrifuged at 3000 g for 15 min at 4 °C to remove platelets. Thrombin was added to the supernatant and incubated at room temperature for 5 min, followed by centrifugation at 10,000 g for 5 min. The supernatant was collected, filtered through a 0.22-μm PVDF membrane, and mixed with ExoQuick reagent (EXOQ5A-1, System Biosciences, USA) for overnight incubation at 4 °C. After centrifugation at 1,500 g for 30 min, the supernatant was discarded. The EV pellet was resuspended in PBS and stored at -80°C until further analysis. To isolate EVs from Schwann cells, cells were cultured in a medium containing 10% exosome-free fetal bovine serum (FBS). After the removal of cellular debris, the cell culture supernatant was filtered using a 0.22-μm PVDF membrane, and then centrifuged at 100,000 g for 90 min. PBS was added to resuspend the pellets, and the solution was centrifuged again. Finally, the pellets were dissolved using PBS and stored at -80 °C. The nanoparticle tracking analysis (NTA) and transmission electron microscopy (TEM) were used to characterize the EVs.
Cell culture and transfection
ENCCs were extracted from the intestines of mouse embryos. The intestinal tissues were incubated in collagenase Ⅳ (C5138-25MG, Sigma, USA) at 37 °C for 30 min. After filtration through a 70-μm mesh, the filtrates were collected and centrifuged at 1,000 g for 5 min. The precipitate was resuspended and cultured in a neural cell-specific medium (CM-M216, Procell, China). All cells were maintained at 37 °C in an incubator with suitable humidity and 5% CO2. Overexpression plasmids were synthesized by GeneChem (Shanghai, China), and Lipofectamine (STEM00003, Thermo Fisher Scientific, USA) was utilized to conduct cell transfections.
Quantitative real-time polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues and cells using TRIzol reagent (Thermo Fisher Scientific, USA) and reverse-transcribed into complementary DNA. The ChamQ SYBR qPCR Master Mix (Q311-02-AA Vazyme, China) was used to perform RT-qPCR. The 2–ΔΔCT method was employed to calculate the relative gene expression levels. GAPDH was used as the internal control. Supplementary Table 3 presents the primers used in this study.
Western blotting
RIPA lysis buffer was used to isolate total protein from tissues and cells. The BCA assay was applied to determine protein concentrations. Protein samples were separated on 10% gels and transferred to PVDF membranes. Subsequently, the membranes were blocked with 5% fat-free milk at room temperature for 2 h. The membranes were then treated with primary antibodies (Supplementary Table 4) overnight at 4 °C. An additional 1-h incubation was performed with the corresponding secondary antibodies. Finally, the membranes were analyzed using an exposure meter (Tanon, Shanghai).
EV uptake assay
Prior to the EV uptake assay, EVs were labeled with PKH67 fluorescent dye. After labeling, the suspension was ultracentrifuged at 100,000 g and 4 °C for 90 min. The supernatant (containing free dye) was discarded, and PBS was added to resuspend the pellet, yielding PKH67-fluorescently labeled EVs. EVs (plasma-EVs 20 μg/mL, SC-EVs 10 μg/mL) were then added to ENCCs (5×104) for a 24-h incubation period. The fluorescent distribution of PKH67 was visualized using a laser confocal microscope.
Ethynyl deoxyuridine (EdU) assay
The EdU assay was conducted using an EdU kit (Ribob Biotechnology, China). After being cultured in EdU media for 2 h, cells were fixed with 4% PFA, followed by incubation using glycine (2 mg/mL) and 0.5% TritonX-100. Subsequently, the cells were treated with Apollo or Hoechst 33342 in the dark. EdU-positive cells were observed under a microscope (Zeiss, Germany).
Transwell assay
Approximately 2×105 cells were seeded into an Transwell chamber with serum-free media. The lower chamber was supplemented with complete media. After 24 h, the migrated cells were fixed with 4% PFA and stained with crystal violet. Cell images were captured using a microscope (Nikon, Japan).
Methylated RNA immunoprecipitation (MeRIP)-qPCR
Total RNA was isolated from the cells using TRIzol reagent (Thermo Fisher Scientific, USA). The RNA was incubated in an immunoprecipitation buffer containing the m6A antibody for 12 h at 4 °C. The mixture was subjected to immunoprecipitation and elution. After normalization to the input, the m6A modification level in each group was detected using RT-qPCR.
Actinomycin D (ActD) assay
Cells were seeded and treated with actinomycin D (Med Chem Express, USA) for 0, 2, 4, and 6 h. Total RNA was extracted at each time point and subjected to RT-qPCR to quantify the expression levels.
Animal experiments
Ednrb−/− C57BL/6 J mouse models of HSCR were generated by Biocytogen Co. (Beijing, China). Wild type (WT) C57BL/6 J mice were obtained from the Experimental Animal Center of Nanjing Medical University. All the mice were housed under specific pathogen-free conditions with free access to food and water. The animal experiments were approved by the Institutional Animal Care and Use Committee of the Nanjing Medical University (IACUC-2205048). Schwann cells were isolated from the colon tissues of Ednrb−/− and Ednrb+/− mice, and cultured in Dulbecco’s modified Eagle medium (Gibco, USA). The WT mice were treated with EV-NC or EV-MIR22HG every day for a period of E9.5-14.5 days via the tail vein. At E16.5, the intestinal tissues form each group were harvested for the whole-mount staining. Gastrointestinal motility was assessed at postnatal day 14 by detecting the excretion time of feces and glass beads.
Statistics and Reproducibility
GraphPad Prism 8 (GraphPad Software, USA) was used for statistical analyses. The data are presented as the “mean ± SD”. Unpaired two-tailed Student’s t-test was used for two-group comparisons. For multigroup comparisons, a one-way analysis of variance (ANOVA) and Tukey’s post-hoc test were used. To evaluate the diagnostic value of plasma EV-derived MIR22HG for HSCR, receiver operating characteristic (ROC) curve analysis was performed using GraphPad Prism 8, based on the expression levels of MIR22HG in plasma EVs isolated from children with HSCR and controls. Results with P < 0.05 were considered to be statistically significant.
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files). The source data behind the graphs in the paper can be found in Supplementary Data. The uncropped and unedited WB images are shown in Supplementary Fig. 10. All other data are available from the corresponding author on reasonable request.
References
Fung, C. & Vanden Berghe, P. Functional circuits and signal processing in theenteric nervous system. Cell Mol. Life Sci. 77, 4505–4522 (2020).
Nagy, N. & Goldstein, A. M. Enteric nervous system development: A crest cell’sjourney from neural tube to colon. Semin Cell Dev. Biol. 66, 94–106 (2017).
Baker, P. A., Ibarra-Garcia-Padilla, R., Venkatesh, A., Singleton, E. W. & Uribe, R. A. In toto imaging of early enteric nervous system development reveals that gutcolonization is tied to proliferation downstream of Ret. Development 149 dev200668 (2022).
Butler Tjaden, N. E. & Trainor, P. A. The developmental etiology and pathogenesis of Hirschsprung disease. Transl. Res 162, 1–15 (2013).
Tang, C. S., Karim, A., Zhong, Y., Chung, P. H. & Tam, P. K. Genetics of Hirschsprung’s disease. Pediatr. Surg. Int 39, 104 (2023).
Karim, A., Tang, C. S. & Tam, P. K. The emerging genetic landscape of hirschsprung disease and its potential clinical applications. Front Pediatr. 9, 638093 (2021).
Luzon-Toro, B. et al. What is new about the genetic background of Hirschsprung disease? Clin. Genet. 97, 114–124 (2019).
Nagy, N. & Goldstein, A. M. Endothelin-3 regulates neural crest cell proliferationand differentiation in the hindgut enteric nervous system. Dev. Biol. 293, 203–217 (2006).
Nagy, N. et al. Sonic hedgehog controls enteric nervous system development by patterning the extracellular matrix. Development 143, 264–275 (2016).
Jia, H. M., Han, X. F., Bai, Y. Z. & Wang, W. L. Expression and significance of Notch-1 and Jagged-2 in patients with Hirschsprung disease. Zhonghua Wei Chang Wai Ke Za Zhi 14, 768–771 (2011).
Kapoor, A. et al. Population variation in total genetic risk of Hirschsprung disease from common RET, SEMA3 and NRG1 susceptibility polymorphisms. Hum. Mol. Genet 24, 2997–3003 (2015).
Diposarosa, R., Bustam, N. A., Sahiratmadja, E., Susanto, P. S. & Sribudiani, Y. Literature review: enteric nervous system development, genetic and epigenetic regulation in the etiology of Hirschsprung’s disease. Heliyon 7, e07308 (2021).
Sun, Q. et al. Mechanism of endogenous peptide PDYBX1 and precursor protein YBX1 in Hirschsprung’s disease. Neurosci. Bull. 40, 695–706 (2023).
Yi, Y. W. et al. Advances in analysis of biodistribution of exosomes by molecular imaging. Int. J. Mol. Sci. 21, 65 (2020).
Kuang, Y. et al. Adipose-derived mesenchymal stem cells reduce autophagy in stroke mice by extracellular vesicle transfer of miR-25. J. Extracell. Vesicles 10, e12024 (2020).
Hermann, D. M., Xin, W., Bahr, M., Giebel, B. & Doeppner, T. R. Emerging rolesofextracellularvesicle-associated non-coding RNAs in hypoxia: Insights from cancer, myocardial infarction and ischemic stroke. Theranostics 12, 5776–5802 (2022).
Ozawa, P. M. M. et al. Identification of miRNAs enriched in extracellular vesicles derived from serum samples of breast cancer patients. Biomolecules 10, 150 (2020).
Dobra, G. et al. Small extracellular vesicles isolated from serum may serve as signal-enhancers for the monitoring of CNS tumors. Int. J. Mol. Sci. 21, 5359 (2020).
Fang, K. et al. Serum-derived exosomes-mediated circular RNA ARHGAP10 modulates the progression of non-small cell lung cancer through the miR-638/FAM83F Axis. Cancer Biother Radiopharm. 37, 96–110 (2022).
Gao, L. et al. Cardio-renal exosomes in myocardial infarction serum regulate proangiogenic paracrine signaling in adipose mesenchymal stem cells. Theranostics 10, 1060–1073 (2020).
Nieland, L., Morsett, L. M., Broekman, M. L. D., Breakefield, X. O. & Abels, E. R. Extracellular vesicle-mediated bilateral communication between glioblastoma and astrocytes. Trends Neurosci. 44, 215–226 (2021).
Wan, C., Stowell, M. H. B. & Shen, J. Progress and gaps of extracellular vesicle-mediated intercellular cargotransfer in the central nervous system. Commun. Biol. 5, 1223 (2022).
Seetharaman, S. & Etienne-Manneville, S. Cytoskeletal crosstalk in cell migration. Trends Cell Biol. 30, 720–735 (2020).
Tokuraku, K., Kuragano, M. & Uyeda, T. Q. P. Long-range and directional allostery of actin filaments plays important roles in various cellular activities. Int. J. Mol. Sci. 21, 3209 (2020).
Deng, X. et al. MIR22HG inhibits breast cancer progression by stabilizing LATS2 tumor suppressor. Cell Death Dis. 12, 810 (2021).
Dudok, J. J., Sanz, A. S., Lundvig, D. M. & Wijnholds, J. MPP3 is required for maintenance of the apical junctional complex, neuronal migration, and stratification in the developing cortex. J. Neurosci. 33, 8518–8527 (2013).
Huang, H. et al. Publisher Correction: Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 22, 1288 (2020).
Zhang, L., Li, C. & Su, X. Emerging impact of the long noncoding RNA MIR22HG on proliferation and apoptosis in multiple human cancers. J. Exp. Clin. Cancer Res 39, 271 (2020).
Li, R. et al. Long non-coding RNA Mir22hg-derived miR-22-3p promotes skeletal muscle differentiation and regeneration by inhibiting HDAC4. Mol. Ther. Nucleic Acids 24, 200–211 (2021).
Ge, Y., Liu, L., Luo, L., Fang, Y. & Ni, T. MIR22HG Aggravates Oxygen-Glucose Deprivation and Reoxygenation-Induced Cardiomyocyte Injury through the miR-9-3p/SH2B3 Axis. Cardiovasc Ther. 2022, 7332298 (2022).
Hwang, S. & Kapur, R. P. Advances and Pitfalls in the Diagnosis of Hirschsprung Disease. Surg. Pathol. Clin. 13, 567–579 (2020).
Espinosa-Medina, I. et al. Dual origin of enteric neurons in vagal Schwann cell precursors and the sympathetic neural crest. Proc. Natl Acad. Sci. USA 114, 11980–11985 (2017).
Windster, J. D. et al. Human enteric glia diversity in health and disease: new avenues for the treatment of hirschsprung disease. Gastroenterology, 168, 965–979 (2024).
Xia, B. et al. Mechanical stimulation of Schwann cells promote peripheral nerve regeneration via extracellular vesicle-mediated transfer of microRNA 23b-3p. Theranostics 10, 8974–8995 (2020).
Sun, J. et al. Down-regulation miR-146a-5p in Schwann cell-derived exosomes induced macrophage M1 polarization by impairing the inhibition on TRAF6/NF-kappaB pathway after peripheral nerve injury. Exp. Neurol. 362, 114295 (2023).
Uesaka, T. et al. Enhanced enteric neurogenesis by Schwann cell precursors in mouse models of Hirschsprung disease. Glia 69, 2575–2590 (2021).
Soret, R. et al. Glial Cell-Derived Neurotrophic Factor Induces Enteric Neurogenesis and Improves Colon Structure and Function in Mouse Models of Hirschsprung Disease. Gastroenterology 159, 1824–1838 e1817 (2020).
Zhang, M. et al. Phosphorylation-dependent recognition of diverse protein targets by the cryptic GK domain of MAGI MAGUKs. Sci. Adv. 9, eadf3295 (2023).
Chen, C. et al. Nuclear m(6)A reader YTHDC1 regulates the scaffold function of LINE1 RNA in mouse ESCs and early embryos. Protein Cell 12, 455–474 (2021).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (approval number: 82001590, 82270540), and High-Level Hospital Science and Technology Innovation Support Program Project, Children’s Hospital of Nanjing Medical University (approval number: KJCXQ2024017).
Author information
Authors and Affiliations
Contributions
W.B.T. and H.X.L designed the experiments. Z.K.Z., Q.C.S. and C.L.W. performed the experiments. Z.K.Z. and Q.C.S. analyzed the data and wrote the manuscript. Y.X.Q., Z.C.G., C.X.D. and J.T. revised the manuscript. All authors approved this manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Marcel Tawk and Sheryl Bui for their contribution to the peer review of this work. Primary Handling Editors: Gregory Lavieu and Joao Valente.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhi, Z., Sun, Q., Wang, C. et al. Extracellular vesicle-mediated transfer of MIR22HG inhibits the colonization of enteric neural crest cells in the colon by decreasing MPP3 expression. Commun Biol 8, 1653 (2025). https://doi.org/10.1038/s42003-025-09047-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09047-y