Abstract
Phase separation of biomolecules into distinct material states is expanding our understanding of cellular processes, particularly nuclear chromatin organization linked to gene regulation. Heterochromatin, containing DNA, repressive histone marks, and HP1α protein, functions in gene silencing and telomeric regulation. While HP1α and DNA phase separation contributes to heterochromatin formation, underlying mechanisms remain unclear. Given telomeric RNAs’ proposed role in heterochromatin formation, we investigated interactions between telomeric RNA, DNA, and HP1α. We demonstrate that HP1α phase separates with both Telomeric Repeat containing RNA (TERRA) and DNA. When both nucleic acids are present, HP1α forms multiphase condensates where HP1α-TERRA clusters localize at the HP1α-DNA phase interface. This multiphase architecture results from sequence-specific TERRA-HP1α interactions. TERRA RNA clusters stabilize DNA condensates and modulate their interfacial properties. This work reveals a previously uncharacterized role for TERRA transcripts in regulating HP1α condensates and provides insights into how structured non-coding RNAs influence protein condensate material properties and organization. These findings advance understanding of complex biomolecular interactions governing nuclear compartmentalization through cooperative phase separation mechanisms, highlighting the sophisticated regulatory roles of non-coding RNAs in chromatin organization.
Similar content being viewed by others
Introduction
Phase separation of biomolecules into condensates with distinct material properties and diverse architectures constitutes an emerging paradigm that is increasingly advancing our understanding of fundamental cellular processes across various subcellular compartments, cell types, and organisms.
This is particularly evident within the nucleus where phase separation has been implicated in transcriptional regulation, RNA processing and the material states of chromatin1,2,3,4. Eukaryotic chromatin is a highly organized yet dynamic structure that can undergo significant changes at molecular and structural levels5. Based on the transcriptional status, eukaryotic chromatin is broadly compartmentalized into a relaxed form of chromatin, associated with transcriptional activity called euchromatin, and a condensed form, called heterochromatin, which can be, further subdivided into “facultative” heterochromatin, associated with silencing of cell-specific and developmental-stage specific transcriptional programs; and “constitutive” heterochromatin, which includes hyper-condensed heterochromatin associated with repetitive DNA sequences found at pericentromeric and telomeric regions6,7. Constitutive heterochromatin is characterized by the presence of the heterochromatin protein HP1α and its cognate epigenetic mark—H3K9me38,9, and of the additional repressive histone mark H4K20me3. Specific interactions between the N-terminal chromodomain of HP1α and the H3K9me3 mark are thought to play a role in the localization of HP1α to heterochromatic regions9,10 such as pericentromeric regions and telomeres, where HP1α contributes towards important functions including transcriptional control11, telomeric elongation12 and chromosomal dynamics13. However, recent studies have underscored the importance of the disordered hinge domain, as well as long non-coding RNAs in targeting HP1α to heterochromatic sites at telomeres14,15,16,17. Recent research suggests that HP1α‘s ability to undergo phase separation is crucial for establishing constitutive heterochromatin domains in eukaryotes18,19,20,21. Specifically, the electrostatic interactions between the hinge region and DNA are reported to drive heterochromatin condensate formation18,21. Interestingly, the same hinge region is also shown to have affinity towards RNA molecules7,14,17,22. However, the role of HP1α-RNA interactions in the context of phase separation remains unclear.
Long non-coding RNA molecules in the context of heterochromatin are particularly relevant at telomeric regions. Telomeres, which lie at the termini of linear chromosomes, are specialized capping structures crucial for maintaining genomic stability23,24,25. Telomeres consist of repetitive guanine-rich DNA sequences (TTAGGG in vertebrates) and are highly enriched in heterochromatic components7. These structures protect chromosome ends from being misidentified as DNA double-strand breaks, preventing inappropriate activation of DNA repair mechanisms and chromosomal fusions7,26. Interestingly, while telomeres are enriched in heterochromatic components, sub-telomeric regions (near telomeres) are transcriptionally active and produce long non-coding RNAs called Telomeric Repeat-containing RNA (TERRA)27. These evolutionarily conserved, G-rich (UUAGGG) transcripts play a crucial role in maintaining the heterochromatic state of the telomeric ends28. The abundance of guanine residues in TERRA facilitates the formation of non-canonical secondary structures through Hoogsteen base pairing, resulting in G-quadruplex (G4) configurations29. TERRA transcripts are known to interact with a diverse array of proteins, including chromatin-modifying enzymes and heterochromatin protein 1α (HP1α), suggesting a multifaceted role in chromatin regulation14,22. The presence of TERRA in telomeric nucleoprotein complexes is suggested to act as a docking site for recruiting HP1α and other heterochromatin components27,28,29.
Notably, TERRA is required for the deposition of heterochromatic histone marks at telomeres in vivo30, where it is also found in association with HP1α. Conversely, the spreading of heterochromatin at subtelomeres may result in repression of TERRA transcription, serving as a self-limiting mechanism that prevents unchecked propagation of silent chromatin31. However, the exact biophysical mechanisms via which TERRA can modulate heterochromatic proteins remain elusive. Given that telomeres are uniquely enriched in TERRA transcripts and that HP1α‘s hinge region can bind both DNA and RNA, the telomeric heterochromatin offers an excellent in vitro model to investigate the crosstalk between distinct telomeric heterochromatic components.
Remarkably, we find that HP1α forms multiphase condensates in the presence of DNA and TERRA RNA, with the HP1α-RNA rich phases localizing at the interfaces of larger HP1α-DNA condensates. These TERRA RNA-rich interfacial clusters regulate fusion dynamics, average condensate size, and interfacial properties of the DNA-rich phases, without drastically impacting the internal dynamics of HP1α within condensates. The sequence specificity of TERRA and its interaction with the lysine patch on HP1α appears to be critical for this formation. Our findings suggest a previously uncharacterized role for TERRA RNA in regulating HP1α condensate properties. We suggest that the biological context within the cell’s nucleus may be poised to further tune TERRA’s regulatory capacity by modulating its accessibility, folding, and molecular interactions. This work provides mechanistic insight into how structured non-coding RNAs can influence the material properties and organization of heterochromatin condensates.
Results
HP1α forms multiphase condensates in the presence of DNA and Telomeric Repeat-containing RNA
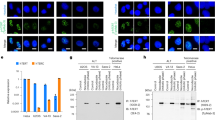
It has been previously reported that TERRA co-localizes with heterochromatic regions at telomeres32 and serves as recruitment for the heterochromatic protein HP1α in eukaryotic cells31. Using a combination of RNAScope-based in situ hybridization and immunofluorescence for HP1α, in immortalized BV2 microglial cells, we identified TERRA foci which co-localized with HP1α within the nuclei of these cells (Supplementary Fig. 1), thereby supporting the concept that at discrete loci in cells, TERRA is found in regions enriched with heterochromatic histone marks and HP1α.
In order to gain mechanistic insight into the molecular interactions underlying this colocalization, we leveraged an in vitro reconstitution system to examine how defined RNA sequences influence HP1α–DNA phase behavior. Specifically, we introduced the telomeric repeat-containing RNA, TERRA45, into our HP1α condensate system. While HP1α and DNA formed large spherical condensates (Fig. 1a) as shown previously18,21, HP1α and TERRA 45 RNA formed much smaller clusters under a range of RNA concentrations (Fig. 1b and Supplementary Fig. 2a). Intriguingly, when all three components were combined, HP1α formed multiphase condensates (Fig. 1c and Supplementary Fig. 2b, SI video 1) (4kbp and 2.5kbp DNA tested). Confocal imaging revealed that interfacial condensates were enriched in TERRA45 and HP1α while the larger/inner phase was enriched in HP1α and DNA (Supplementary Fig. 2c). 3D reconstruction of these multiphasic condensates revealed that smaller (1–3 µm) RNA-containing phases were adsorbed on the interfaces of larger HP1α-DNA condensates (Fig. 1d, e). We confirmed that multiphases formed irrespective of the order of addition of each component (Supplementary Fig. 2d–f), and upon decreasing RNA concentration tenfold to 500 nM (Supplementary Fig. 2g). Finally, since HP1α, TERRA, and local DNA concentrations vary in response to cellular and developmental cues, we tested their interplay at 10 μM HP1α (within the physiological range)18,33, and found that increasing TERRA45 relative to DNA shifted the system from DNA-rich to multiphase (Supplementary Fig. 3a), and eventually to RNA-rich single-phase organization, suggesting that the TERRA-to-DNA ratio may influence HP1α-TERRA-DNA assembly.
a Confocal images of condensates comprising 20 µM HP1α-alexa488 (green) and 20 nM 4kbp DNA-YOYO3 (magenta). Scale bar - 5 µm. b Confocal images of condensates comprising 20 µM HP1α-alexa488 (green) and 5 µM TERRA45 RNA-Tamra (magenta). Scale bar - 5 µm. c Multiphases observed for HP1+TERRA+DNA with TERRA-rich phases localized on the interfaces of HP1α-DNA droplets. Confocal images of 20 µM HP1α-alexa488 (green) and 20 nM 4kbp DNA in the presence of 5 µM TERRA45 (magenta). Scale bar - 5 µm. d 3D reconstruction confocal images of 20 µM HP1α (green), 20 nM 4kbp DNA and 5 µM TERRA45(magenta) indicating that TERRA-rich phases localize to the interfaces. The top panels are a maximum projection of Z-stack through the condensate. The middle panels are a single XZ plane through the same condensate and the bottom panels and a single XY plane through the same condensate. Scale bar - 5 µm. e Line profile for images in (c) indicating the colocalization of HP1α-TERRA45 at interfaces.
Sequence specific interactions between TERRA RNA and HP1α drive immiscibility
To gain further insight into the apparent orthogonality of HP1α interactions with TERRA (RNA) vs DNA, and to see if the multiphase formation was specific to the sequence, topology (parallel vs. antiparallel G4), and length of this long non-coding RNA, we next examined a series of RNA sequences (Supplementary Table) and their ability to promote phase separation and/or alter phase behavior.
To see if multiphase assembly was specific to TERRA45, we first looked at two additional TERRA variants of different length, TERRA96 and TERRA22, 96 and 22 nucleotides long, respectively. Multiphases were observed for TERRA96 RNA (Fig. 2a) at lower concentration (0.5 µM) of RNA compared to TERRA45 while the TERRA 22 transcript was not able to form multiphase droplets at comparable (Fig. 2b), or even at 4X higher (20 µM) concentration to compensate for the decreased length (Supplementary Fig. 3b). While this data suggested a potential role for RNA length, other RNAs tested, of length 45nt or longer, including a shuffled TERRA45 sequence (shuff-TERRA45) (Fig. 2c) and unstructured homopolymer (polyU50) (Supplementary Fig. 3c), did not result in the formation of multiphases.
a Confocal images of 20 µM HP1α (green) and 20 nM 4kbp DNA in the presence of 0.5 µM TERRA96 (magenta). Scale bar - 5 µm. b Confocal images of 20 µM HP1α-alexa488 (green) and 20 nM 4kbp DNA in the presence of 5 µM TERRA22 (magenta). Scale bar - 5 µm. c Confocal images of 20 µM HP1α-alexa488 (green) and 20 nM 4kbp DNA in the presence of 5 µM shuff-TERRA45 (magenta). Scale bar - 5 µm. d 1-H NMR spectra displaying G4 signature peaks in 10-12ppm region. e Confocal images of 20 µM HP1α-alexa488 (green) and 20 nM 4kbp DNA in the presence of 5 µM FMR1 RNA (magenta). Scale bar - 5 µm. f Confocal images of 20 µM HP1α-alexa488 (green) and 20 nM 4kbp DNA in the presence of 5 µM mut-TERRA45 (magenta). Scale bar - 5 µm. g Confocal images of 20 µM Mutant HP1α-3K-A (green) and 20 nM 4kbp DNA in the presence of 5 µM TERRA45 (magenta). Scale bar - 5 µm.
Comparing the 1H NMR spectra of TERRA45 and shuff-TERRA45 we observed a near complete loss of signal in the region reporting G-quadruplex structure for shuff-TERRA45 (chemical shifts in the region 10–12.5 ppm) (Fig. 2d). We therefore next asked if G-quadruplex RNA structure was a determining factor for multiphase formation. We found that other G4-containing sequences, with comparable lengths and/or G4 content (Supplementary Table), such as fmr1 (Fig. 2e), Map1b34(Supplementary Fig. 3d) and Sc-1-double (Supplementary Fig. 3e), were however, not able to generate multiphases. Furthermore, since HP1α has been shown to interact with nucleic acids with parallel G4 configurations, we noted that FMR1 showed prominent imino signals in the parallel G4 region (~11.0–11.5 ppm, Fig. 2d) while Sc1 and Sc1-double presented peaks spanning both the typical parallel region and slightly beyond (Supplementary Fig. 3f), indicating hybrid G4 topologies. Despite forming stable G-quadruplex structures - whether parallel or hybrid in nature - none of these RNAs aside from Telomeric RNAs (TERRA 45 and TERRA96) supported multiphase formation with HP1α under our conditions. Additionally, mutating select G’s in TERRA45 (mut-TERRA45), leading to only partial loss of G4 topology, also did not yield multiphases (Fig. 2d, f). This data indicates that G4 topology is not sufficient for generating multiphases, suggesting that sequence-specific interactions with TERRA are necessary to achieve this effect. Supporting this interpretation, we found that the mutant-HP1α(3K-A) in which the three-lysine basic patch within the HP1α hinge domain is mutated to alanines and which has been previously reported to no longer bind TERRA4514, retained the ability to phase separate with DNA, but was not able to form immiscible phases in the presence of TERRA45 (Fig. 2g). We suspect that the inability for TERRA22 to form multiphases may likewise stem from a decrease in binding specificity, as binding to HP1α is known to be enhanced by TERRA sequence length14 Additionally, no multiphase architecture is observed when swapping HP1α for another phase separating RNA binding protein, Fragile X Mental Retardation Protein (FMRP) (Supplementary Fig. 3g).
TERRA45-HP1α interactions lead to the formation of highly stable condensates with decreased dynamics
Interestingly, while the mut-TERRA45 RNA sequence was not able to form immiscible phases with HP1α and DNA, it was able to phase separate with HP1α in the absence of DNA (Fig. 3a, b), similar to TERRA45, but in contrast to other homopolymer or canonical G-quad RNA sequences tested (Fig. 3c). We used FRAP to determine if there were any differences in HP1α dynamics within TERRA45 versus mut-TERRA45 condensates. We found that HP1α was significantly less dynamic in the TERRA45 condensates (t-half = 14.43 ± 6.37, mobile fraction = 0.48 ± 0.05) compared to mut-TERRA45 condensates (t-half = 6.45 ± 4.81, mobile fraction = 0.94 ± 0.03) (Fig. 3d–f). The slow dynamics and low mobile fraction of HP1α in the TERRA45 condensates suggest that a more rigid structure may underlie the immiscibility in the tri-component system. Consistent with this interpretation, we found that in the multiphase system, while the inner phase quickly dissolved upon the addition of 5 M NaCl, the interfacial TERRA45 clusters remained fully intact (Fig. 3g and SI video 2).
a Brightfield images of condensates comprising 20 µM HP1α and TERRA45 across a range of RNA concentrations. Scale – 5 µm. b Brightfield images of condensates comprising 20 µM HP1α and mut-TERRA45 across a range of RNA concentrations. Scale – 5 µm. c Brightfield images showing that HP1α does not phase separate with control RNAs including homopolymers (polyA30, polyU30) and G-quadruplex RNAs (polyG15, Sc1, Map1b). Scale bar - 5 µm. d Time-lapse FRAP images showing HP1α recovery in TERRA45 condensates. Scale bar - 5 µm. e Time-lapse FRAP images showing HP1α recovery in mut-TERRA45 condensates. Scale bar - 5 µm. f FRAP recovery curves comparing HP1α dynamics in TERRA45 versus mut-TERRA45 condensates. Error bars represent SD (n = 3 independent experiments). g Time-lapse images showing differential salt sensitivity of 20 µM HP1α (magenta) condensate phases: TERRA45-rich phases persist while DNA-rich phases dissolve upon addition of 5 M sodium chloride. Scale bar - 10 µm.
TERRA45 RNA clusters stabilize HP1α-DNA condensates
Given their isolation at the interface and their less dynamic and more stable interaction network, we investigated how TERRA45-enriched phases influenced the physical properties of HP1α-DNA condensates. Unlike traditional two-phase systems where distinct proteins segregate to different phases, our system presents a unique scenario where the same protein (HP1α) forms condensates of different viscoelastic properties when complexed with either DNA or TERRA RNA. We found that TERRA45-enriched phases modified the interfacial properties of DNA condensates, as demonstrated by changes in surface wetting contact angles (Fig. 4a). The interfacial contact angle of a droplet on a glass surface provides a quantitative measure of the balance between cohesive and adhesive forces at the droplet-substrate interface. We observed that increasing concentrations of TERRA45 led to a significant increase in the contact angle of HP1α-DNA condensates. Specifically, the contact angle increased from ~117 ± 2.21 ° in the absence of TERRA45 to nearly 157.71 ± 5.10 ° at 5 µM TERRA45 (Fig. 4b). In contrast, mut-TERRA45 (Fig. 4b) or TERRA22 (Fig. 4c) did not lead to an increase in interfacial angles at the same RNA concentrations. This increase in contact angle indicates a reduction in wettability, suggesting that TERRA45-rich phases at the HP1α-DNA condensate interfaces modulate the condensates’ surface interactions and interfacial properties. While the interfacial properties were altered in the presence of TERRA45, FRAP demonstrated that the internal dynamics of HP1α was not significantly impacted by the presence of TERRA-rich interfacial clusters (Fig. 4d).
a 3D confocal images (indicating surface contact angles) for 20 µM HP1α-alexa594 (magenta), 20 nM 4kbp DNA at varying RNA concentrations for TERRA45 (top) and mutant45(bottom). Scale bar = 5 µm. b Comparison of average contact angles of 20 µM HP1α-20nM 4kbpDNA across increasing concentrations of TERRA45 versus mutant45. Error bars indicate SD across 5 independent experiments. (Statistical analysis was performed using two-way ANOVA followed by Tukey’s multiple comparisons test. TERRA45 showed concentration-dependent increases in contact angles (0 µM vs 1 µM: p < 0.0001; 0 µM vs 5 µM: p < 0.0001; 1 µM vs 5 µM: p = 0.0043). Mut-TERRA45 did not show significant increase in contact angles (0 µM vs 5 µM: p = 0.5095)). c Interfacial angles of 20 µM HP1α, 20 nM 4kbp DNA condensates in the presence of 5 µM TERRA45 versus mut-TERRA45 or TERRA22 at the same RNA (5 µM) concentration. Error bars indicate SD (n = 5 independent experiments). (Statistical analysis was performed using two-way ANOVA followed by Tukey’s multiple comparisons test. TERRA45 showed significant increase in contact angle compared to TERRA22 p < 0.0001 while mut-TERRA45 did not show significant increase p = 0.1379). d Recovery in HP1α-alexa488 fluorescence with and without interfacial TERRA45 clusters for 20 µM HP1α, 40 nM 2.5kbp DNA, 5 µM TERRA45 (N = 3 independent experiments). e Confocal timelapse images showing the fusion of inner DNA –rich phases in the presence of TERRA45 for 20 µM HP1α-alexa488 (green), 20 nM 4kbp DNA and 5 µM TERRA45. f Confocal timelapse images showing the arrested fusion of inner DNA–rich phases in the presence of TERRA45 when the condensates interact via TERRA-rich phases for 20 µM HP1α-alexa488 (green), 20 nM 4kbp DNA and 5 µM TERRA45.
Additionally, confocal imaging timelapses revealed that while droplets were still capable of fusion when they came into contact within a cluster-free region (Fig. 4e), the presence of RNA-enriched interfacial phases created physical barriers that hindered droplet fusion (Fig. 4f SI Video 3). This reduction in droplet coarsening is further quantified by the measured decrease in droplet size as a function of TERRA45 concentration (Fig. 5a, b). In contrast, mut-TERRA45 did not significantly alter the average condensate size (Fig. 5c, d).
a Confocal images of 20 µM HP1α-alexa488 (green), 20 nM 4kbp DNA and TERRA45 showing the reduction in average condensate size with increasing concentration of TERRA45. Scale bar - 5 µm. b Comparison of average condensate size in the presence of TERRA45 at 1 µM and 5 µM RNA concentration (N = 3 independent experiments) (Kolmogorov–Smirnov (KS) test 0 µM vs 1 µM: p = 0.0107;1 µM vs 5 µM: p < 0.0001). c Confocal images of 20 µM HP1α-alexa488 (green), 20 nM 4kbp DNA and mut-TERRA45 showing no reduction in average condensate size with increasing concentration of mut-TERRA45. Scale bar - 5 µm. d Comparison of average condensate size in the presence of mut-TERRA45 at 1 µM and 5 µM RNA concentration (N = 3 independent experiments) (Kolmogorov–Smirnov (KS) test 0 µM vs 1 µM: p = 0.0692;1 µM vs 5 µM: p = 0.1279).
Discussion
TERRA sequences are evolutionarily conserved long non-coding RNAs proposed to serve as scaffolding elements involved in the recruitment, stabilization and spread of heterochromatic domains. Alterations to TERRA transcript cellular concentrations, localization or the heterochromatic state of telomeres are hallmarks of several pathologies, including cancer and premature aging syndromes28,35,36,37,38. Despite their demonstrated significance, the precise functional role of TERRA at telomeric regions has remained poorly understood6,16,39,40 Our findings that TERRA long non-coding RNA can phase separate with HP1α in vitro and form interfacial, immiscible clusters with HP1α-DNA condensates, identifies a new potential mechanism by which TERRA transcripts may modulate heterochromatin dynamics at telomeres. Importantly, our data provide a mechanistic framework to the concept that the unique sequence and structure of TERRA, which is critical for maintaining telomere integrity41,42,43, is also required for the observed immiscibility. Future studies will be needed to validate this framework and directly demonstrate the existence of such multiphasic organization at telomeric ends in vivo.
This work adds to the increasingly diverse set of multiphase architectures reported for condensates44 including immiscible nested phases45,46, and those that involve unique condensate interfaces47,48,49. The interfacial components and physical properties of interfaces have been shown to not only control condensate stability and dynamics but also create unique microenvironments that can selectively partition biomolecules50,51,52,53 regulate reaction kinetics, and fine-tune the material properties of the condensates, ultimately influencing diverse cellular processes from gene regulation to structural regulation of heterochromatin. The interfacial clusters we observe here are strikingly reminiscent of recent work from the Seydoux lab on P granules which found that MEG3 proteins from solid-like clusters enriched at the interface of a liquid-like PGL-3 core, acting as Pickering-like agents47. In the case of the P granule model, the immiscible phases are driven by two different proteins, whereas in our system, distinct nucleic acids drive the immiscibility with a single protein, HP1α, that strongly partitions to both phases. Interestingly, a recent computational study demonstrates that a single “shared” component capable of partitioning into both phases can act as a tunable surfactant - modulating interfacial interactions to trigger wetting transitions and thereby direct multiphase organization within condensates54. Consistent with these predictions, our system - where the same scaffold protein localizes to both core and shell—exhibits precisely the wettability shifts and hierarchical layering modeled by Li and Jacobs, providing experimental validation for their computational framework.
Our observation of unique and immiscible HP1α condensates with DNA and TERRA RNA raises interesting questions about the potential role of phase separation in modulating the regional dynamics of constitutive heterochromatin domains. While it is acknowledged that post-translational modifications of HP1α play an important role in determining its localization in heterochromatic domains during development and differentiation55,56,57 our data suggest that its differential phase behavior with DNA and TERRA RNA may also underlie its context-dependent functions at chromosomal ends. Prior studies in Drosophila have suggested several roles for HP1α within these regions, ranging from: protective telomere capping (a function related to direct binding of HP1α to telomeric DNA) to silencing of telomeric transposons (due to the interaction with the H3K9me3 heterochromatic mark)58, and more recently to binding and stabilizing nascent RNAs59. Our data provide the intriguing possibility that those distinct functions of HP1α could be mediated by the different properties of distinct condensates. Furthermore, additional factors may modulate the multiphase assembly: HP1β, which competes with HP1α for chromatin binding, could alter condensate composition through heterodimerization or displacement. hnRNPA1, known to bind TERRA and promote condensate liquefaction60, may regulate RNA-driven dynamics. Telomere-protecting factor (POT1)61, which binds both telomeric DNA and TERRA, could influence co-condensate formation and compartmentalization. Together, these factors are likely to modulate the material properties and functional outcomes of HP1α-TERRA-DNA assemblies in cells.
TERRA is known to recruit heterochromatin-specific factors to telomeric regions, thereby contributing to the stability of constitutive heterochromatic regions at telomeres30,31. At the same time, the spread of repressive histone marks into subtelomeric regions can reduce TERRA expression, suggesting a built-in feedback mechanism where TERRA both helps establish and is regulated by heterochromatin. This regulatory loop appears to be disrupted in some cancers, potentially contributing to telomeric instability during carcinogenesis34,35,62,63. Our in vitro system addresses the molecular interactions among naked DNA, HP1α, and structured TERRA RNA and identifies the presence of interfacial clusters that stabilize DNA-rich condensates. Thus, our reductionist approach enables delineation of the biophysical parameters governing HP1α-mediated phase separation at heterochromatic sites, rather than addressing their functionality in vivo. This approach distinguishes between two key interaction modes: the electrostatic HP1α-DNA interactions with established roles in condensate formation, and the specific interactions between HP1α and TERRA RNA mediated by a lysine-rich basic patch within the disordered region. These findings highlight how molecular grammars within intrinsically disordered domains can significantly modulate condensate miscibility and material properties46,64,65. This framework provides the necessary resolution to elucidate how distinct nucleic acid clients, DNA versus TERRA RNA, differentially modulate the miscibility and material properties of HP1α condensates. In vivo, HP1α‘s interactions are influenced by additional factors such as nucleosomes bearing H3K9me3 marks and shelterin proteins like TRF2 and crowded nuclear environment which will likely modulate HP1α‘s phase separation and condensate material properties. Future investigations that integrate chromatinized templates, shelterin proteins, and other telomere-associated factors will be essential to fully elucidate the role of multiphasic phase separation of HP1α with DNA and TERRA at telomeric ends. Extending this mechanistic framework into living cells, particularly through super-resolution imaging approaches, will enable direct evaluation of whether multiphasic organization underlies telomeric compartmentalization in vivo. Together, this work not only advances insights into telomere biology and heterochromatin domain formation but also highlights the potential role for non-coding RNAs in shaping condensate properties and expands the current understanding of the molecular drivers of condensate immiscibility.
Materials and methods
Materials
Recombinantly expressed (bacterial) and purified non-phosphorylated HP1α (MW = 22 kDa) and Mutant-HP1α(3K-A) (lyophilized against PBS buffer) from Biomatik Corporation (www.biomatik.com) (Ontario, Canada). Protein was resuspended in ultrapure water (0.5 mg/ml) and refolded following denaturation and reconstitution. The protein was initially solubilized in a buffer containing 6 M guanidine hydrochloride (GdHCl) at 1 mg/mL (350 mM KCl, 20 mM HEPES, 1 mM DTT, pH 7.4) and loaded into a 3000 MWCO dialysis cassette. Dialysis was performed overnight at 4 °C against buffer containing 350 mM KCl, 20 mM HEPES, 1 mM DTT, and 1 M GdHCl. Residual denaturant was then removed by an additional overnight dialysis at 4 °C into buffer lacking GdHCl (350 mM KCl, 20 mM HEPES, 1 mM DTT, pH 7.4). Finally, the protein solution was exchanged into storage buffer (150 mM KCl, 20 mM HEPES, 1 mM DTT, pH 7.4) supplemented with 10% glycerol and stored at −80 °C. Protein aliquots (with 10% glycerol) were flash-frozen 200 μL aliquots and stored at −80 °C.
dsDNA of various lengths (product catalog - NoLimits #SM1571), fluorescent labeling reagents including Alexa-488 C5-Maleimide and Alexa-594 C5-Maleimide, along with the Nucleic Acid Dimer Sampler Kit (YOYO-3 iodide), dialysis cassettes (Slide-A-Lyzer G2, 3 kDa molecular weight cutoff) were procured from Thermo Fisher scientific and used as per the manufacturer’s instructions. Fluorescently labeled and unlabeled RNAs including TERRA (Sequences- Supplementary Table 1) were obtained from Integrative DNA Technologies (IDT) and were resuspended in IDTE buffer (1X TE solution - 10 mM Tris, 0.1 mM EDTA) at desired concentrations. Potassium chloride and Pluronics F127 were purchased from Sigma Aldrich (St. Louis, MO). HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) was obtained from Fisher Scientific (Pittsburgh, PA).
BV2 cells were a gift from Dr. Allison Goat (Icahn School of Medicine).
Amino acid sequences of wild-type and Mutant-HP1α (3K-A) are as follows
Wild-type HP1α (1–191) – MGKKTKRTAD SSSSEDEEEY VVEKVLDRRV VKGQVEYLLK WKGFSEEHNT WEPEKNLDCP ELISEFMKKY KKMKEGENNK PREKSESNKR KSNFSNSADD IKSKKKREQS NDIARGFERG LEPEKIIGAT DSCGDLMFLM KWKGTDEADL VLAKEANVKC PQIVIAFYEE RLTWHAYPED AENKEKETAK S.
Mutant- HP1α (3K-A) (1-191) – MGKKTKRTAD SSSSEDEEEY VVEKVLDRRV VKGQVEYLLK WKGFSEEHNT WEPEKNLDCP ELISEFMKKY KKMKEGENNK PREKSESNKR KSNFSNSADD IKSAAAREQS NDIARGFERG LEPEKIIGAT DSCGDLMFLM KWKGTDEADL VLAKEANVKC PQIVIAFYEE RLTWHAYPED AENKEKETAK S.
HP1α labeling
HP1 was fluorescently labeled with Alexa488/594-C5-Maleimide. Briefly, resuspended HP1 was buffer exchanged into 70 mM KCl, 20 mM HEPES, 1 mM TCEP, pH 7.4 buffer before setting up the reaction. Protein was labeled using Invitrogen Alexa dyes as per manufacturer’s instructions. Approximately 50–100 µM HP1 was mixed with tenfold molar excess solution of dye. The reaction was incubated for 2 h at room temperature and protected against light. The reaction was quenched by adding excess (10 mM) DTT and labeled protein was separated from free dye using 3 kDa molecular weight cut off dialysis cassette against dialysis buffer (300 mM KCl, 20 mM HEPES, 1 mM DTT, pH 7.4). Protein was buffer exchanged and concentrated using 3 kDa MWCO Amicon ultra-centrifugal filter unit (in 70 mM KCl, 20 mM HEPES, 1 mM DTT, pH 7.4) buffer and flash frozen aliquots were stored at –80 °C. ~0.1% labeled protein was diluted with unlabeled protein and used for FRAP and florescence imagining experiments.
Condensate preparation
Prior to condensate preparation HP1α was filtered using 0.22 µm filtered and was buffer exchanged into reaction buffer – 70 mM KCl, 20 mM HEPES, 1 mM DTT, pH 7.4. The protein was concentrated to desired concentration using 3-kDa cutoff Amicon ultra-centrifugal unit. Microscope slide was cleaned with 70% ethanol and treated with Pluronics F-127 for 20 min and washed with deionized water and dried before incubating the samples.
Condensates were prepared by mixing required amounts of RNAs as well as DNA with HP1α. Final reaction volume was fixed at 7 µL, and all the measurements were performed at 23 °C (CherryTemp, CherryBiotech) at 1 h. incubation.
DIC and confocal imaging
DIC imaging was performed using a wide-field Axio Observer 7 Inverted Microscope (Zeiss) with a ×40 or ×63 numerical aperture (NA) Plan-Apochromat air objective. Confocal imaging was performed at 1 h incubation using a Marianas Spinning Disc confocal microscope (Intelligent Imaging Innovations). The imaging setup included a spinning disc confocal head (CSU-X1; Yokagawa) and ×63/1.4-NA Plan-Apochromat (oil immersion) objective. A solid-state laser (LaserStack) was used to select the wavelength of choice – 488 nm line was used to excite Alexa488, Dylight594, or TAMRA594 was excited using 561 nm line. Emissions were collected using a 440-/521-/607-/700-nm quad emission dichroic and 525-/530-nm emission filter. Images were acquired using a Prime sCMOS camera (Photometrics) controlled by Slidebook 6 (Intelligent Imaging Innovations). Images were processed using ImageJ (FiJi).
Fluorescence recovery after photobleaching (FRAP)
FRAP assays were performed was fluorescently labeled HP1α. A small amount (0.1–1%) protein labeled with fluorophore (Alexa488) was combined with unlabeled protein before inducing condensate formation. The final concentration of the protein was kept constant at 20 µM with the concentration of RNA or DNA variable. Upon condensate formation the samples were allowed to settle for 1 h. following which a small area (0.5–1.0 µm) was photobleached and fluorescence recovery was tracked by capturing a series of images on the confocal setup. FRAP images were acquired using a spinning disc confocal microscope (Intelligent Imaging Innovations). Images were acquired at a 200-ms interval with 50–100 ms exposure time. Fluorescence recovery was mapped as a function of time and the resulting data was fit using the following using exponential function of the form:
ImageJ (FiJi) was used to process images and data was analyzed using MATLAB to obtain the mobile fraction (I0) and half-time of recovery [ln(2)/β].
In situ hybridization with RNAScope
Mouse microglial BV2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS; HyClone) and 100 µ/mL penicillin-streptomycin (Pen/Strep; Gibco) at 37 °C in a humidified incubator with 5% CO2. Cells grown in chamber slides were fixed with freshly prepared 4% paraformaldehyde (PFA), dehydrated through a graded ethanol series (50, 70, 100%), and stored at –20 °C until further use. In situ hybridization combining immunohistochemistry was performed using RNAScope probes and reagents from Advanced Cell Diagnostics (ACD), following the manufacturer’s instruction with minor modification. Briefly, endogenous peroxidase activity was quenched with hydrogen peroxide reagent for 10 min. Immunocytochemistry was then carried out using an antibody against HP1-a (Abcam, ab109028;1:500), incubated overnight at 4 °C. After post-fixation, samples were subjected to a diluted protease digestion for 10 min at room temperature. RNAScopeTM Probe-Hs-TELO, targeting Terra, was hybridized for 2 h at 40 °C in a humidity-controlled oven (HybEZ II; ACDbio). Amplification was achieved through the sequential application of proprietary AMP reagents. Signal detection was performed using probe-specific horseradish-peroxidase-based amplification and visualized with Opal dyes (Perkin Elmer), diluted 1:1500. Slides were then incubated with secondary antibody, counterstained with DAPI, and mounted using Prolong Gold Antifade (Thermofisher). Imaging was conducted using a Zeiss LSM-880 microscope. Distribution of HP1-a and TERRA were analyzed using ImageJ (NIH).
Statistics and reproducibility
All quantitative data were analyzed using standard statistical approaches as indicated in the figure legends. Unless otherwise stated, experiments were performed using at least three independent biological replicates, each comprising separately prepared protein, DNA, or RNA samples. Data are presented as mean ± standard deviation (s.d.) unless otherwise specified. Statistical analyzes were carried out using GraphPad Prism 10.3.1. The statistical tests used and the number of replicates (n) are specified in the corresponding figure legends.
Fluorescence microscopy and phase separation assays were independently reproduced in at least three separate experiments, and representative images are shown. All biochemical reconstitutions, binding, and partitioning assays were repeated a minimum of three times with consistent outcomes. For qualitative experiments (e.g., imaging-based condensate morphology), reproducibility was confirmed across independent protein and nucleic acid preparations.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
References
Peng, L., Li, E. M. & Xu, L. Y. From start to end: Phase separation and transcriptional regulation. Biochim Biophys. Acta Gene Regul. Mech. 1863, 194641 (2020).
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K. & Sharp, P. A. A phase separation model for transcriptional control. Cell 169, 13–23 (2017).
Shao, W. et al. Phase separation of RNA-binding protein promotes polymerase binding and transcription. Nat. Chem. Biol. 18, 70–80 (2021).
Zidovska, A. Chromatin: liquid or solid? Cell 183, 1737–1739 (2020).
Misteli, T. Beyond the sequence: cellular organization of genome function. Cell 128, 787–800 (2007).
Bizhanova, A. & Kaufman, P. D. Close to the edge: heterochromatin at the nucleolar and nuclear peripheries. Biochim. Biophys. Acta Gene Regul. Mech. https://doi.org/10.1016/j.bbagrm.2020.194666 (2021).
Schoeftner, S. & Blasco, M. A. A ‘higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J. 28, 2323–2336 (2009).
Nishibuchi, G. & Nakayama, J. I. Biochemical and structural properties of heterochromatin protein 1: understanding its role in chromatin assembly. J. Biochem. 156, 11–20 (2014).
Lachner, M., O’Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001).
Nishibuchi, G. et al. N-terminal phosphorylation of HP1α increases its nucleosome-binding specificity. Nucleic Acids Res. 42, 12498–12511 (2014).
Schoelz, J. M. & Riddle, N. C. Functions of HP1 proteins in transcriptional regulation. Epigenetics Chromatin 15, 1–15 (2022).
Chow, T. T. et al. Local enrichment of HP1alpha at telomeres alters their structure and regulation of telomere protection. Nat. Commun. 9, 1–13 (2018).
Strom, A. R. et al. Hp1α is a chromatin crosslinker that controls nuclear and mitotic chromosome mechanics. eLife 10, e63972 (2021).
Roach, R. J. et al. Heterochromatin protein 1α interacts with parallel RNA and DNA G-quadruplexes. Nucleic Acids Res. 48, 682–693 (2020).
Meehan, R. R., Kao, C. F. & Pennings, S. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 22, 3164–3174 (2003).
Maison, C. et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30, 329–334 (2002).
Muchardt, C. et al. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep. 3, 975–981 (2002).
Keenen, M. M. et al. HP1 proteins compact dna into mechanically and positionally stable phase separated domains. eLife 10, 1–30 (2021).
Larson, A. G. et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017).
Strom, A. R. et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017).
Deshpande, P., Prentice, E., Vidal Ceballos, A., Casaccia, P. & Elbaum-Garfinkle, S. Epigenetic marks uniquely tune the material properties of HP1α condensates. Biophys. J. 123, 1508–1518 (2024).
De Silanes, I. L., D’Alcontres, M. S. & Blasco, M. A. TERRA transcripts are bound by a complex array of RNA-binding proteins. Nat. Commun. 1, 33 (2010).
O’Sullivan, R. J. & Karlseder, J. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 11, 171–181 (2010).
Blackburn, E. H., Epel, E. S. & Lin, J. Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science 350, 1193–1198 (2015).
Blackburn, E. H. Structure and function of telomeres. Nature 350, 569–573 (1991).
De Lange, T. How telomeres solve the end-protection problem. Science 326, 948–952 (2009).
Azzalin, C. M., Reichenbach, P., Khoriauli, L., Giulotto, E. & Lingner, J. Telomeric repeat-containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801 (2007).
Montero, J. J., Lopez De Silanes, I., Grana, O. & Blasco, M. A. Telomeric RNAs are essential to maintain telomeres. Nat Commun. 7, 1–13 (2016).
Xu, Y., Kaminaga, K. & Komiyama, M. G-quadruplex formation by human telomeric repeats-containing RNA in Na + solution. J. Am. Chem. Soc. 130, 11179–11184 (2008).
Montero, J. J. et al. TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat. Commun. 9, 1–14 (2018).
Deng, Z., Norseen, J., Wiedmer, A., Riethman, H. & Lieberman, P. M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 35, 403–413 (2009).
Chu, H. P. et al. TERRA RNA antagonizes ATRX and protects telomeres. Cell 170, 86–101.e16 (2017).
Larson, A. G. et al. Phase separation in heterochromatin. Nature 547, 236–240 (2017).
Vidal Ceballos, A., Geissmann, A., Favaro, D. C., Deshpande, P. & Elbaum-Garfinkle, S. RNA guanine content and G-quadruplex structure tune the phase behavior and material properties of biomolecular condensates. Sci. Rep. 15, (2025).
Arnoult, N., Van Beneden, A. & Decottignies, A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α. Nat. Struct. Mol. Biol. 19, 948–956 (2012).
Bettin, N., Pegorar, C. O. & Cusanelli, E. The emerging roles of TERRA in telomere maintenance and genome stability. Cells 8, 246 (2019).
Shay, J. W. Role of telomeres and telomerase in aging and cancer. Cancer Discov. 6, 584–593 (2016).
Shay, J. W. & Wright, W. E. Telomeres and telomerase: three decades of progress. Nat. Rev. Genet. 20, 299–309 (2019).
Towbin, B. D., Gonzalez-Sandoval, A. & Gasser, S. M. Mechanisms of heterochromatin subnuclear localization. Trends Biochem. Sci. 38 https://doi.org/10.1016/j.tibs.2013.04.004 (2013).
Babu, A. & Verma, R. S. Chromosome structure: euchromatin and heterochromatin. Int. Rev. Cytol. 108, 1–60 (1987).
Xu, Y., Suzuki, Y., Ito, K. & Komiyama, M. Telomeric repeat-containing RNA structure in living cells. Proc. Natl Acad. Sci. USA 107, 14579–14584 (2010).
Bryan, T. M. G-quadruplexes at telomeres: friend or foe? Molecules 25, 3686 (2020).
Mei, Y. et al. TERRA G-quadruplex RNA interaction with TRF2 GAR domain is required for telomere integrity. Sci. Rep. 11, 1–14 (2021).
Fare, C. M., Villani, A., Drake, L. E. & Shorter, J. Higher-order organization of biomolecular condensates. Open Biol. 11 (2021).
Lafontaine, D. L. J., Riback, J. A., Bascetin, R. & Brangwynne, C. P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 22, 165–182 (2020).
Fisher, R. S. & Elbaum-Garfinkle, S. Tunable multiphase dynamics of arginine and lysine liquid condensates. Nat. Commun. 11, 1–10 (2020).
Folkmann, A. W., Putnam, A., Lee, C. F. & Seydoux, G. Regulation of biomolecular condensates by interfacial protein clusters. Science 373 (2021).
Yu, H. et al. HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science 371, eabb4309 (2021).
Jain, S. et al. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 (2016).
Choi, C. H., Lee, D. S. W., Sanders, D. W. & Brangwynne, C. P. Condensate interfaces can accelerate protein aggregation. Biophys. J. 123, 1404–1413 (2024).
Dai, Y. et al. Interface of biomolecular condensates modulates redox reactions. Chem 9, 1594–1609 (2023).
Linsenmeier, M. et al. The interface of condensates of the hnRNPA1 low-complexity domain promotes formation of amyloid fibrils. Nat. Chem. 15, 1340–1349 (2023).
Posey, A. E. et al. Biomolecular condensates are characterized by interphase electric potentials. J. Am. Chem. Soc. 146, 47 (2024).
Li, T. & Jacobs, W. M. Predicting the morphology of multiphase biomolecular condensates from protein interaction networks. PRX Life 2 (2024).
Panteleeva, I. et al. HP1alpha guides neuronal fate by timing E2F-targeted genes silencing during terminal differentiation. EMBO J. 26, 3616–3628 (2007).
Cammas, F., Janoshazi, A., Lerouge, T. & Losson, R. Dynamic and selective interactions of the transcriptional corepressor TIF1 beta with the heterochromatin protein HP1 isotypes during cell differentiation. Differentiation 75, 627–637 (2007).
Yahi, H. et al. Differential cooperation between heterochromatin protein HP1 isoforms and MyoD in myoblasts. J. Biol. Chem. 283, 23692–23700 (2008).
Perrini, B. et al. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol. Cell 15, 467–476 (2004).
Casale, A. M., Cappucci, U., Fanti, L. & Piacentini, L. Heterochromatin protein 1 (HP1) is intrinsically required for post-transcriptional regulation of Drosophila Germline Stem Cell (GSC) maintenance. Sci. Rep. 9, 1–12 (2019).
Ghosh, M. & Singh, M. Structure specific recognition of telomeric repeats containing RNA by the RGG-box of hnRNPA1. Nucleic Acids Res. 48, 4492–4506 (2020).
Flynn, R. L., Chang, S. & Zou, L. RPA and POT1: friends or foes at telomeres? Cell Cycle 11, 652–657 (2012).
Kroupa, M. et al. TElomeric repeat-containing RNA (TERRA): Physiological functions and relevance in cancer. Front. Oncol. 12, 913314 (2022).
Oliva-Rico, D. et al. Methylation of subtelomeric chromatin modifies the expression of the lncRNA TERRA, disturbing telomere homeostasis. Int J. Mol. Sci. 23, 3271 (2022).
Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016).
Mittag, T. & Pappu, R. V. A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol. Cell 82, 2201–2214 (2022).
Acknowledgements
The authors would like to acknowledge Dr. Denize Favaro and the Biomolecular NMR facility at the ASRC for assistance with NMR experiments; and the Epigenetics Core at the ASRC for assistance This work was supported by NIH grant n. R01AG092250 to SEG and NIH grant n. R35NS111604 to PC.
Author information
Authors and Affiliations
Contributions
P.D., P.C., and S.E.-G. conceived the experiments and wrote/edited the manuscript. NMR experiments were performed by A.G. Cell experiments were performed by H.-J.P. and J.L. All other experiments and analysis were performed by P.D.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors Laura Rodríguez Pérez.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Deshpande, P., Geissmann, A., Park, HJ. et al. Telomeric RNA and HP1α form interfacial clusters that stabilize HP1α-DNA condensates. Commun Biol 8, 1815 (2025). https://doi.org/10.1038/s42003-025-09173-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09173-7