Abstract
Neuronal reprogramming using adeno-associated viruses with a GFAP mini-promoter offers a promising strategy for astrocyte-to-neuron conversion; however, specificity remains a challenge due to off-target transgene expression in endogenous neurons. To address this issue, here we incorporated microRNA-124 target sequences (124T) into a transcriptional cassette containing the GFAP mini-promoter and Ascl1SA6, a potent reprogramming transcription factor. Lineage tracing via Slc1a3-CreERT and Aldh1l1-CreERT2, used to pre-label astrocytes prior to conversion, confirmed the glial derivation of reprogrammed neuron-like cells, even with 124T. Single-cell transcriptomics identified a transitional cluster emerging from a proliferative astrocyte population with low GSK3 signaling, which branched towards hybrid neuronal and oligodendrocyte fates. Pseudotime trajectory analysis revealed that Ascl1SA6 drives rapid neuronal transitions, whereas 124T delays conversion and introduces lineage bifurcation. Ascl1SA6 favors a GABAergic interneuron-like identity, while Ascl1SA6 -124T biases fate transitions towards an oligodendrocyte-like fate, and to a lesser extent, glutamatergic neuronal-like cells. SeeSawPred linked these distinct trajectories to transcription factor shifts, including Foxo1 in neuron-like cell fates and Stat1 in oligodendrocyte lineages. 124 T thus effectively detargets endogenous neurons, refining target cell specificity, while further guiding reprogramming outcomes. This approach establishes a foundation for precision reprogramming platforms aimed at restoring specific neural cell types.

Similar content being viewed by others
Introduction
Age-related neurological disorders are marked by progressive neuronal loss or dysfunction, impairing critical daily functions such as movement, speech, and cognition. As global populations continue to age, the burden of these conditions is increasing, yet effective treatments remain limited. Since dying neurons are only replaced in rare circumstances in the brain1,2, developing neuronal replacement strategies is a pressing clinical challenge. One promising approach is direct neuronal reprogramming, or the conversion of somatic cells, such as astrocytes, into neurons using lineage-converting, proneural transcription factors (TFs)3,4. This approach leverages the brain’s own endogenous cellular reservoirs and mimics innate regenerative processes observed in species like teleost fish5.
An expanding body of preclinical evidence supports the therapeutic potential of astrocyte-to-neuron conversion across multiple neurological conditions, including Parkinson’s disease6, stroke7,8, and other neurodegenerative conditions6,9,10,11,12,13. However, despite irrefutable evidence for astrocyte-to-neuron conversion in vitro3, the direct demonstration of in vivo lineage conversion has been challenging, owing in part to technical challenges and concerns surrounding the specificity of gene delivery vehicles14,15. Adeno-associated viruses (AAVs) are the gold standard for gene delivery as they are well tolerated, with low immunogenicity and cytotoxicity, and they promote high levels of ectopic gene expression16. However, early claims of astrocyte-to-neuron conversion using AAVs carrying a GFAP mini-promoter have been disputed since GFAP mini-promoter vectors exhibit off-target transgene activation in endogenous neurons when coupled with neurogenic TFs, confounding lineage attribution14,17,18,19,20,21. However, the lack of GFAP promoter specificity does not negate the capacity of brain glia to be converted to neurons, as shown definitively using retroviruses in vivo3, and supported by in vitro studies in purified astrocytes2,4.
The neurotransmitter identity of reprogrammed neurons is critical for circuit repair. Ascl1 is a proneural gene encoding a basic-helix-loop-helix (bHLH) TF that promotes GABAergic neuronal fates during forebrain development22,23 and is also later involved in oligodendrocyte lineage specification24,25,26 (Fig. 1a). This neuronal fate specificity is recapitulated in early postnatal astrocytes converted to induced neurons (iNeurons) in vitro27. However, the function of Ascl1 and other proneural TFs is tightly regulated by external signals, for instance, by direct phosphorylation by proline (P)-directed serine/threonine (S/T) kinases (e.g., ERKs, GSK3, CDKs)28,29,30,31,32,33,34,35,36,37,38. Since phosphorylation of bHLH TFs modulates their lineage-inductive capacity, here, we used Ascl1SA6, in which serines in the six SP sites were converted to alanines (Fig. 1a). SA mutations increase the neurogenic capacity of proneural bHLH TFs in the embryonic mouse brain, during primary neurogenesis in Xenopus, and in cancer cells28,29,33,34,35,36,37. Additionally, Neurog2SA9 and Ascl1SA6 are more efficient at neuronal conversion in vitro and in vivo39,40,41.
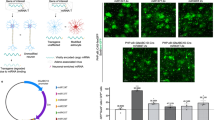
a Ascl1 can specify a GABAergic neuron or OPC fate, with ERK-mediated phosphorylation biasing neural progenitor cells to select an OPC fate. ERK phosphoacceptor sites, which are serines or threonines adjacent to prolines, were mutated to alanines to generate Ascl1SA6. BioRender was used to create the schematics. b Astrocytes were pre-labeled using Slc1a3-CreERT;Rosa-zsGreen mice that were administered 4-OHT four times, separated by a day per injection. At 3 days after the last injection, AAVs were administered. BioRender was used to create the schematics. c Analysis of zsGreen (zsG) and mCherry (mCh) expression at 21 days post AAV5-GFAP-mCherry administration into Slc1a3-CreERT;Rosa-zsGreen mice. Scale bars = 100 µm. d Quantification of the percentage of GFAP+ cells that were zsGreen+, and the percentage of mCherry+ cells that were zsGreen+. e–h Marker analysis of Slc1a3-CreERT;Rosa-zsGreen motor cortices transduced with mCherry control or Ascl1SA6 vectors. Co-expression of mCherry and zsGreen were analyzed with ASCL1 (e), GFAP (f), DCX (g) or NEUN (h). White arrows indicate mCherry+zsGreen+; yellow arrows indicate mCherry-negative/zsGreen+. Scale bars = 100 µm. The percentage of marker expression in mCherry+ zsGreen+ cells was quantified (n = 3 animals each vector). Mean values and error bars representing the standard error of the mean (s.e.m.) were plotted. p-values were calculated with an unpaired t-test.
To address limitations in astrocyte-specific targeting using AAV-GFAP vectors, we incorporated four copies of the microRNA-124 (miR-124) target sequence (124T) into the 3′ untranslated region of a transcriptional cassette encoding the neurogenic TF Ascl1SA6. miR-124 is a neuronal-specific microRNA that plays a central role in driving neurogenesis, and its presence in neurons suppresses REST and other anti-neurogenic programs42. We report here that 124T-sites added to the AAV-GFAP-Ascl1SA6 vector effectively suppressed transgene expression in endogenous neurons while preserving Ascl1SA6 expression in astrocytes. Lineage tracing using Slc1a3-CreERT and Aldh1l1-CreERT2 systems confirmed that reprogrammed neurons originated from astrocytic glia, even in the context of 124T detargeting. However, while Ascl1SA6 preferentially drives astrocytic conversion towards GABAergic neuronal-like cell fates, inclusion of 124T sites attenuated reprogramming kinetics and introduced lineage bifurcation, redirecting cells towards more glutamatergic neuronal and oligodendrocyte-like cell identities. These altered outcomes are linked to distinct TF programs, highlighting the influence of post-transcriptional modulation on fate specification. Collectively, our findings establish a refined gene delivery platform that enhances cellular precision and reveals how targeted transcriptional modifications reshape the lineage trajectories of glial conversion.
Results
Ascl1 SA6 converts pre-labeled astrocytes to neuron-like cells
During embryonic development of the telencephalon, Ascl1 drives the differentiation of either GABAergic neurons or oligodendrocyte precursor cells (OPCs), depending on developmental time and phosphorylation status by proline-directed serine/threonine kinases, such as ERK (Fig. 1a)28. To counteract the pro-OPC influence of ERK signaling on Ascl1’s proneural function, we mutated all six serine residues adjacent to prolines to alanines, creating a phosphorylation-resistant variant, Ascl1SA6 (Fig. 1a)39. To assess the reprogramming capacity of Ascl1SA6, we employed an AAV5-GFAP vector to drive its expression, including an mCherry reporter (Fig. 1b).
Notably, the astroglial origin of neurons generated using AAV-GFAP promoter-based vectors have been questioned due to off-target expression of reprogramming TFs in endogenous neurons14,17,18,19,20. Thus, to test whether any neuron-like cells induced by Ascl1SA6 expression were astrocyte-derived, we performed lineage tracing by pre-labeling astrocytes using Slc1a3-CreERT;Rosa-zsGreen mice (Fig. 1b). To activate CreERT, we administered 4-hydroxytamoxifen (4-OHT) intraperitoneally every other day for four total doses prior to viral transduction. Three days after 4-OHT administration, AAV5-GFAP-mCherry vectors were stereotactically delivered into the motor cortex (Fig. 1b).
At 21 days post infection (dpi), 29% of GFAP+ astrocytes were zsGreen+, likely reflecting incomplete recombination and/or molecular heterogeneity among astrocyte populations (Fig. 1c, d)43. Of the mCherry+ transduced cells, 18% were pre-labeled zsGreen+ astrocytes, which became the focus of subsequent analyses (Fig. 1c, d). To assess whether Ascl1SA6 could induce neuronal reprogramming in these pre-labeled astrocytes, AAV5-GFAP-mCherry (hereafter, mCherry) or AAV5-GFAP-Ascl1SA6-v2a-mCherry (hereafter, Ascl1SA6) vectors were delivered into the motor cortex of 16-week-old Slc1a3-CreERT;Rosa-zsGreen mice. At 21 dpi with Ascl1SA6, ASCL1 expression was detected in 50.3 ± 7.7% of mCherry+ transduced cells that were also zsGreen-expressing (Fig. 1e). In the control condition, all zsGreen+mCherry+ transduced cells retained GFAP expression, whereas only 17.8 ± 9.7% of mCherry+zsGreen+ cells co-expressed GFAP after Ascl1SA6 transduction (Fig. 1f). Instead, 63.9 ± 7.3% of mCherry+zsGreen+ cells in the Ascl1SA6-condition initiated expression of DCX, an immature neuronal marker (Fig. 1g), and 46.3 ± 6.7% mCherry+zsGreen+ cells expressed NEUN, a marker of mature neurons (Fig. 1h).
To further validate the astrocytic origins of Ascl1SA6-converted neuronal-like cells, we implemented an alternative lineage-tracing strategy, crossing Aldh1l1-CreERT2 mice with a Rosa-tdTomato reporter line. CreERT2 recombinase was activated in Aldh1l1+ astrocytes by administering tamoxifen in the chow for seven days, and three days later, AAV5-GFAP vectors expressing either Emerald GFP (EmGFP) alone or Ascl1SA6 were delivered to the motor cortex (Supplementary Fig. 1a). At 21 dpi, histological analysis revealed that Ascl1SA6 induced expression of the neuronal marker NEUN in 9.23 ± 0.49 of EmGFP+ astrocytes per mm2, significantly more than observed in EmGFP controls (Supplementary Fig. 1b, c). Taken together, these results confirm that Ascl1SA6 reprograms a subset of lineage-traced astrocytes towards a neuronal fate, as evidenced by downregulation of astrocytic gene expression and activation of neuronal markers in targeted astrocytes.
Incorporation of miR-124 target sites reduces transgene expression in endogenous neurons
Despite encouraging reprogramming outcomes, off-target transgene expression in endogenous neurons remains a persistent challenge when utilizing AAV vectors in which the GFAP promoter drives neurogenic TF expression14,17,18,19,20. A previous study demonstrated that incorporating four copies of a neuron-specific microRNA-124 (miR-124) target site, hereafter referred to as 124 T, into the 3’UTR of a GFAP-driven GFP reporter, effectively reduces neuronal expression in vitro44. To test whether this strategy could similarly suppress off-target neuronal expression of GFAP-Ascl1SA6 vectors in vivo, we incorporated four-124T repeats into the 3’UTR of mCherry (mCh) in control (mCh-124T) and Ascl1SA6 (Ascl1SA6-mCh-124T) vectors (Supplementary Fig. 2a). As an additional control, the directional 124 T sequences were reversed to generate Ascl1SA6-mCh-124T-R, which should not bind miR-124 (Supplementary Fig. 2a). All constructs were cloned into AAV2/5-GFAP backbones and delivered into the motor cortex of adult C57BL/6 mice (Supplementary Fig. 2b). At 21 dpi, ASCL1 was expressed in 85.80 ± 2.82% of mCherry+ cells transduced with Ascl1SA6-mCh-124T, and 68.39 ± 6.25% with Ascl1SA6-mCh-124T-R (Supplementary Fig. 2c, f). Expression of the astrocytic marker, GFAP, was reduced in Ascl1SA6-mCh-124T (46.73 ± 6.13%) and Ascl1SA6-mCh-124T-R (22.56 ± 1.08%) transduced cells relative to the mCh-124T control vector (96.83 ± 1.9%), but to a lesser extent when the 124T sequence was in the correct orientation (Supplementary Fig. 2d, f). Consistently, the neuronal marker NEUN was detected in only 44.54 ± 4.71% of Ascl1SA6-mCh-124T-transduced cells versus 81.29 ± 1.37% in Ascl1SA6-mCh-124T-R conditions, the latter increase possibly due to the reported reduction in gene expression when incorporating 124 T sites44 (Supplementary Fig. 2e, f).
Given the promising utility of the 124 T strategy, we assessed its impact on neuronal conversion in Slc1a3-CreERT;Rosa-zsGreen mice pre-labeled for astrocytic identity via four injections of 4-OHT (Fig. 2a, b). Three days following the final injection, AAV5-GFAP vectors encoding mCh-124T, Ascl1SA6-mCh-124T, or Ascl1SA6-mCh-124T-R were delivered intracranially into the motor cortex (Fig. 2a–c), and cellular marker expression was evaluated at 21 dpi. Focusing on transduced mCherry+ cells that were also zsGreen+, ASCL1 was expressed in comparable numbers of cells after transduction with Ascl1SA6-mCh-124T (59.08 ± 1.1%) and Ascl1SA6-mCh-124T-R (55.58 ± 1.22%) (Fig. 2d, h). Astrocytic marker GFAP expression was similarly reduced in mCherry+zsGreen+ cells following transduction with Ascl1SA6-mCh-124T (4.93 ± 0.14%) and Ascl1SA6-mCh-124T-R (2.56 ± 0.56%) (Fig. 2e, i). However, expression of DCX, an immature neuronal marker, was more modest in mCherry+zsGreen+ cells in the correctly oriented 124T construct (23.11 ± 3.6%) compared to the reversed sequence (46.30 ± 6.7%) (Fig. 2f, j). Similarly, fewer mCherry+zsGreen+ cells expressed NEUN, a mature neuronal marker, in the correctly oriented 124T construct (42.2 ± 0.44%) compared to the reversed sequence (56.55 ± 1.88%) (Fig. 2g, k). Relative to Ascl1SA6 lacking the 124T sequences (Fig. 1h), Ascl1SA6-mCh-124T induced neuronal expression in approximately half as many mCherry+zsGreen+ cells, possibly due to reduced transgene expression, as reported with 124 T inclusion in vitro44. Nevertheless, these findings demonstrate that the AAV-GFAP-Ascl1SA6 vector can effectively reprogram lineage-traced astrocytes into neuron-like cells and suggest that 124 T sequences function in vivo to detarget endogenous neurons.
a Pre-labeling of astrocytes in Slc1a3-CreERT;Rosa-zsGreen mice treated with 4-OHT four times, separated by a day per injection. At 3 days after the last injection, AAVs were administered, and brains were collected after 21 dpi. BioRender was used to create the schematics. b Slc1a3-CreERT mediated recombination of the Rosa-zsGreen locus. BioRender was used to create the schematics. c AAV-GFAP vectors incorporating four copies of the miR-124 target sites (‘124T’) in the 3’UTR of mCherry, either in the correct orientation or reversed. The three constructs utilized were abbreviated mCh-124T, Ascl1SA6-mCh-124T, and Ascl1SA6-mCh-124T-R. BioRender was used to create the schematics. d–g Labeling of Slc1a3-CreERT;Rosa-zsGreen motor cortices that had been transduced with mCh-124T, Ascl1SA6-mCh-124T, or Ascl1SA6-mCh-124T-R, assayed for co-expression of mCherry and zsGreen with ASCL1 (d), GFAP (e), DCX (f) or NEUN (g). White arrows indicate mCherry+zsGreen+; yellow arrows indicate mCherry-negative/zsGreen+. Scale bars = 100 µm. h–k The percentage of mCherry+zsGreen+ cells that co-expressed ASCL1 (h), GFAP (i), DCX (j) and NEUN (k) (n = 3 each vector). Mean values and error bars representing the standard error of the mean (s.e.m.) were plotted. Statistical significance was assessed by a one-way ANOVA with Tukey’s post hoc multiple comparisons test.
Single-cell transcriptomics identifies a transitional cell cluster with bifurcation toward neuronal and oligodendrocyte lineages
To investigate the molecular features of neuronal conversion and profile the transcriptomes of reprogrammed cells, we performed single-cell RNA sequencing (scRNA-seq). For this purpose, we generated a new series of AAV5-GFAP constructs by replacing the mCherry coding sequence with iCre, maintaining the established naming convention (Fig. 3a). Each iCre construct was co-delivered with an AAV5-FLEX-GFP reporter into the motor cortex of C57BL/6 mice to facilitate identification of transduced regions, even at longer time points post-transduction (Fig. 3b, c). At 7 and 50 dpi, GFP-positive motor cortex tissue was microdissected (Fig. 3c, d), dissociated into single cells, and processed for library preparation and sequencing. Following quality control filtering, we retained 2671, 12,434, and 1972 high-quality cells from the iCre-124T, Ascl1SA6-iCre, and Ascl1SA6-iCre-124T conditions at 7 dpi, respectively (Supplementary Fig. 3a–e). At 50 dpi, the corresponding numbers of retained cells were 7169, 5133, and 12,610, respectively (Supplementary Fig. 3a–e). The resulting disparity in cell counts across conditions, and particularly within certain populations (e.g., only 20 cells undergoing conversion at 7 dpi for Ascl1SA6-iCre-124T), limited our ability to perform balanced subsampling analyses. Nevertheless, the dataset preserved similar cellular representation across time points and constructs, permitting meaningful comparative analyses of transcriptional trajectories, lineage bias, and molecular signatures of conversion, despite imbalances in cell yield. Finally, as part of additional quality control assessments, Gapdh expression, a constitutively expressed reference gene, was comparable across all samples (Supplementary Fig. 3f). Furthermore, iCre transcripts were detected across all experimental conditions, although substantial variability in expression levels among samples precluded a reliable assessment of the impact of the 124T sequences on transgene expression (Supplementary Fig. 3g, h).
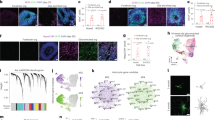
a AAV-GFAP vectors incorporating four copies of the miR-124 target sites in the 3’UTR of iCre in the correct orientation. The three constructs utilized were abbreviated iCre-124T, Ascl1SA6-iCre, and Ascl1SA6-iCre-124T. BioRender was used to create the schematics. b, c A FLEX-GFP reporter system used to identify transduced cells (b). Time points of scRNA-seq analyses at 7 and 50 dpi (c). BioRender was used to create the schematics. d Motor cortex collected 21 days after co-transduction with Ascl1SA6-iCre and FLEX-GFP, showing expression of GFP in the transduced area of the motor cortex. Scale bar = 100 µm. e UMAP plot of combined scRNA-seq data sets from day 7 and day 50 post-infection (n = 3 mice/construct/time point). f Subclustering of the neural cell-specific region revealed distinct cell populations, including astrocytes (clusters 1–3, 9), oligodendrocytes (clusters 4, 8), neuron-like cells (cluster 5), an undetermined group (cluster 6), and endogenous neurons (cluster 7). g Cellular proportion shifts, comparing control to Ascl1SA6, or control to Ascl1SA6-124T at 50 dpi, and plotting Log2 fraction differences. Red dots show statistically significant increases in cellular populations, with an FDR < 0.05 and Log2FD > 1.5. h Proportion of cells in clusters 1-9 that were in G1, S or G2/M-phases of the cell cycle.
Integration of all scRNA-seq samples and subsequent clustering led to the identification of 17 unique groups (Fig. 3e), which were mostly present in all samples (Supplementary Fig. 3a–d). Cluster annotation was performed using cell type-specific markers obtained from the CellMarker 2.0 database45. All major brain cell types were identified, including astrocytes, oligodendrocytes and neurons, as well as endothelial cells and immune cells, such as microglia, B-cells, T-cells and macrophages (Fig. 3e). The prevalence of immune cells was in keeping with the disruption of brain tissue by the microsurgery, and the adaptive immune response of the brain to AAVs46. In a subclustering analysis of neural cells, cluster 6 was positioned at the intersection of astrocyte-associated clusters (1, 2, 3, and 9), oligodendrocyte-associated clusters (4 and 8), and neuron-associated clusters (5 and 7) in the UMAP space (Fig. 3f). Cluster 6 could not be identified since it displayed high levels of heterogeneity in marker expression, so we labeled it as “Undetermined” (Fig. 3f). Notably, this “Undetermined” cluster was not present in the control iCre-124T samples at either 7 or 50 dpi, and its location was consistent with the expected transcriptional profile of cells undergoing astrocyte-to-neuron or astrocyte-to-oligodendrocyte conversion.
To assess shifts in cell-type proportions, we compared cluster abundances between experimental and control groups and evaluated statistical significance using a permutation-based test (5000 iterations), as previously described47. The analysis revealed no significant changes in cell proportions for most clusters at both 7- and 50-days post transduction (Fig. 3g, Supplementary Fig. 4a, b). However, when differences were detected, they predominantly involved neuron- and oligodendrocyte-related clusters and occurred more frequently in the Ascl1SA6 and Ascl1SA6-124T experimental conditions (Fig. 3g). These shifts were especially pronounced at 50 dpi within neuron-like clusters, representing the primary population that was the product of fate conversion, further reinforcing the conclusion that successful reprogramming events had taken place (Fig. 3g). Notably, the x-axis of the plot depicts fold change in cell proportions. In cases where neuron-like cells were absent in controls, the fold change is mathematically infinite, causing those data points to appear at the edge of the plot, a visual reflection of the proportional shift when comparing zero to non-zero populations (Fig. 3g, Supplementary Fig. 4a, b).
Unique cell cycle and molecular signatures of astrocytes in cluster 3
We were intrigued by the observation that astrocytes in cluster 3 exhibited an enhanced capacity for neuronal conversion. During cortical development, neurogenic divisions are accompanied by an extended G1 phase, which provides a transcriptionally permissive window prior to DNA replication, facilitating chromatin engagement by neurogenic TFs and the initiation of lineage-specific gene expression48,49. Similarly, during fibroblast to motor neuron conversion in vitro, elevated proliferation rates support higher rates of Neurog2-induced neuronal conversion50. To evaluate whether astrocyte reprogramming potential was associated with cell cycle phase in vivo, we assigned cells to G1, S, or G2/M phases based on scoring with 43 markers for S phase and 54 markers for G2/M phase; cells with low scores for both phases were categorized as G1 (Fig. 3h). A Fisher’s exact test comparing astrocytes from cluster 3 to those in clusters 1 and 2 revealed a significant decrease in the proportion of G1-phase cells and concomitant increase in S and G2/M phases in cluster 3 (Fig. 3h). These findings suggest that proliferative activity, and progression into S- or G2/M-phases, may enable the transcriptional reconfiguration required for neuronal fate conversion.
To further investigate molecular features underlying this heightened reprogramming potential of cluster 3 astrocytes, we examined WNT signaling, a pathway known to regulate neurogenesis in postnatal and adult brains51,52. Notably, neurogenic induction in pluripotent stem cells can be enhanced by CHIR99021, a potent GSK3 inhibitor that activates WNT signaling4. Furthermore, high levels of GSK3 suppress Neurog2-mediated neurogenesis during embryonic development29. We therefore analyzed the expression of GSK3 downstream targets using feature plots and found that relative to clusters 1 and 2, cluster 3 astrocytes exhibited reduced expression of several key GSK3-regulated genes, including Dpysl2, Clasp1, Apc, Nfkb1, Tsc1, Tsc2, Creb1, and Ctnnb1 (Supplementary Fig. 5). Taken together, these data suggest that astrocytes in cluster 3 possess a distinctive cell cycle profile and a reduced GSK3 target gene expression signature, both of which may underpin their enhanced susceptibility to neuronal reprogramming.
Cell fate conversion involves several hybrid cell types
To further assess differences across the scRNA-seq datasets, we first overlaid UMAP embeddings of iCre control and Ascl1SA6 or Ascl1SA6-124T conditions at both 7- and 50 dpi; however, gross differences were not detectable at this global level (Supplementary Fig. 6a–d). We therefore focused subsequent analyses on subclusters corresponding to astrocytes, neurons, oligodendrocytes and undetermined populations, in which reprogramming events were expected to occur (Fig. 3f). To examine marker heterogeneity at finer resolution, we curated a panel of cell-type-specific genes and calculated scores for individual cells based on the geometric mean of non-zero expression values. Marker sets included astrocytic (Fabp7, Gfap, Aqp4, Slc1a2), mature oligodendrocytic (Mbp, Mog, Mag, Opalin), newly-formed oligodendrocytes (Cnp, Plp1, Foxo4, Galc), GABAergic (Pvalb, Gad1, Gad2, Npy), and glutamatergic (Slc17a7, Slc17a6, Gap43, Camk2a) neurons (Supplementary Fig. 6e–h). Cells with score differentials below 0.2 between the top two identity groups were designated as “hybrids” (Fig. 4a–d).
a, b Cluster assignments, including hybrid cell types, with a label indicating the centroid of each cell score distribution in a UMAP plot (a), or as a percentage of cells in each cluster with the assigned identity (b), focusing on 7 dpi. c, d Cluster assignments, including hybrid cell types with a label indicating the centroid of each cell score distribution shown in a UMAP plot (c), or as a percentage of cells in each cluster with the assigned identity (d), focusing on 50 dpi.
At 7 dpi, cluster 6 (undetermined) appeared transcriptionally heterogeneous, consistent with a transitional state possibly undergoing astrocyte-to-neuron or astrocyte-to-oligodendrocyte conversion events (Fig. 4a, b). In parallel, cluster 5, annotated as Neuron-like, was predominantly composed of GABAergic and astrocyte-GABAergic hybrid cells, with approximately 25% retaining astrocytic signatures (Fig. 4a, b). By 50 dpi, cluster 6 had largely shifted toward an oligodendrocyte identity, coinciding with a decline in the neuronal trajectory emanating from the same region (Fig. 4c, d). This shift may reflect either a slower trajectory into oligodendrocytes or a more rapid neuronal conversion that relocates reprogrammed cells from cluster 6 to cluster 5. Supporting this latter possibility, cluster 5 at 50 dpi displayed a reduced proportion of astrocytes and a corresponding increase in GABAergic-scored cells and hybrids thereof. Importantly, the spatial distribution of cell scores revealed that cells with GABAergic and astrocyte-GABAergic hybrid identities were enriched within the zone predicted to harbor conversion activity (Fig. 4c, d).
Marker-based scoring revealed that Cluster 7 initially comprises a dominant population of endogenous GABAergic neurons, which undergo substantial cell loss over time. At day 7, GABAergic cells (including hybrids) represented the majority of this cluster (Fig. 4a, b), but by day 50, their numbers and relative proportions declined markedly (Fig. 4c, d). This shift in cell-type composition is most likely attributed to decreased GABAergic neuronal survival in the sorted cell pool, rather than transdifferentiation. Additionally, classification noise arising from non-specific markers became more pronounced at later time points due to lower cell counts, resulting in a more heterogeneous cell identity distribution within the cluster (Fig. 4c, d). Together, these results demonstrate that combining subclustering with marker-based scoring enhances resolution of fate transitions, and pinpoints cluster 5 as a likely endpoint of successful astrocyte-to-neuron reprogramming.
Incorporation of 124T sequences alters neuronal transcriptional profiles during astrocyte reprogramming
To assess whether 124T sequences function as neuronal detargeting elements, we compared differentially expressed genes (DEGs) across cell clusters following transduction with either Ascl1SA6 or Ascl1SA6-124T vectors, relative to controls (Supplementary Fig. 7a–h). At 7 dpi, Ascl1SA6-transduced samples exhibited transcriptional changes in astrocytes, converting cells, mature neurons, and to a lesser extent, newly generated and mature oligodendrocytes (Fig. 5a; Supplementary Data 1). In contrast, Ascl1SA6-124T transduction induced DEGs exclusively in astrocytes, consistent with effective detargeting of endogenous neurons (Fig. 5a; Supplementary Data 2). Although DEG numbers declined in the Ascl1SA6 group by 50 dpi, transcriptional changes persisted in astrocytes and converting cells (Fig. 5b; Supplementary Data 3). However, by 50 dpi, transcriptomic divergence became more pronounced in the Ascl1SA6-124T condition, notably within astrocytes and converting neurons, suggestive of a lag in the onset of gene expression (Fig. 5b; Supplementary Data 4). Interestingly, both conditions showed enrichment of DEGs in mature oligodendrocytes at this later stage, suggesting a potential bifurcating lineage trajectory (Supplementary Data 3 and 4). Collectively, these results support robust neuronal detargeting effects mediated by the 124T sequences.
a, b Total number of DEGs in each cell type after transduction with Ascl1SA6 (purple bars) or Ascl1SA6-124T (blue bars) at day 7 (a) and day 50 (b). c, d Log2FC in gene expression, comparing iCre to Ascl1SA6-iCre (c) or iCre to Ascl1SA6-iCre-124T (d). Genes were categorized as associated with neurons, astrocytes or glycolysis. e, f Motor cortices transduced with mCh-124T, Ascl1SA6-mCh, or Ascl1SA6-mCh-124T were assayed for co-expression of mCherry with NPY (e), or NLGN1 (f). White arrows indicate marker+mCherry+; yellow arrows indicate marker-negative/mCherry+. Scale bars = 100 µm. The percentage of marker expression in mCherry+ cells was quantified (n = 3 animals each vector). Mean values and error bars representing the standard error of the mean (s.e.m.) were plotted. Statistical significance was assessed by a one-way ANOVA with Tukey’s post hoc multiple comparisons test.
To investigate Ascl1SA6-driven astrocyte-to-neuron conversion, we performed gene ontology (GO) enrichment analysis of DEGs at 7 dpi. Compared to iCre controls, Ascl1SA6-iCre samples showed significant enrichment of Biological Process terms related to neurogenesis, including “positive regulation of nervous system development”, “synaptic assembly”, and “positive regulation of axonogenesis” (Supplementary Fig. 7a, b). Further analysis of the astrocyte cluster revealed increased expression of genes linked to neuronal differentiation and decreased expression of astrocytic markers and glycolytic genes (Fig. 5c), a metabolic shift consistent with transition toward a neuronal phenotype53. In contrast, at 7 dpi, astrocytes transduced with Ascl1SA6-iCre-124T exhibited enrichment in “regulation of membrane potential” terms (Supplementary Fig. 7c, d) and also upregulated neuron-associated genes while suppressing astrocytic and glycolytic signatures in the astrocytic cluster (Fig. 5d). At 50 dpi, both constructs induced immune-related gene expression, indicative of inflammatory responses to reprogramming (Supplementary Fig. 7e–h).
Focusing on neuronal genes turned on in astrocytes, Ascl1SA6 induced expression of both GABAergic interneuron-specific genes (Sp8, Npy, Pvalb, Sox6) and pan-neuronal genes (Dll1, Dcx, Map2, Rnd3, Id1, Grik2, Gria2) (Fig. 5c). Of note, Dll1 and Rnd3 are known Ascl1 target genes, supporting a direct mechanistic link to reprogramming. In contrast, Ascl1SA6-124T preferentially activated genes linked to a glutamatergic identity, including Grik2, Kcnd2, Nlgn1, Nrxn1, Ncam2, Npas3, Syn2, Gria2, Map2, Nrcam, suggesting a bias toward excitatory neuronal reprogramming (Fig. 5d). Notably, other genes induced by Ascl1SA6-124T, such as Sox6, Sox5, and Tle4, have context-dependent roles in inhibitory interneuron specification, indicating partial overlap in transcriptional programs during early neuronal fate transitions.
To validate differential marker induction, we transduced brains with mCh-124T control, Ascl1SA6-mCh, or Ascl1SA6-mCh-124T vectors and analyzed them at 21 dpi. In line with the scRNA-seq results, Ascl1SA6 robustly induced NPY expression (Fig. 5e), whereas Ascl1SA6-124T preferentially activated NLGN1 (Fig. 5f). However, induction of these target genes was not exclusive; both constructs activated both markers, albeit to a lesser degree with one construct or the other. In contrast, SOX6, which was upregulated by Ascl1SA6 and Ascl1SA6-124T in the scRNA-seq datasets to a similar extent, was also induced in a similar number of cells at the protein level (Supplementary Fig. 8). These findings are consistent with the high prevalence of hybrid cell types following astrocytic transduction with Ascl1SA6 or Ascl1SA6-124T. In summary, the 124T sequence effectively detargets endogenous neurons, fine-tuning the site of transgene expression, while simultaneously altering the transcriptional identity of reprogrammed cells. This modulation drives a distinct reprogramming trajectory by preventing activation of ectopic genes in endogenous neurons and enhancing excitatory neuronal signatures.
124T sequences delay conversion kinetics and drive lineage bifurcation during neuronal reprogramming
To investigate the dynamics of cellular conversion driven by Ascl1SA6 and Ascl1SA6-124T, we performed pseudotime trajectory analysis on scRNA-seq datasets. At 7 dpi following Ascl1SA6-iCre transduction, pseudotime ordering revealed a continuum from astrocytes at low (early) pseudotime values, through undetermined intermediate cells, to neuron-like cells at higher (later) pseudotime values (Fig. 6a). By 50 dpi, both undetermined and neuron-like populations were no longer detectable (Supplementary Fig. 9a), despite continued expression of iCre transcripts (Supplementary Fig. 3g, h), suggesting either selective cell loss or convergence toward endogenous neuronal identity.
a, b Pseudotime trajectory analyses, originating in the astrocyte cluster, of Ascl1SA6-iCre-transduced cells at 7 dpi (a) and Ascl1SA6-iCre-124T-transduced cells at 50 dpi (b). c TF analysis of scRNA-seq data using SeeSawPred reveals Foxo1 enrichment in neuron-like cells and Stat1/Nr3c1 enrichment in oligodendrocyte-like cells.
In contrast, Ascl1SA6-iCre-124T transduction resulted in only a sparse population of undetermined cells, and no neuron-like cells, at 7 dpi (Supplementary Fig. 9b). However, by 50 dpi, both populations emerged, and pseudotime trajectories revealed a bifurcation into either oligodendrocyte or neuron-like lineage, with oligodendrocytes occupying the highest pseudotime values, indicative of more advanced or terminal states (Fig. 6b). These findings indicate that Ascl1SA6 drives rapid neuronal-like conversion, while Ascl1SA6-124T delays fate acquisition and expands lineage outcomes.
scRNA-seq analysis reveals a lineage bifurcation into oligodendrocyte and neuronal trajectories upon incorporation of 124T sequences, indicating that target astrocytes retain a degree of multipotency and that Ascl1SA6 alone does not consistently drive neurogenesis. To investigate the transcriptional mechanisms underlying this divergence, we employed SeeSawPred, a computational tool designed to identify TFs governing binary cell fate decisions54. In alignment with trajectory inference, Foxo1 expression was enriched in neuron-like cells (Fig. 6c). Foxo1 is expressed in neural progenitor cells during the period of neurogenesis, although its expression declines upon neuronal differentiation, in part due to targeting by the neurogenic miR-955. Conversely, Stat1 and Nr3c1 were preferentially expressed in oligodendrocyte-like populations, consistent with the known role of cytokine signaling and downstream STAT-TFs in glial maturation and the regulation of myelin-associated genes56. These findings suggest that astrocyte reprogramming outcomes may remain plastic, with terminal identity influenced by intrinsic transcriptional states and potentially modulated by extrinsic environmental cues.
Taken together, our results confirm that Ascl1SA6 is a potent driver of astrocyte-to-neuron conversion, rapidly inducing a neuron-like identity. Importantly, addition of the 124T modification to Ascl1SA6 prevents ectopic transgene activation in endogenous neurons when employing the GFAP promoter, and markedly reconfigures the reprogramming trajectory, delaying fate transitions, and promoting a bifurcation into neuronal and oligodendrocyte lineages.
Discussion
In this study we demonstrate that Ascl1SA6 is a potent neurogenic TF that drives astrocyte-to-neuron reprogramming in the adult brain, converting lineage-traced astrocytes into neuron-like cells expressing PVALB, GAD1, and NPY. This phenotype mirrors that of fast-spiking interneurons previously shown to be induced by Ascl1SA6 in proliferating, early postnatal astrocytes via retroviral delivery40. Notably, NPY is frequently co-expressed with PVALB during development or in specific physiological states, reinforcing an inhibitory interneuron identity of the reprogrammed neuron-like cells. Among three astrocyte clusters, only one progressed into a transitional state enriched for cells in S- and G2/M-phases and marked by low GSK3 target gene expression, a molecular and cellular signature that may enhance reprogramming responsiveness. Incorporating 124T sites into the Ascl1SA6 transcriptional cassette effectively suppressed transgene expression in endogenous neurons, refining cell-type specificity. However, beyond detargeting, 124T appears to delay reprogramming kinetics and redirects converting cellular trajectories towards both glutamatergic neuron-like and oligodendrocyte-like fates. This bifurcation is substantiated by transcriptional profiling and SeeSawPred analysis, which reveal distinct TF programs along each lineage. These results underscore Ascl1SA6’s capacity to induce subtype-specific neural cell identities and demonstrate how regulatory sequence modifications can broaden glial lineage potential. For instance, the ability to generate both oligodendrocytes and neurons could enhance the therapeutic relevance of neuronal reprogramming in treating conditions involving neuronal loss and demyelination.
Based on our scRNA-seq analyses, we identified three transcriptionally distinct astrocyte clusters, among which cluster 3 showed the highest propensity for neuronal reprogramming. This cluster exhibited reduced expression of GSK3 target genes compared to clusters 1 and 2, a molecular profile that may facilitate neuronal conversion, given GSK3’s established role in repressing proneural TF activity29 and in suppressing neuronal morphogenesis57. Indeed, pharmacological or genetic inhibition of GSK3 enhances fibroblast-to-neuron conversion4, and facilitates reprogramming by neurogenic TFs such as Neurog258,59. Thus, attenuated GSK3 signaling in cluster 3 astrocytes may create a more permissive cellular environment for reprogramming. Intriguingly, cell cycle phase analysis revealed that reprogrammable astrocytes in cluster 3 were enriched in S and G2/M phases. This transition likely reflects a transition from G₀ quiescence to an actively proliferative state, rather than implicating particular phases as intrinsically favorable for neuronal conversion. Supporting this interpretation, increased proliferative capacity of target cells has recently been shown to enhance the efficiency of neuronal reprogramming in vitro50. Notably, these findings diverge from the well-established cell cycle length hypothesis in developmental neurogenesis, which posits that a prolonged G₁ phase enhances the differentiation capacity of neural progenitor cells49. Dissecting the mechanistic distinctions between embryonic neurogenesis and induced neuronal reprogramming remains a key future challenge.
One intriguing aspect of our study was the transcriptomic shift observed in induced neurons generated with Ascl1SA6 compared to those produced using the Ascl1SA6-124T construct. A similar phenomenon was reported in a separate study employing Neurod1-based reprogramming, in which the inclusion of 124T sites in the 3′ UTR of Neurod1 led to a marked shift in neuronal subtype specification, from a predominantly glutamatergic profile with Neurod1 alone to a higher proportion of GABAergic neurons with Neurod1-124T60. This altered fate outcome was attributed to reduced Neurod1 expression levels due to miR-124-mediated post-transcriptional suppression60. Although we did not observe a statistically significant decrease in Ascl1SA6-iCre transgene expression when incorporating 124T sites, variability across our three in vivo replicates may have obscured expression-level effects. Nevertheless, it is well-established that proneural TF dosage can influence neuronal fate decisions. For instance, Neurog2 expression at endogenous levels, achieved via knock-in into the Ascl1 locus in ventral telencephalic progenitors, fails to redirect these cells, normally committed to a GABAergic fate, towards a glutamatergic identity61. In contrast, Neurog2 overexpression via in utero electroporation under the control of a strong CAG promoter successfully reprograms ventral telencephalic progenitors toward a glutamatergic lineage62, illustrating the importance of TF dosage in directing neuronal specification.
The ability of the 124T sites not only to detarget neurons, but also to redirect cell fates towards glutamatergic and oligodendrocyte fates was unexpected. One possibility is that the 124T sites added to the 3′UTR of Ascl1SA6 may function as a microRNA sponge, sequestering endogenous miR-124 and thereby impairing its ability to repress native target genes. This sponge effect could occur if the 124T sites were present in sufficient copy number and affinity to competitively bind miR-124, diverting it from its physiological targets. This mechanism has precedent: circular RNAs such as circHIPK3 and circMMP9 have been shown to act as sponges for miR-124 in cancer and neurodevelopmental contexts, resulting in the upregulation of target genes like AQP3, CDK4, and AURKA, which are normally repressed by miR-124 and play roles in proliferation and migration63,64. Consequently, in the presence of supernumerary 124T sites, genes normally silenced by miR-124, including those inhibiting neuronal differentiation, may become derepressed, potentially influencing the fate or identity of reprogrammed cells. For instance, miR-124 inhibits REST and CoREST, lifting repression on neuronal genes65. If this effect were lost through 124T sponge effects, then a shift to the oligodendrocyte lineage could be facilitated. Notably, Ascl1 itself can upregulate miR-124 in lung carcinomas66, and miR-124 overexpression has been shown to reprogram astrocytes in vitro67, highlighting the important role of this molecular switch in neuronal conversion.
Sponge-based interference may also disrupt the “fate erasure” that miR-124 normally orchestrates during neuronal reprogramming68. In this process, miR-9/9* and miR-124 initiate neuronal conversion by first silencing somatic identity, repressing gene networks involved in cell cycle, adhesion, metabolism, and chromatin organization, before activating neuronal transcriptional programs68. Key steps in this erasure process include repression of TFs such as KLF4 and KLF5, and activation of chromatin remodelers such as the small nuclear RNA 7SK, which facilitates neuronal gene accessibility. miR-124 also governs early metabolic reprogramming critical to the neurogenic cascade69. Thus, if 124 T sites act as a sponge in our constructs, they may limit the functional pool of miR-124, allowing residual somatic gene networks to persist and potentially skewing the reprogramming trajectory. Thus, even without a measurable reduction in transgene expression, the functional availability of miR-124 may be compromised, leading to indirect effects on gene networks that are sensitive to miR-124 regulation and contributing to the altered transcriptomic profiles and fate outcomes observed with Ascl1SA6-124T.
Collectively, our findings position Ascl1SA6 as a potent and programmable effector of astrocyte-to-neuron conversion and demonstrate that strategic placement of miR-124 target sequences within the 3′UTR modulates transcriptional specificity, refines lineage trajectory, and alters glial reprogramming potential in the adult brain.
Methods
Animals
All animal procedures were approved by the Sunnybrook Research Institute Animal Care Committee (ACC 21-757 and 16-769) or the University of Toronto ACC (AUP 20012508) in agreement with the Guidelines of the Canadian Council of Animal Care (CCAC). Mice were purchased from The Jackson Laboratory, Bar Harbor, ME, USA, and included Slc3a1-CreERT (RRID:IMSR_JAX:012586), Rosa-zsGreen (RRID:IMSR_JAX:007906), Aldh1l1-CreERT2 (RRID:IMSR_JAX:029655) and Ai14 (B6.Cg-Gt(ROSA)26Sor tm14(CAG-tdTomato)Hze/J) (RRID:IMSR_JAX:007914) strains. All transgenic mice were maintained on a C57BL/6J background (RRID:IMSR_JAX:000664). PCR genotyping was conducted according to Jackson Laboratory protocols. The following PCR primers were used: Rosa-zsGreen: wild-type forward: 5’-CTG GCT TCT GAG GAC CG-3’; wild-type reverse: 5’-AAT CTG TGG GAA GTC TTG TCC-3’; mutant forward: 5’-ACC AGA AGT GGC ACC TGA C-3’; mutant reverse: 5’-CAA ATT TTG TAA TCC AGA GGT TGA-3. PCR cycles were: 94 °C 2 min, 10x (94 °C 20 s, 65 °C 15 s *−0.5c per cycle decrease, 68 °C 10 s), 28x (94 °C 15 s min, 60 °C 15 s, 72 °C 10 s), 72 °C 2 min Slc1a3-cre/ERT wild-type Locus F: 5’-CTAGGCCACAGAATTGAAAGATCT-3’, Slc1a3-CreERT wild-type Locus R 5’-GTAGGTGGAAATTCTAGCATCATCC-3’. Slc1a3-CreERT mutant Locus F: 5’-ATACCGGAGATCATGCAAGC-3’. Slc1a3-CreERT mutant Locus R: 5’GGCCAGGCTGTTCTTCTTAG-3’. Cycles MT + WT: JAX: 94 °C 2 min, 10x (94 °C 20 s, 65 °C 15 s [*−0.5 °C per cycle], 68 °C 10 s), 28x (94 °C 15 s, 60 °C 15 s, 72 °C 10 s), 72 °C 2 min Amplicon Sizes: MT: ~200 bp; WT: 324 bp. Cre-transgene F: 5ʹ-TTGCCAGGATCAGGGTTAAAG-3ʹ. Cre-transgene R: 5ʹ-TCATCCTTAGCGCCGTAAATC-3ʹ. PCR Cycling Conditions: 94 °C 3 min, 35x (94 °C 30 s, 58 °C 30 s, 72 °C, 1 min), 72 °C 10:00. Amplicon Size: Cre+: 300 bp. tdTomato wild-type F: 5ʹ-AAGGGAGCTGCAGTG GAGTA-3ʹ. tdTomato wild-type R: 5ʹ-CCGAAAATCTGTGGGAAGTC-3ʹ. tdTomato mutant R: 5ʹ-GGCATTAAAGCAGCGTATCC-3ʹ. tdTomato R: 5ʹ-CTGTTCCTGTACGGCATGG-3ʹ. PCR Cycling conditions same as for Slc1a3-CreERT described above. Amplicon Sizes: MT: 196 bp; WT: 297 bp.
Tamoxifen-induced CreERT activation
To activate Slc1a3-CreERT, 100 μg of 4-hydroxy-tamoxifen (4-OHT) was administered every second day via intraperitoneal injection for a total of four injections. Three days later, AAVs were transduced. To lineage trace Aldh1l1+ astrocytes, 6-month-old male Aldh1l1-CreERT2 mice were fed ~250 mg/kg of tamoxifen (TAM) in the form of Tamoxifen chow (6 kg of Catalog #314.46, Teklad Diets, Indianapolis, IN, USA formulated with 350 g of Tamoxifen (0.5% tamoxifen in sucrose, Catalog #T5648, Sigma-Aldrich, St Louis, MO, USA)) for seven days, followed by a 3-day wash-out period.
Adeno-associated viruses (AAVs)
All pAAV-GFAPshort vectors included a 681 bp (gfaABC(1)D) modified GFAP promoter70. The generation of AAV2/5-GFAP-mCherry and AAV2/5-GFAP-Ascl1SA6-t2a-mCherry vectors was previously described39. We outsourced to GenScript to generate the following five new AAVs: (1) pAAV-GFAPshort-mCherry-miR-124 target sites (x4), designated mCh-124T; (2) pAAV-GFAPshort-Ascl1SA6-mCherry-miR-124 target sites (x4), designated Ascl1SA6-mCh-124T; (3) pAAV-GFAPshort-Ascl1SA6-mCherry-miR-124 target sites (x4) reversed in orientation, designated Ascl1SA6-mCh-124T-R; (4) pAAV-GFAPshort-iCre-miR-124 target sites (x4), designated iCre-124T; and (5) pAAV-GFAPshort-Ascl1SA6-iCre-miR-124 target sites (x4), designated Ascl1SA6-iCre-124T. Sequence information for the four miR-124 target sites was previously described44. After cloning, all AAVs were packaged into an AAV5 capsid by VectorBuilder Inc (Chicago, IL, USA). Titers for all viruses are provided in Supplementary Table 1. For the iCre constructs, AAVs were co-transduced with pAAV-FLEX-GFP (Catalog # 28304, Addgene) packaged in capsid 5. As a control for Supplementary Fig. 1, AAV2/5-GFAPshort-EmGFP (cloned and packaged by VectorBuilder Inc.) was injected into the left burr hole.
Intracranial injections
AAVs were injected in the motor cortex bilaterally using a stereotax with the following coordinates: anterior-posterior (AP) + 2.15, medial-lateral (ML) ± 1.7, dorsal-ventral (DV): −1.7 as previously described39. For Supplementary Fig. 1, the coordinates were slightly different (AP + 2.2 mm, ML ± 1.7 mm, DV −1.7). Briefly, mice were anaesthetized with isoflurane (2%, 1 L/min; Fresenius Kabi, CP0406V2) and administered Tramadol-HCL (20 mg/kg; Chiron, RxN704598), Baytril® (2.5 mg/kg; Bayer, 02249243), and saline (0.5 ml, Braun, L8001). To access the motor cortex, a burr hole was drilled through the skull using a dental drill (P/N 8177 #77, 0.018”, David Kopf Instruments, Tujunga, CA, USA), and bregma and lambda coordinates were landmarked using a stereotaxic instrument. Each motor cortex was injected with 4.8 × 109 genome copies (GC) in a 1 μL volume at a speed of 0.1–0.2 µl/min over 5–10 min, using a 5 μl Hamilton syringe with 33-gauge or 26-gauge needle (Hamilton, 7803-07, The Hamilton Company, Reno, NV, USA), followed by a 5-min rest before the needle was removed.
Tissue processing and immunostaining
Mice were perfused prior to dissection. Briefly, animals were terminally anesthetized with Avertin prepared by dissolving 1.25% (w/v)2,2,2-tribromoethanol (Catalog #T48402, Sigma-Alrich) and 2.5% (v/v) 2-methyl-2-butanol (Catalog #152463, Sigma-Aldrich), or with ketamine (75 mg/kg, Narketan, 0237499) and xylazine (10 mg/kg, Rompun, 02169592). Once anesthetized, mice were perfused transcardially with a 20-fold amount (compared to blood volume) of ice-cold saline (0.9% NaCl, Braun, L8001) that was delivered with a peristaltic pump at 10 ml/min, followed by 4% paraformaldehyde (PFA, Electron Microscopy Sciences, 19208) diluted in 1X phosphate buffer saline (PBS, Wisent, 311-011-CL) for 5 min. Brains were drop-fixed overnight in 4% PFA at 4 °C or for 2 hours in 4% PFA at 4 °C. Brains were then washed three times for 10 min each in 1X PBS, cryoprotected in 20–30% sucrose/1X PBS overnight at 4 °C, and blocked and frozen directly, or frozen in O.C.T™ (Tissue-Tek®, Sakura Finetek U.S.A. Inc., Torrance, CA). Brains were sectioned coronally and cut at 10 μm or 20 μm (only Supplementary Fig. 1) on a Leica CM3050 cryostat (Leica Microsystems Canada Inc., Richmond Hill, ON, Canada) or a Thermo Scientific HM525 NX Cryostat (Waltham, MA, USA) and mounted on Fisherbrand™ Superfrost™ Plus Microscope Slides (Thermo Fisher Scientific, 12-550-15).
Immunostaining
Cryosections were hydrated with 250 µL of PBS (1X) for 5 min and then were permeabilized with 0.3% Triton X-100 (Catalog #TRX506.500, BioShop, Burlington, ON, CA) in 1X PBS for 30 min. Cryosections were then blocked for 1 h at room temperature in blocking buffer containing 10% horse serum (HS, Wisent, 065-150) and 0.1% Triton X-100 (Sigma, T8787) in PBS (PBST). For Supplementary Fig. 1, the blocking buffer was 5% normal goat serum (NGS, Catalog #005-000-121 Jackson Immunoresearch Labs, West Grove, PA, USA), 1% bovine serum albumin (BSA, Catalog #A7906-50G, Sigma-Aldrich) and 0.3% Triton X-100 in 1X PBS. Slides were incubated at 4 °C overnight with primary antibodies diluted in blocking solution as follows: rabbit anti-ASCL1 (1:500, Abcam #74065, rabbit anti-DCX (1:500, Abcam #ab18723), rat anti-GFAP (1:500, ThermoFisher #13-0300), mouse anti-NEUN (1:500, Millipore Sigma #MAB377), rabbit anti-NEUN (Sigma-Aldrich #ABN78), rabbit anti-NPY (1:500, Invitrogen PA5-85762), mouse anti-Neuroligin 1/NLGN1 (1:500, Novus #NBP2-42192), and rabbit anti-SOX6 (1:300, Protein Tech 14010-1-AP). The next day, cryosections were washed three times for 10 min each in PBS or in 0.1% PBST and incubated with 1:500–1:1000 dilutions of species-specific secondary antibodies all from Invitrogen Molecular Probes™ for 1 h at room temperature. Secondary antibodies conjugated to Alexa647 goat-anti-rat (A21247), donkey-anti-rabbit (A31573), goat-anti-mouse (A21235), and goat-anti-rabbit (A21245). Slides were washed three times in PBS and counterstained with 1:10,000 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen, D1306). Finally, cryosections were washed three times in PBS and mounted in Aqua-polymount (Polysciences Inc., 18606-20) or in Fluorescence Mounting Medium (Catalog #S302380-2, Agilent, Santa Clara, CA, USA).
Tissue processing for scRNAseq
Brains were harvested into cold 1X PBS 7- and 50-days post AAV injection. Using a brain slicer, brains were cut into coronal slices and the injected region in the cortex was microdissected. Dissected tissue was dissociated into single cell suspension according to the manufacturer’s instructions using Worthington Papain System kit (Worthington, LK003150). Using a Dead cell removal kit (130-090-101, Miltenyi Biotec), dead cells were removed following the manufacturer’s instructions. Trypan blue staining was used to determine cell viability, and samples with 90% viable cells were processed for analysis. Single-cell libraries were prepared using the Chromium Next GEM Single Cell 5’ Reagent Kit v2 (10X Genomics, Cat # 1000263), following the manufacturer’s protocol. Briefly, cell suspensions were prepared at a concentration of 1 × 10⁶ cells/mL, and 16,500 cells were loaded into each lane of a Chromium GEM Chip K for an expected recovery of 10,000 cells. Gel Bead-in-Emulsions (GEMs) were generated using the Chromium Controller (Wang Lab, Biological Sciences Platform of Sunnybrook Research Institute) followed by cDNA generation and amplification. For each sample, a total of 50 ng of cDNA was used to generate libraries according to the manufacturer’s instructions. Equal molar quantities of each library were sequenced on the Illumina NovaSeq 6000 platform using the S4 flow cell system (The Centre for Applied Genomics, The Hospital for Sick Children, Toronto).
scRNA-seq data analysis
scRNA-seq data were analyzed using R (v.4.4.1). Quality control, data clustering and visualization were performed using the Seurat (v.5.1.0) packages. Data preprocessing, clustering, and visualization were performed with the Seurat (v5.1.0) package, following standard guidelines from the Seurat integration tutorial. Cross-condition dataset integration was achieved using Canonical Correlation Analysis (CCA), which effectively minimized batch effects between control and experimental samples.
Dimensionality reduction was performed via Principal Component Analysis (PCA), and the top 30 principal components were used for Uniform Manifold Approximation and Projection (UMAP) embedding and graph-based clustering. UMAPs were generated from integrated data, ensuring that spatial relationships were not biased by experimental conditions, thereby highlighting biologically meaningful differences.
To quantify shifts in cell-type abundance between conditions, we used scProportionTest (R package) to perform a permutation-based statistical comparison of cluster proportions47. We added iCre and zsGreen coding sequences to the reference mouse genome (refdata-gex-mm10-2020-A_zsgreen_iCre) so that AAV transduced cells could be assigned. Statistically significant increases in cellular populations had a false discovery rate (FDR) < 0.05 and a log2 fold-difference (Log2FD) > 1.5.
To visualize spatial expression patterns of key genes, we utilized FeaturePlot from the Seurat package, overlaying normalized gene expression values onto UMAP embeddings. Color gradients were adjusted to improve contrast and interpretability. Genes selected for visualization included canonical lineage markers and candidate regulators identified from differential expression and TF enrichment analyses.
Cell cycle phase classification was performed using Seurat’s Cell Cycle Scoring functionality, which assigns G1, S, or G2/M phases based on curated marker sets (43 S-phase genes and 54 G2/M-phase genes). Cells with low scores in both categories were assigned to G1 (https://satijalab.org/seurat/articles/cell_cycle_vignette.html).
Differential gene expression analysis was conducted using DElegate (v1.2.1), and clusterProfiler (v4.12.6) was used to perform pathway enrichment and Gene Ontology (GO) analysis. For pseudotime trajectory inference, we applied Monocle3 (v1.3.7) to model dynamic lineage progression.
To identify TFs involved in fate bifurcations, we employed SeeSawPred, a computational tool designed to detect mutually inhibitory TF pairs using pseudobulk scRNA-seq data. This platform (https://seesaw.lcsb.uni.lu/webapp/) enabled inference of key TFs driving binary lineage decisions based on the expression divergence observed in our dataset.
Imaging, quantification
All images were taken using a Leica DMi8 Inverted Microscope (Leica Microsystems CMS, 11889113) or Zeiss Z1 Observer/Yokogawa spinning disk (Carl Zeiss) microscope with a 20X objective. One exception is Supplementary Fig. 1, for which images were taken using Axiovision software (version 4.8.10) on a Zeiss Observer D1 inverted microscope at 20x magnification.
Imaging and image processing
Images were analyzed using QuPath Software (v0.5.1)71. For each mouse, cells were analyzed from three serial sections. Counts were standardized to the number of transduced cells, or to the counting area, as indicated in the y-axis for each graph. For Supplementary Fig. 1, to account for differences in tamoxifen labeling between mice, counts were normalized to the number of tdTomato labeled cells/mm2. Adobe Photoshop under the terms of a Creative Cloud subscription was used to prepare figures and a license from BioRender was used to create the schematics.
Statistics and reproducibility
Data was plotted and analyzed using GraphPad Prism 9.2.0 Software to confirm that they met the assumptions of parametric testing. Comparisons between two groups were performed using an unpaired two-tailed t-test, and between three groups, a one-way ANOVA followed by Tukey’s multiple comparisons tests was performed. Significance was defined as p-values less than 0.05. Mean values and error bars representing the standard error of the mean (s.e.m.) were plotted.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Data section. The numerical source data for all graphs are provided in Supplementary Data 5. All data for the scRNAseq dataset is publicly available and deposited in the Gene Expression Omnibus (GEO accession number GSE288733).
References
Grade, S. & Gotz, M. Neuronal replacement therapy: previous achievements and challenges ahead. NPJ Regen. Med. 2, 29 (2017).
Barker, R. A., Gotz, M. & Parmar, M. New approaches for brain repair-from rescue to reprogramming. Nature 557, 329–334 (2018).
Bocchi, R., Masserdotti, G. & Götz, M. Direct neuronal reprogramming: fast forward from new concepts toward therapeutic approaches. Neuron https://doi.org/10.1016/j.neuron.2021.11.023 (2021).
Vasan, L., Park, E., David, L. A., Fleming, T. & Schuurmans, C. Direct neuronal reprogramming: bridging the gap between basic science and clinical application. Front Cell Dev. Biol. 9, 681087 (2021).
Pushchina, E. V., Kapustyanov, I. A. & Kluka, G. G. Adult neurogenesis of teleost fish determines high neuronal plasticity and regeneration. Int. J. Mol. Sci. https://doi.org/10.3390/ijms25073658 (2024).
Qian, H. et al. Reversing a model of Parkinson’s disease with in situ converted nigral neurons. Nature 582, 550–556 (2020).
Irie, T. et al. Direct neuronal conversion of microglia/macrophages reinstates neurological function after stroke. bioRxiv https://doi.org/10.1101/2021.09.26.461831 (2021).
Livingston, J. M. et al. Ectopic expression of Neurod1 is sufficient for functional recovery following a sensory–motor cortical stroke. Biomedicines 12, 663 (2024).
Lentini, C. et al. Reprogramming reactive glia into interneurons reduces chronic seizure activity in a mouse model of mesial temporal lobe epilepsy. Cell Stem Cell https://doi.org/10.1016/j.stem.2021.09.002 (2021).
Zhang, L. et al. Development of neuroregenerative gene therapy to reverse glial scar tissue back to neuron-enriched tissue. Front Cell Neurosci. 14, 594170 (2020).
Tai, W. et al. In vivo reprogramming of NG2 glia enables adult neurogenesis and functional recovery following spinal cord injury. Cell Stem Cell 28, 923–937 e924 (2021).
Rivetti di Val Cervo, P. et al. Induction of functional dopamine neurons from human astrocytes in vitro and mouse astrocytes in a Parkinson’s disease model. Nat. Biotechnol. 35, 444–452 (2017).
Zhou, H. et al. Glia-to-neuron conversion by CRISPR-CasRx alleviates symptoms of neurological disease in mice. Cell 181, 590–603.e516 (2020).
Wang, L. L. et al. Revisiting astrocyte to neuron conversion with lineage tracing in vivo. Cell 184, 5465–5481 e5416 (2021).
Rao, Y. et al. NeuroD1 induces microglial apoptosis and cannot induce microglia-to-neuron cross-lineage reprogramming. Neuron 109, 4094–4108.e4095 (2021).
Wang, D., Tai, P. W. L. & Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 18, 358–378 (2019).
Xie, Y., Zhou, J., Wang, L. L., Zhang, C. L. & Chen, B. New AAV tools fail to detect Neurod1-mediated neuronal conversion of Müller glia and astrocytes in vivo. EBioMedicine 90, 104531 (2023).
Le, N., Appel, H., Pannullo, N., Hoang, T. & Blackshaw, S. Ectopic insert-dependent neuronal expression of GFAP promoter-driven AAV constructs in adult mouse retina. Front Cell Dev. Biol. 10, 914386 (2022).
Puglisi, M. et al. Comparing viral vectors and fate mapping approaches for astrocyte-to-neuron reprogramming in the injured mouse cerebral cortex. Cells https://doi.org/10.3390/cells13171408 (2024).
Guo, Z. et al. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell 14, 188–202 (2014).
Enbar, T. et al. Regionally distinct GFAP promoter expression plays a role in off-target neuron expression following AAV5 transduction. Sci. Rep. 14, 31583 (2024).
Casarosa, S., Fode, C. & Guillemot, F. Mash1 regulates neurogenesis in the ventral telencephalon. Development 126, 525–534 (1999).
Horton, S., Meredith, A., Richardson, J. A. & Johnson, J. E. Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor MASH1. Mol. Cell Neurosci. 14, 355–369 (1999).
Parras, C. M. et al. Mash1 specifies neurons and oligodendrocytes in the postnatal brain. EMBO J. 23, 4495–4505 (2004).
Parras, C. M. et al. The proneural gene Mash1 specifies an early population of telencephalic oligodendrocytes. J. Neurosci. 27, 4233–4242 (2007).
Han, S. et al. Proneural genes define ground-state rules to regulate neurogenic patterning and cortical folding. Neuron 109, 2847–2863.e2811 (2021).
Heinrich, C., Gotz, M. & Berninger, B. Reprogramming of postnatal astroglia of the mouse neocortex into functional, synapse-forming neurons. Methods Mol. Biol. 814, 485–498 (2012).
Li, S. et al. RAS/ERK signaling controls proneural genetic programs in cortical development and gliomagenesis. J. Neurosci. 34, 2169–2190 (2014).
Li, S. et al. GSK3 temporally regulates neurogenin 2 proneural activity in the neocortex. J. Neurosci. 32, 7791–7805 (2012).
Hand, R. et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron 48, 45–62 (2005).
Ge, W. et al. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc. Natl Acad. Sci. USA 103, 1319–1324 (2006).
Sun, Y. et al. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell 104, 365–376 (2001).
Ali, F. et al. Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development 138, 4267–4277 (2011).
Ali, F. R. et al. Dephosphorylation of the proneural transcription factor ASCL1 re-engages a latent post-mitotic differentiation program in neuroblastoma. Mol. Cancer Res. 18, 1759–1766 (2020).
Azzarelli, R., Hardwick, L. J. & Philpott, A. Emergence of neuronal diversity from patterning of telencephalic progenitors. Wiley Interdiscip. Rev. Dev. Biol. 4, 197–214 (2015).
Azzarelli, R. et al. ASCL1 phosphorylation and ID2 upregulation are roadblocks to glioblastoma stem cell differentiation. Sci. Rep. 12, 2341 (2022).
Hindley, C. et al. Post-translational modification of Ngn2 differentially affects transcription of distinct targets to regulate the balance between progenitor maintenance and differentiation. Development 139, 1718–1723 (2012).
Lende-Dorn, B. A., Atkinson, J. C., Bae, Y. & Galloway, K. E. Chemogenetic tuning reveals optimal MAPK signaling for cell-fate programming. Cell Rep. 44, 116226 (2025).
Ghazale, H. et al. Ascl1 phospho-site mutations enhance neuronal conversion of adult cortical astrocytes in vivo. Front Neurosci. 16, 917071 (2022).
Marichal, N. et al. Reprogramming astroglia into neurons with hallmarks of fast-spiking parvalbumin-positive interneurons by phospho-site-deficient Ascl1. Sci. Adv. 10, eadl5935 (2024).
Pereira, A. et al. Direct neuronal reprogramming of mouse astrocytes is associated with multiscale epigenome remodeling and requires Yy1. Nat. Neurosci. 27, 1260–1273 (2024).
Lee, S. W., Oh, Y. M., Lu, Y. L., Kim, W. K. & Yoo, A. S. MicroRNAs overcome cell fate barrier by reducing EZH2-controlled REST stability during neuronal conversion of human adult fibroblasts. Dev. Cell 46, 73–84.e77 (2018).
Pestana, F., Edwards-Faret, G., Belgard, T. G., Martirosyan, A. & Holt, M. G. No longer underappreciated: the emerging concept of astrocyte heterogeneity in neuroscience. Brain Sci. https://doi.org/10.3390/brainsci10030168 (2020).
Taschenberger, G., Tereshchenko, J. & Kügler, S. A microRNA124 target sequence restores astrocyte specificity of gfaABC1D-driven transgene expression in AAV-mediated gene transfer. Mol. Ther. Nucleic Acids 8, 13–25 (2017).
Zhang, X. et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 47, D721–D728 (2019).
Ciesielska, A. et al. Cerebral infusion of AAV9 vector-encoding non-self proteins can elicit cell-mediated immune responses. Mol. Ther. 21, 158–166 (2013).
Miller, S. A. et al. LSD1 and aberrant DNA methylation mediate persistence of enteroendocrine progenitors that support BRAF-mutant colorectal cancer. Cancer Res. 81, 3791–3805 (2021).
Hardwick, L. J., Ali, F. R., Azzarelli, R. & Philpott, A. Cell cycle regulation of proliferation versus differentiation in the central nervous system. Cell Tissue Res. 359, 187–200 (2015).
Calegari, F., Haubensak, W., Haffner, C. & Huttner, W. B. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J. Neurosci. 25, 6533–6538 (2005).
Wang, N. B. et al. Proliferation history and transcription factor levels drive direct conversion to motor neurons. Cell Syst. 16, 101205 (2025).
Lie, D. C. et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature 437, 1370–1375 (2005).
Zhang, L., Yang, X., Yang, S. & Zhang, J. The Wnt /β-catenin signaling pathway in the adult neurogenesis. Eur. J. Neurosci. 33, 1–8 (2011).
Turner, D. A. & Adamson, D. C. Neuronal-astrocyte metabolic interactions: understanding the transition into abnormal astrocytoma metabolism. J. Neuropathol. Exp. Neurol. 70, 167–176 (2011).
Hartmann, A., Okawa, S., Zaffaroni, G. & Del Sol, A. SeesawPred: a web application for predicting cell-fate determinants in cell differentiation. Sci. Rep. 8, 13355 (2018).
Kim, D. Y., Hwang, I., Muller, F. L. & Paik, J. H. Functional regulation of FoxO1 in neural stem cell differentiation. Cell Death Differ. 22, 2034–2045 (2015).
Miron, V. E. Microglia-driven regulation of oligodendrocyte lineage cells, myelination, and remyelination. J. Leukoc. Biol. 101, 1103–1108 (2017).
Zhang, J., Yang, S.-G. & Zhou, F.-Q. Glycogen synthase kinase 3 signaling in neural regeneration in vivo. J. Mol. Cell Biol. https://doi.org/10.1093/jmcb/mjad075 (2023).
Zhang, L. et al. Small molecules efficiently reprogram human astroglial cells into functional neurons. Cell Stem Cell 17, 735–747 (2015).
Liu, M. L. et al. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 4, 2183 (2013).
Mseis-Jackson, N. et al. Dynamic regulation of NeuroD1 expression level by a novel viral construct during astrocyte-to-neuron reprogramming. bioRxiv https://doi.org/10.1101/2025.02.17.638625 (2025).
Parras, C. M. et al. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 16, 324–338 (2002).
Kovach, C. et al. Neurog2 simultaneously activates and represses alternative gene expression programs in the developing neocortex. Cereb. Cortex 23, 1884–1900 (2013).
Chen, G., Shi, Y., Liu, M. & Sun, J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 9, 175 (2018).
Wang, R. et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol. Cancer 17, 166 (2018).
Gu, X. et al. Molecular mechanisms involved in the regulation of neurodevelopment by miR-124. Mol. Neurobiol. 60, 3569–3583 (2023).
Enokido, T. et al. Distinct microRNA signature and suppression of ZFP36L1 define ASCL1-positive lung adenocarcinoma. Mol. Cancer Res. 22, 29–40 (2024).
Papadimitriou, E. et al. A miR-124-mediated post-transcriptional mechanism controlling the cell fate switch of astrocytes to induced neurons. Stem Cell Rep. 18, 915–935 (2023).
Cates, K., Yuan, L., Yang, Y. & Yoo, A. S. Fate erasure logic of gene networks underlying direct neuronal conversion of somatic cells by microRNAs. Cell Rep. https://doi.org/10.1016/j.celrep.2024.115153 (2025).
Son, G. et al. miR-124 coordinates metabolic regulators acting at early stages of human neurogenesis. Commun. Biol. 7, 1393 (2024).
Lee, Y., Messing, A., Su, M. & Brenner, M. GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia 56, 481–493 (2008).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Acknowledgements
We thank Oliver Kent (adMare BioInnovations) for critical reading of the manuscript. The authors acknowledge the support of the Sunnybrook Research Institute (SRI) Genomics Core Facility, the SRI Histology Core Facility (Petia Stefanova), and Hon Leong for microscopy access. This work was supported by a Government of Canada’s New Frontiers in Research Fund (NFRF) Transformation grant (NFRFT-2022-00327) to C.S., C.M.M., and A.d.S., funded through the three federal research funding agencies (CIHR, NSERC, and SSHRC). This study was also supported by a contractual agreement with adMare BioInnovations to C.S. and C.M.M.
Author information
Authors and Affiliations
Contributions
H.G. conceptualized the study, conducted data curation and formal analysis, carried out the investigation, data visualization, and validation, developed the methodology, and drafted, wrote, and edited the manuscript. M.P., S.J., and A.d.S. performed the single-cell RNA sequencing (scRNAseq) data analysis and contributed to writing, reviewing, and editing the corresponding manuscript sections. K.C. and C.W. processed tissue samples for the scRNAseq experiments and revised the related Methods section. L.V. and L.Z. assisted with tissue dissection and sample preparation for the scRNAseq procedures. J.W.H. and C.M.M. designed and performed the Aldh1l1-lineage tracing experiments. C.S., C.M.M and A.d.S. secured funding, oversaw project administration and resource allocation, contributed to conceptualization, supervised and validated the data, and was actively involved in drafting, reviewing, and editing the manuscript. All authors contributed to the manuscript and approved the final version for submission.
Corresponding authors
Ethics declarations
Competing interests
Provisional patent 509459-US. Submitted Jun 9, 2021. Conversion application submitted Jun 9, 2022. Method and compositions for neuronal reprogramming.
Peer review
Peer review information
Communications Biology thanks Jason Weick, C. Justin Lee and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editor: Benjamin Bessieres. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ghazale, H., Parga Pazos, M., Jung, S. et al. MicroRNA-mediated neuronal detargeting alters astrocyte cell fate conversion trajectories in vivo. Commun Biol 8, 1750 (2025). https://doi.org/10.1038/s42003-025-09169-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09169-3