Abstract
Although the cyprinid zebrafish has been widely used to study gametogenesis and gonadal differentiation, it has remarkable limitations, such as the loss of sex determinants in laboratory strains, a juvenile ovary stage in both sexes, and oocyte defects generally leading to sex reversal. Here, we develop a novel small cyprinid model for studying gonadal differentiation and gametogenesis, Juji (Gobiocypris rarus), which possesses a genetic sex determination system. We generate a transgenic line of Juji, Tg(ddx4:EGFP-UTRddx4), enabling lifetime visualization of germline development, and find that the dimorphic expression of ddx4:EGFP, rather than primordial germ cell number, at early stages determines the sexual fate of adults. RNA-seq analysis of juvenile ovaries and testes prior to germ cell differentiation identifies sexually dimorphic genes and signaling pathways. Several key factors, such as foxl2l and dmrt1, which drive gonadal differentiation, are among the sexually dimorphic genes. Knockout of dmrt1 leads to sex reversal in juvenile males, with all mutants developing into females. Furthermore, transplanting germline stem cells from genetic males into dmrt1 mutants results in all transplants developing into females, highlighting the essential Sertoli-cell-specific role of dmrt1 in triggering spermatogenesis. Our study establishes Juji as a promising model for investigating sex differentiation, gametogenesis, and gonadal development.
Similar content being viewed by others
Introduction
Gonadal differentiation is the process by which a sexually undifferentiated gonad develops into a functional testis or ovary. This process is regulated by multiple factors, including sex-determining factors and their downstream gene regulatory networks1. Over the last several decades, zebrafish (Danio rerio), as a representative cyprinid fish, has emerged as a popular model organism for the study of vertebrate gametogenesis and gonadal differentiation2,3,4. Most zebrafish undergo a transient “juvenile ovary” stage during early sex differentiation, with some individuals progressing to develop mature ovaries, while others undergo sex reversal to become male5,6. Laboratory strains of zebrafish lack a genetic sex determinant7,8, and germ cells play a vital role in sex differentiation, since a sufficient number of primordial germ cells (PGCs) are required for female development9,10,11,12. Generally, depletion of germ cells at different stages leads to masculinization of zebrafish13,14. Therefore, zebrafish are not considered an ideal model for studying oocyte development and sex differentiation, as their developmental trajectory obscures the primary mechanisms driving ovarian versus testicular commitment15. Consequently, there is an urgent need to develop an alternative cyprinid model with a genetic sex determination system.
The Chinese rare minnow (Gobiocypris rarus, G. rarus, called Xiyou Juji in Chinese, hereafter referred to as “Juji”) is a small freshwater cyprinid fish native to Sichuan Province, China, with adult lengths ranging from 38 to 85 mm. Juji has been widely used as an animal model to study molecular toxicology16,17,18, since it is easily bred in laboratory conditions and is highly sensitive to many environmental pollutants. In addition, the whole genome of Juji has been sequenced19, and Juji shows strong reproductive capacity, as it matures at 3-4 months, and a pair of adults usually spawns 300-400 eggs every four days. Unlike zebrafish, Juji possesses a genetic sex determination mechanism19, further enhancing its utility as a model for studying sex differentiation and reproductive biology. Recently, we transplanted Juji germline stem cells (GSCs) or induced primordial germ cells (iPGCs) into germ cell-depleted zebrafish hosts, and Juji mature sperm could be successfully produced by the zebrafish hosts20,21, indicating that Juji might be an ideal model to study germ cell biology. In order to fully understand the molecular and cellular processes governing sex differentiation and germ cell development in this species, it is urgent to establish a germline-labeling system that can visualize germ cells during development10. Ddx4, also known as Vasa, is an ATP-dependent RNA helicase that belongs to the DEAD-box protein family22. ddx4 is a conserved gene expressed specifically in the germ cells of nearly all sexually reproducing organisms, including humans23,24,25. In teleosts, promoters of ddx4 orthologs from various species have been extensively employed to drive germ cell-specific expression of reporter genes to label their germ cells26,27,28,29. These lines serve as powerful tools for investigating germ cell migration and development, providing critical insights into the molecular and cellular mechanisms underlying reproductive biology.
In this study, we cloned the promoter and 3′-UTR of Juji ddx4 and successfully generated transgenic Juji, Tg(ddx4:EGFP-UTRddx4), which specifically labels germ cells from the gastrulation stage to adulthood. Utilizing this transgenic line, we systematically characterized the gonadal development process in Juji and demonstrated that sex could be reliably determined based on distinct fluorescence patterns as early as 20 days post-fertilization (dpf), which represent the dimorphic expression of ddx4. Additionally, we performed a comprehensive RNA sequencing (RNA-seq) analysis of Juji gonads at 20 dpf, and identified the regulatory network governing sex differentiation, including some key regulatory factors driving gonadal differentiation, such as foxl2l3,30, amh31,32,33 and dmrt133,34,35. We generated dmrt1 mutants using CRISPR/Cas9-mediated gene editing and demonstrated the indispensable role of dmrt1 in male sexual differentiation and testicular development. Furthermore, we employed germ cell transplantation techniques to demonstrate that somatic dmrt1 deficiency drives male GSCs to differentiate into oocytes, thereby establishing that somatic cell fate reversal can dictate germ cell fate determination. In summary, our findings establish a comprehensive framework for elucidating the mechanisms of sex differentiation and gametogenesis in Juji, significantly enhancing its value as a model organism for reproductive biology research.
Results
Dimorphic expression of ddx4:EGFP at 20 dpf but not early PGC number determined sex differentiation of Juji
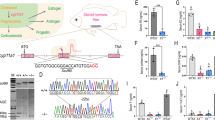
In order to trace the germline development of Juji, we have generated a transgenic line Tg(ddx4:EGFP-UTRddx4) to visualize the germ cells from PGC stage to mature gamete stage. First, we cloned a 5.4 kb genomic fragment upstream of the start codon, encompassing the core promoter region of 3.5 kb upstream of the transcription start site, and a 0.6 kb fragment downstream of the stop codon containing the 3′-UTR as well. These fragments were subsequently used to construct the transgenic vector pTol2(ddx4:EGFP-UTRddx4) for injection (Fig. 1A). In F0 injected embryos at 8 dpf, specific EGFP signal was observed in RFP-UTRnanos3-labeled PGCs (Fig. 1B). Through selective breeding of EGFP-positive adults exhibiting gonadal-specific fluorescence, we successfully established the stable transgenic line Tg(ddx4:EGFP-UTRddx4), which could precisely label germ cells in Juji (Fig. 1, C and D).
A Schematic diagram of the pTol2(ddx4:EGFP-UTRddx4) vector. The vector contained, from 5′ to 3′, the Tol2 transposon element, a 5.4 kb promoter region from ddx4, EGFP, a 0.6 kb 3′-untranslated region (UTR) sequence from ddx4, and another Tol2 transposon element. B Representative fluorescent detection results at 8 dpf after co-injecting pTol2(ddx4:EGFP-UTRddx4) plasmid and RFP-UTRnanos3 mRNA into 1-cell stage embryos. (Left) EGFP; (Middle) RFP; (Right) merged. The ratio represents the number of fluorescence-positive larvae relative to the total count. Scale bar: 200 µm. C Representative EGFP signal from offspring produced by crossing Tg(ddx4:EGFP-UTRddx4) females with wild-type males at different stages. White arrows indicate labeled PGCs. Scale bars: 200 µm, 250 µm, 250 µm, and 750 µm, respectively. D Representative EGFP signal from offspring produced by crossing Tg(ddx4:EGFP-UTRddx4) males with wild-type females at 3.5dpf and 25 dpf. Scale bars: 200 µm and 750 µm. E Representative individuals of PGCs-rich and PGCs-less larvae at 6 dpf. Scale bars: 200 µm. F Frequency distribution of embryos based on the number of PGCs at 6 dpf. The left of the blue dashed line represents the PGCs-less group, while the right of the red dashed line represents the PGCs-rich group. G Sex ratio in PGCs-less, PGCs-moderate, and PGCs-rich groups after maturation (n = 15 PGCs-less, n = 26 PGCs-moderate, n = 15 PGCs-rich).
Continuous fluorescence tracing demonstrated that in embryos derived from crosses between Tg(ddx4:EGFP-UTRddx4) females and wild-type males, maternally deposited EGFP was initially distributed ubiquitously throughout the embryo (Fig. 1C, panels a and b). Starting from the 90%-epiboly stage, PGCs became clearly detectable with stronger EGFP fluorescence (Fig. 1C, panel b). By 15 dpf, maternally derived EGFP outside the PGCs was entirely cleared (Fig. 1C, panel c). However, by 25 dpf, only half of the individuals still had EGFP signal. (Fig. 1C, panel d). This suggests that maternally derived EGFP could effectively label germ cells before 25 dpf. In contrast, in progeny from crosses between Tg(ddx4:EGFP-UTRddx4) males and wild-type females, EGFP signals could firstly be observed in PGCs at 3.5 dpf, and about half of these individuals exhibited specific EGFP signals (Fig. 1D, panels a and b). These results demonstrated that the Tg(ddx4:EGFP-UTRddx4) line could serve as a reliable tool for monitoring germ cell development in Juji.
To evaluate the potential correlation between PGC abundance and sex differentiation in Juji, we quantified the PGC number of Tg(ddx4:EGFP-UTRddx4) larvae at 6 dpf (Fig. 1, E and F). The larvae were divided into three groups: those with more than 25 PGCs (PGCs-rich group), those with fewer than 19 PGCs (PGCs-less group), and those with 19-25 PGCs (PGCs-moderate group) (Fig. 1, E and F). These groups of fish were reared to sexual maturity separately. Notably, no significant differences in sex ratios were observed among the three groups (Fig. 1G). These results demonstrated that the PGC numbers at early developmental stage do not influence sex differentiation in Juji.
Meanwhile, we evaluated the EGFP intensity of Tg(ddx4:EGFP-UTRddx4) larvae at 10 dpf, 15 dpf, and 20 dpf, respectively (Fig. 2A). Compared to 10 dpf and 15 dpf, larvae at 20 dpf exhibited a marked elevation in EGFP intensity, accompanied by substantial inter-individual variability within the population, including the emergence of individuals with exceptionally strong EGFP signals. Based on these divergences, individuals at 20 dpf were categorized into a Bright group, characterized by high fluorescence intensity, and a Faint group, exhibiting lower intensity (Fig. 2, A and B). A significant difference in fluorescence intensity was detected between these groups (Fig. 2C). Subsequent analysis of adult sex revealed that all Bright individuals developed into females, whereas all Faint individuals developed into males (Fig. 2D). Furthermore, fluorescence intensity among the previously defined groups—PGCs-rich, PGCs-moderate, and PGCs-less groups—exhibited no differences at 20 dpf (Fig. 2E). These findings further confirmed that the abundance of PGCs during early developmental stages does not influence sex differentiation in Juji, and demonstrated that the sexual dimorphic expression of ddx4 in Juji could be detected as early as 20 dpf, with significantly higher expression levels in females compared to males.
A Statistical analysis of fluorescence intensity at 10 dpf, 15 dpf and 20 dpf. (n = 30 for each group, one-way ANOVA; *: P < 0.05; ****: P < 0.0001). The up of the red dashed line represents the Bright larvae, while the down of the blue dashed line represents the Faint larvae. B Representative individuals of Bright and Faint larvae at 20 dpf. Scale bars: 500 µm. C Fluorescence intensity of Bright and Faint groups at 20 dpf (n = 8 Bright, n = 8 Faint, ****: P < 0.0001 by unpaired two-tailed Student′s t-test, mean ± SEM). D Sex ratio in Bright and Faint groups after maturation (n = 8 Bright, n = 8 Faint). All Bright fish developed into females, and all Faint fish developed into males. E Fluorescence intensity of PGCs-less, PGCs-moderate, and PGCs-rich groups at 20 dpf. (n = 15 PGCs-less, n = 26 PGCs-moderate, n = 15 PGCs-rich, one-way ANOVA, mean ± SEM; ns: Not significant). F Ovarian development tracing and ddx4 expression of Bright Juji. (a1-f1): Representative EGFP signals of Bright female larvae at 20 dpf to 120 dpf. Scale bars: 500 µm, 750 µm, 1 mm, 1.5 mm, 2 mm, and 3 mm, respectively. (a2-f2): EGFP signals (green) in the gonads of female larvae at 20 dpf to 120 dpf. Nuclei were stained with DAPI (gray). Scale bars: 15 µm, 30 µm, and 60 µm, respectively. (a3-f3): Anti-Ddx4 signals (magenta) in the gonads of female larvae at 20 dpf to 120 dpf. Nuclei were stained with DAPI (gray). Scale bars: 15 µm, 30 µm, and 60 µm, respectively. G Testicular development tracing and ddx4 expression of Faint Juji. (a1-f1): Representative EGFP signals of Faint male larvae at 20 dpf to 120 dpf. Scale bars: 500 µm, 750 µm, 1 mm, 1.5 mm, 2 mm, and 3 mm, respectively. (a2-f2): EGFP signals (green) in the gonads of male larvae at 20 dpf to 120 dpf. Nuclei were stained with DAPI (gray). Scale bars: 15 µm and 30 µm, respectively. (a3-f3): Anti-Ddx4 signals (magenta) in the gonads of male larvae at 20 dpf to 120 dpf. Nuclei were stained with DAPI (gray). Scale bars: 15 µm and 30 µm, respectively. H Proportional distribution of ovarian germ cell types from 20 dpf to 120 dpf. The numbers of germ cells analyzed are shown in parentheses above each bar (Five fields of view were analyzed per developmental stage). GSC, germline stem cell; oGSC, ovarian germline stem cell; IA, stage IA oocyte; IB, stage IB oocyte; II, stage II oocyte; III, stage III oocyte; IV, stage IV oocyte. I Proportional distribution of testicular germ cell types from 20 dpf to 120 dpf. The numbers of germ cells analyzed are shown in parentheses above each bar (Five fields of view were analyzed per developmental stage). GSC, germline stem cell; tGSC, testicular germline stem cell; SPG, spermatogonia; SPC, spermatocyte; SPD, spermatid; SPZ, spermatozoa. J EGFP intensity profiles of testicular germ cells. (n = 10 for each group, one-way ANOVA, mean ± SEM; *: P < 0.05; ****: P < 0.0001).
Characterization of germ cell development of Juji
Using the Tg(ddx4:EGFP-UTRddx4) fish, we continuously monitored the germ cell development in female and male Juji from the undifferentiated stage at 20 dpf to sexual maturity at 120 dpf. At 20 dpf, female Juji displayed strong endogenous EGFP fluorescence in the gonads (Fig. 2F, panel a1). Immunostaining revealed that EGFP is ubiquitously distributed throughout the cytoplasm of germ cells (Fig. 2F, panel a2), whereas endogenous Ddx4 localizes predominantly around the nuclei, with all EGFP-positive cells exhibiting Ddx4 signal (Fig. 2F, panel a3). At this stage, the germ cells exhibited prominent nucleoli and remained as undifferentiated germline stem cells (Fig. S1A). At 25 dpf, the gonads noticeably enlarged (Fig. 2F, panel b1), accompanied by rapid increasing of germ cell number (Fig. 2F, panels b2 and b3). At this time point, early meiotic oocytes at the stage IA could be observed (Fig. S1B), indicating the initiation of germ cell differentiation. By 30 dpf, the gonads further enlarged (Fig. 2F, panel c1), and stage IB oocytes could be detected (Fig. 2F, panels c2 and c3; Fig. S1C). At 60 dpf, the gonads exhibited pronounced enlargement (Fig. 2F, panel d1), with a significant increase in the number of stage IB oocytes (Fig. 2F, panels d2 and d3; Fig. S1D). By 90 dpf, the gonads underwent rapid expansion (Fig. 2F, panel e1), and stage II oocytes, characterized by the accumulation of cortical alveoli, were observed (Fig. 2F, panels e2 and e3; Fig. S1E). Finally, at 120 dpf, the gonads further enlarged (Fig. 2F, panel f1), and stage III-IV oocytes, containing yolk granules, were detected (Fig. 2F, panels f2 and f3; Fig. S1F), marking the attainment of sexual maturity. Throughout ovarian development of Juji, EGFP is continuously expressed from ovarian germline stem cells (oGSCs) to stage IV oocytes.
In contrast to the ovary, the EGFP signal in the male was notably dimmer and exhibited a thinner profile, with no rapid expansion observed prior to 60 dpf (Fig. 2G, panels a1 to f1). From 20 dpf to 30 dpf, germ cells in the testis remained in GSC (1-2 cells) (Fig. 2G, panels a2 to c2; a3 to c3; Fig. S1, G to I), indicating that early testicular germ cells did not undergo rapid proliferation but rather appeared to be in a state of mitotic arrest—similar to what is observed in mammals. Unlike ovarian germ cells, which initiate meiosis at 25 dpf, testicular germ cells did not enter meiosis before 60 dpf (Fig. 2, H and I). At 60 dpf, testicular germ cells drastically increased in number, and reached spermatogonia (SPG) (>4 cells) (Fig. 2G, panels d2 and d3; Fig. S1J). Spermatogenesis proceeded rapidly thereafter. By 90 dpf, the testis had fully matured and produced a large amount of sperm (Fig. 2I; Fig. S1K). At this stage, EGFP labeled different types of germ cells from testicular germline stem cell (tGSC) to mature sperm (Fig. 2G, panels e2 and e3). By 120 dpf, the proportion of sperm had increased markedly (Fig. 2G, panels f2 and f3; Fig. S1L). Notably, germ cells exhibited the highest EGFP fluorescence intensity at the germline stem cell stage, with a progressive and significant decline observed throughout their developmental trajectory (Fig. 2J).
Overall, while both the testis and ovary only contained GSCs at 20 dpf, significant divergence in germ cell development progressions was observed at 25 dpf. Ovarian germ cells underwent differentiation and initiated meiosis, whereas testicular germ cells did not differentiate. These findings suggest that the period between 20 dpf and 25 dpf represents a critical window for the initiation of sex differentiation in Juji.
Global identification of sexually dimorphic genes (SDGs) at 20 dpf
At 20 dpf, although no morphological differences were apparent between male and female germ cells, the differential EGFP fluorescence pattern in the Tg(ddx4:EGFP-UTRddx4) fish indicated that sex differentiation was poised to initiate in Juji. To identify key genes involved in sex differentiation, we performed RNA sequencing on fish at 20 dpf.
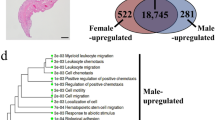
Principal component analysis (PCA) revealed a clear separation between the female and male groups (Fig. 3A), demonstrating distinct gene expression profiles between groups. In total, 123 genes were significantly differentially expressed between the two groups, with 84 genes highly expressed in female and 39 genes with significantly lower expression than those in male (Fig. 3B). Analysis of sex differentiation-related genes showed that genes associated with testicular development, such as dmrt1, amh, star, and cyp17a136,37,38,39,40, were significantly highly expressed in male. In contrast, genes involved in female sex determination, germ plasm assembly, and oogenesis, including foxl2, foxl2l, ddx4, dnd1, and zar122,41,42,43,44,45, were highly expressed in the female (Fig. 3, B and C; Fig. S2). Further analysis of meiosis-related genes revealed that genes involved in synaptonemal complex formation, recombination, and DNA damage repair, such as sycp1, sycp3, syce1, rad21l1, dmc1, and rad51b46,47,48,49,50,51,52, were significantly highly expressed in the female (Fig. 3D). These results suggested that during the critical period of sex differentiation, male expressed high levels of genes associated with testicular development and spermatogenesis, whereas female exhibited elevated expression of genes involved in ovarian development, germ plasm formation, and oogenesis. Moreover, the high expression of meiosis-related genes in female implied that germ cells in the ovary were already poised to enter meiosis.
A PCA of the female and male groups transcriptomes at 20 dpf (n = 3 for each group). B Volcano plot showing differentially expressed genes (DEGs) between female and male groups (Fold change calculated as female/male). C Expression heatmap showing representative genes related to sex differentiation. D Expression heatmap showing representative genes related to meiosis. E GO enrichment analysis of genes highly expressed in female. F KEGG enrichment analysis of genes highly expressed in female. G GO enrichment analysis of genes highly expressed in male. H KEGG enrichment analysis of genes highly expressed in male.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses further revealed that genes highly expressed in females were enriched in terms such as meiotic cell cycle, oogenesis, P granule formation, synaptonemal complex assembly, and meiotic recombination checkpoint signaling (Fig. 3, E and F). Conversely, genes highly expressed in males were significantly enriched in terms like steroid biosynthesis and male sex determination (Fig. 3, G and H). Collectively, these findings underscore that the dimorphic expression of genes and pathways related to meiosis and gonadal differentiation during this critical developmental window orchestrated the proper sex differentiation in Juji.
Sex-specific expression of foxl2l, amh and dmrt1 in juvenile gonads at 20 dpf
In vertebrates, although species-specific sex-determining genes vary, many genes and signaling pathways involved in sex differentiation are highly conserved1. In teleosts, key regulators such as foxl2l, amh, and dmrt1 have been identified as crucial players in gonadal differentiation53. The presence of these orthologs in Juji and their sexually dimorphic expression during gonadal differentiation (Fig. 3, B and C) suggested that they would also play fundamental roles in sex differentiation in Juji.
To investigate their roles, we performed single-molecule fluorescence in situ hybridization (smFISH) to detect the expression of foxl2l, amh, and dmrt1 in the gonads of Juji at 20 dpf. In these experiments, the endogenous EGFP in Tg(ddx4:EGFP-UTRddx4) fish and ddx4 probes were used to label germ cells, and co-staining was performed with probes for foxl2l, amh, and dmrt1. The forkhead box transcription factor Foxl2l (or Foxl3) acts as a germ cell-intrinsic sex determinant in medaka30,54,55. In medaka, the loss of foxl2l (also known as foxl3) permits ovarian germ cells to undergo spermatogenesis, leading to the production of functional sperm in the ovary30. In Nile tilapia (Oreochromis niloticus), foxl3 was reported to play a similar role56. In zebrafish, the absence of foxl2l results in a failure of oogenesis entry, with all mutants developing into males3. In Juji, foxl2l is specifically expressed in female germ cells-with high expression in cystic germ cells and no detectable expression in gonadal somatic cells or the testis (Fig. 4A and B), which is in accordance with quantitative PCR results (Fig. 4C). Anti-Müllerian hormone (Amh), a member of the transforming growth factor-β (TGF-β) superfamily57, is well known for its role in male sex differentiation across vertebrates32,58,59. In zebrafish and medaka, loss of amh leads to female-biased sex differentiation, arrested oogenesis in females, and testicular enlargement with spermatogenic arrest in males31,33,60,61,62. In Juji, amh is predominantly expressed in gonadal somatic cells, with particularly high expression in testicular Sertoli cells, while only a subset of ovarian granulosa cells exhibits weak amh expression (Fig. 4D and E), which is in accordance with quantitative PCR results (Fig. 4F). Doublesex and mab-3-related transcription factor 1 (Dmrt1) is a highly conserved regulator of male sex differentiation across a wide range of species, from nematodes to mammals1,63. In medaka, loss of dmrt1 results in complete sex reversal of XY individuals to females34,64,65. In zebrafish, the loss of dmrt1 results in a severe female-biased sex ratio and the male exhibits retarded gonadal development33,35. In Juji, dmrt1 was highly expressed in testicular somatic cells, and also exhibit some expression in spermatogenic cells, while its expression in the ovary is nearly undetectable (Fig. 4G to I). These results indicate that dmrt1 likely plays an important role in testicular development and spermatogenesis in Juji.
A, D, and G smFISH detection of ddx4 (cyan), foxl2l, amh, and dmrt1 (magenta) expression in female and male at 20 dpf. Germ cells are labeled with EGFP (green). Nuclei were stained with DAPI (gray). Scale bars: 30 µm and 15 µm, respectively. B, E, and H Analysis of foxl2l, amh, and dmrt1-positive cell fluorescence intensity in female and male at 20 dpf (n = 10 for each group, unpaired two-tailed Student′s t-test, mean ± SEM). C, F, and I qPCR analysis of foxl2l, amh, and dmrt1 mRNA expression in female and male at 20 dpf. (n = 3 for each group, unpaired two-tailed Student′s t-test, mean ± SEM).
Together, these findings revealed the potential roles of the key genes during Juji sex differentiation. foxl2l appears to promote oogenesis in females, while amh and dmrt1 are likely key regulators of testicular development and spermatogenesis. Furthermore, this work confirms the functional conservation and predictive validity of SDGs in Juji, solidifying its utility as a robust experimental platform for investigating mechanisms of sex differentiation, gonadal development, and gametogenesis.
Dmrt1 deficiency prevented spermatogenesis in Juji
As dmrt1 is specifically expressed in testis during sex differentiation and expressed in both gonadal somatic cells and germ cells, it is an ideal target for further investigation. Using the CRISPR/Cas9 system, we knocked out dmrt1 in Juji. We established two nonsense mutant lines carrying an 8 bp deletion and a 13 bp deletion that caused premature translational termination (Fig. 5A). The mutations produced two truncated forms of Dmrt1 that lack a complete functional domain, likely leading to loss of function (Fig. 5B). To assess the impact of dmrt1 deletion on sex differentiation, we dissected dmrt1-/- adults at 150 dpf (Fig. 5C). Unlike wild-type controls, all dmrt1-/- individuals exclusively exhibited ovarian structures. The ovarian structure and oogenesis in dmrt1-/- fish were comparable to those observed in wild-type individuals. Then, we compared the sex ratios of adult fish at 150 dpf across different genotypes. All dmrt1-/- fish developed as females, whereas wild-type and heterozygous dmrt1+/- siblings both exhibited a sex ratio of approximately 45% males (Fig. 5D). Additionally, eggs produced by dmrt1-/- fish could be normally fertilized by wild-type (WT) sperm (Fig. 5E), and the fecundity was similar to that of WT females (Fig. 5F). These findings indicate that dmrt1 is essential for testicular development in Juji, but is not required for ovarian development.
A Schematic of dmrt1 gene structure with 5 exons and CRISPR/Cas9-induced mutation. Underlined sequence is CRISPR site, and protospacer adjacent motif (PAM) sequence is shown in red. Mutant line with an 8 bp deletion was generated for phenotype analysis. B Schematic of wild-type and mutant Dmrt1 protein structures. DM, DM DNA binding domain; Dmrt1, Double-sex mab3 related transcription factor 1 domain. Arrowhead points to the mutation site. C Gross morphology of gonads in dmrt1+/+ females, dmrt1+/+ males and dmrt1-/- fish. Asterisks indicate gonads. dmrt1-/- testis was not found. Scale bars: 5 mm and 4 mm, respectively. D Sex ratio of dmrt1+/+, dmrt1+/- and dmrt1-/- fish after maturation (n = 26 dmrt1+/+, n = 51 dmrt1+/-, n = 25 dmrt1-/-). All dmrt1-/- fish developed into females. E Fertility analysis of female adult fish of different genotypes crossed with WT males (n = 3 for each group, one-way ANOVA exhibited no significant difference, mean ± SEM). The dmrt1-/- females exhibited comparable fertility to WT and dmrt1+/- females. F Fecundity analysis of female adult fish of different genotypes crossed with WT males (n = 3 for each group, one-way ANOVA exhibited no significant difference, mean ± SEM). G Fluorescence intensity distribution in dmrt1+/- and dmrt1-/- mutants at 20 dpf (n = 20 for each group, unpaired two-tailed Student′s t-test; ns: Not significant). H Gonadal EGFP fluorescence in dmrt1-/- mutants at 30 dpf, 60 dpf, and 90 dpf. Scale bars: 1 mm, 2 mm, and 2 mm, respectively. I Fluorescence intensity of Bright and Faint groups in dmrt1-/- mutants at 90 dpf (n = 5 for each group, unpaired two-tailed Student′s t-test, mean ± SEM; ns: Not significant). J Histological analysis of gonads in dmrt1-/- fish at different ages (30 dpf, n = 5; 60 dpf, n = 7; 90 dpf, n = 6; 120 dpf, n = 10; 150 dpf, n = 7). IA, stage IA oocyte; IB, stage IB oocyte; II, stage II oocyte; III, stage III oocyte; IV, stage IV oocyte; GSC, germline stem cell. Scale bars: 25 µm, 50 µm, and 200 µm, respectively.
Leveraging the Tg(ddx4:EGFP-UTRddx4) transgenic line, we monitored the gonadal development in dmrt1-/- Juji at different developmental stages. At 20 dpf, both Bright and Faint groups were present in dmrt1-/- mutants, showing no significant difference in fluorescence intensity compared to dmrt1+/- controls (Fig. 5G; Fig. S3A, B), indicating that dmrt1 mutation did not affect gonadal differentiation at this stage. In subsequent developmental stages, the fluorescence intensity between these two groups of dmrt1-/- mutants still showed significant differences at 30 dpf and 60 dpf (Fig. 5H; Fig. S3C), but disappeared at 90 dpf (Fig. 5H, I), indicating that the Faint group have been sex-reversed into females after 30 dpf. Histological analysis further demonstrated the sex reversal progress of the dmrt1-/- mutants between 30 dpf and 150 dpf (Fig. 5J; Fig. S3D). At 30 dpf, about half of gonads displayed ovaries containing stage IB oocytes, while others were full of GSCs—similar to dmrt1+/- controls. At 60 dpf, all the gonads turned to be ovary-type, with approximately 30% individuals exhibiting delayed germ cell development. From 90 dpf to 120 dpf, in the developmentally delayed ovaries, numerous stage IB oocytes were already present, which, together with surrounding somatic cells, formed primary growth (PG) follicles. This is consistent with the increased fluorescence intensity in the Faint group. At 150 dpf, all dmrt1-/- ovaries had fully matured and were fertile (Fig. 5E, F). We then analyzed the offspring of sex-reversed females from dmrt1-/- and wild-type males. Contrary to the expected 100% male offspring under the ZW system (Fig. S3E), the offspring included both females and males (Fig. S3F). In summary, loss of dmrt1 resulted in all Juji developing as females, demonstrating that dmrt1 is indispensable for male development and spermatogenesis.
Testicular somatic cell-specific function of dmrt1 determined the male fate of germ cells
As mentioned above, dmrt1 is expressed in both testicular somatic cells and germ cells and plays an essential role in male development. In addition, the number of PGCs during early developmental stages in Juji did not influence sexual differentiation, which suggested that the driving force of sex differentiation originates from gonadal somatic cells rather than germ cells. So, we hypothesize that the somatic deficiency of dmrt1 is sufficient to induce feminization.
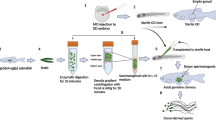
To confirm this hypothesis, we performed germ cell transplantation. Testicular germline stem cells (tGSCs) isolated from adult Tg(ddx4:EGFP-UTRddx4) heterozygous testis were transplanted into 6 dpf dmrt1-/- hosts depleted of endogenous germ cells, with dmrt1+/- hosts serving as controls (Fig. 6A). EGFP signal could be visualized to track the colonization and development of donor germ cells (Fig. 6B–E). At 160 dpf, dmrt1+/- hosts exhibiting bright EGFP fluorescence developed as females (Fig. 6F, G). Their mature ovaries produced exclusively EGFP-positive offspring when crossed with wild-type fish (Fig. 6H). While, dmrt1+/- hosts displaying faint fluorescence developed functional testes capable of fertilizing eggs, resulting in approximately 50% EGFP-positive offspring (Fig. 6I–K), likely due to the hemizygous state of the Tg(ddx4:EGFP-UTRddx4) transgene. In contrast, all dmrt1-/- hosts exhibited bright EGFP fluorescence and developed exclusively as females, with their ovaries undergoing normal folliculogenesis and producing 100% EGFP-positive offspring (Fig. 6L–O). Fertilization rates of dmrt1-/- oocytes were comparable to those of dmrt1+/- females (Fig. 6P). The maternal EGFP signal confirmed that all eggs originated from Tg(ddx4:EGFP-UTRddx4) donor-derived germ cells rather than endogenous host cells. These findings demonstrate that dmrt1-deficient somatic cells could drive testicular germline stem cells to differentiate into oocytes, highlighting that gonadal somatic cell fate reversal could reprogram germ cell fate determination.
A Schematic of tGSCs transplantation. All dmrt1-/- host gonads developed ovaries. B dnd MO injected controls. Scale bar: 300 µm. C–E Fluorescence imaging of EGFP-positive hosts at 6 dpf, 10 dpf, and 30 dpf, respectively. tGSCT: Testicular germline stem cell transplantation. Scale bars: 300 µm and 750 µm, respectively. F, I, and L Fluorescence imaging of host gonads at 160 dpf. Scale bars: 3 mm. G, J, and M Histological analysis of host gonads at 160 dpf. Scale bars: 200 µm, 50 µm, and 200 µm, respectively. H, K, and N Offspring of hosts at 10 dpf. At this stage, the fluorescence observed in female offspring still includes maternal contribution, while the fluorescence in male offspring is solely derived from zygotic expression. Scale bars: 500 µm. O Sex ratio of dmrt1+/- and dmrt1-/- hosts after maturation (n = 15 dmrt1+/-, n = 12 dmrt1-/-). P Fertility analysis of female adult hosts of different genotypes crossed with WT males (n = 3 for each group, unpaired two-tailed Student′s t-test exhibited no significant difference, mean ± SEM).
Discussion
As the only native Chinese fish species utilized as an experimental animal, Juji is widely used for chemical and biological testing in wastewater detection16,17,18. In this study, we successfully established a transgenic line of Juji, Tg(ddx4:EGFP-UTRddx4), to achieve lifetime labeling of germ cell from the 90%-epiboly stage to sexual maturity, which provides a powerful tool for studying germ cell development. By using this transgenic line, we isolated juvenile ovaries and testes before the differentiation of germ cells to perform RNA-seq analysis, and identified a large number of sexually dimorphic genes (SDGs) at this stage, including some key genes involved in gonadal differentiation. Gene knockout and germ cell transplantation experiments further expanded the scope of applications for Juji as an experimental animal and validated its potential as a valuable model for investigating sex differentiation and gametogenesis.
Unlike zebrafish and some other teleosts9,10,11,13,14,66,67,68, PGC abundance during the early developmental stages does not influence sex differentiation in Juji. Our observations reveal that the dimorphic expression of ddx4 serves as an indicator of sex differentiation trajectory in Juji. Strong fluorescence is predictive of ovarian development, whereas weak fluorescence suggests testicular development. Leveraging this property, this transgenic line enables live imaging and precise sex identification as early as 20 dpf, thereby overcoming a significant limitation of the Juji model. By the way, the transgenic line provides precise labeling of distinct germ cell populations, making it highly suitable for fluorescence-activated cell sorting (FACS) for the isolation and purification of germ cells with high specificity.
We have thoroughly characterized the gametogenesis and gonadal differentiation of Juji using well-established morphological criteria and an optimized immunofluorescence protocol to identify and quantify distinct germ cell populations in both the ovary and testis10,69. Unlike zebrafish that develop a transient ovarian-like gonad during early development5,70, Juji males do not develop this structure. In Juji, female germ cells undergo a rapid expansion and initiate meiosis at 25 dpf. In contrast, male germ cells expanded slowly during 20-30 dpf, remaining exclusively at the SPG stage without meiotic initiation until 60 dpf, after which spermatogenesis proceeds rapidly. Although female germ cells initiate meiosis earlier, males attain sexual maturity at a significantly younger age. By 90 dpf, Juji testes contain abundant mature sperm, whereas females appear to exhibit full-grown oocytes after 120 dpf.
Using Tg(ddx4:EGFP-UTRddx4) line, we have globally identified sex differentiation-related genes before gonadal differentiation at 20 dpf, a stage at which no morphological distinctions in germ cells were observed between the female and male. At this stage, there are sexually dimorphic expression patterns of genes and pathways involved in oogenesis, meiosis, and spermatogenesis. These findings suggest that the coordinated regulation of these gene networks is the intrinsic driving force underlying the orderly progression of sex differentiation in Juji. Based on this analysis, we have investigated the expression patterns and potential roles of several key genes related to sex differentiation. foxl2l was specifically expressed in ovarian germ cells, suggesting its involvement in the autonomous regulation of oogenesis, which aligns with observations in other species30,54,55,56. The high expression of amh in Sertoli cells indicated its role in spermatogenesis31,32,33. Moreover, the pro-male factor dmrt1 was only expressed in males, and in both gonadal somatic cells and germ cells, implying its multiple roles in testicular development33,34,35. Besides, given the incomplete genome annotation of Juji, our transcriptomic analysis has also identified many unannotated genes that may play crucial roles in sex determination and gonadal differentiation. Further studies are required to illustrate their functions in sex differentiation. However, not all genes related to sex differentiation exhibited sexually dimorphic expression at this stage, such as cyp19a1. Their expression levels remain low in both females and males at 20 dpf, as they often exert their functions during later developmental stages71,72. This is biologically reasonable as cyp19a1 expression typically increases later during ovarian differentiation, similar to patterns observed in medaka and zebrafish where early expression differences are minimal71,72.
Furthermore, employing CRISPR/Cas9-mediated gene knockout and GSC transplantation approaches, we have demonstrated that the somatic function of dmrt1 was essential for male development. Two ovarian phenotypes, displaying normal and delayed oogenesis, were observed in dmrt1-/- mutants, which may be attributed to different genetic backgrounds. In addition, using GSC transplantation, we have confirmed that all the transplanted male GSCs developed into oocytes in germ cell-depleted dmrt1-/- hosts, suggesting that somatic dmrt1 loss is sufficient to drive feminization. Nevertheless, the possible contribution of germ cell-specific dmrt1 function requires further investigation. Moreover, the results from crossing the sex-reversed females with WT males are contradictory to the previous hypothesis that Juji follows a ZW sex determination system19, since the offspring did not yield exclusively ZZ male offspring (Fig. S3F). This suggests that Juji may follow another type of sex determination, such as XX/XY system or environmental sex determination. Overall, our findings provide new insights into the sex determination mechanism of Juji.
In summary, our work demonstrates the potency of Juji as an animal model for investigating conserved and species-specific mechanisms of sex differentiation and gametogenesis. Furthermore, this transgenic line enables real-time tracking of germ cells during surrogate reproduction, serving as a reliable tool for investigating the colonization and development of donor germ cells in hosts, which hold significant potential for advancing this biotechnology.
Methods
Fish and maintenance
All experiments in this study were conducted using Juji obtained from the Institute of Hydrobiology, Chinese Academy of Sciences (Wuhan, China). All Juji were maintained and reared at the China Zebrafish Resource Center of the National Aquatic Biological Resource Center (CZRC-NABRC, Wuhan, China). All experiments were conducted in full accordance with animal care and use guidelines with ethical approval by the Institutional Animal Care and Use Committee of the Institute of Hydrobiology, Chinese Academy of Sciences (protocol number IHB2014-006). We have complied with all relevant ethical regulations for animal use.
Generation of Transgenic Fish
The ddx4 mRNA sequence of Juji was obtained from NCBI and aligned to identify its corresponding genomic region. The promoter region of ddx4 was predicted upstream using Promoter-2.0. Based on this prediction, primer pairs were designed to amplify a 5.4 kb promoter region, encompassing exon 2 and its upstream sequence. The forward primer was 5′-GGTGGTATGGGGAATGGTTTG-3′, and the reverse primer was 5′-TCCCAGTCGTCCATGTTGAGC-3′. Additionally, primer pairs were designed to amplify the 0.6 kb untranslated region (UTR) of ddx4. The forward primer was 5′-CAGCAGATGATGAGGAATGGG-3′, and the reverse primer was 5′-GCCATCTATGACAGCCTGATC-3′. These two fragments were inserted upstream and downstream of EGFP, respectively, to generate the transgenic plasmid pTol2(ddx4:EGFP-UTRddx4). Plasmid construction was performed using the In-Fusion Cloning Kit (TaKaRa), and all constructs were validated by Sanger sequencing.
To generate transgenic Juji Tg(ddx4:EGFP-UTRddx4), a mixture containing 50 ng of the transgenic construct pTol2(ddx4:EGFP-UTRddx4), 200 pg of transposase mRNA, and 100 pg of RFP-UTRnanos3 mRNA was microinjected into the animal pole of 1-cell stage embryos. Fluorescent screening of F0 embryos was performed using a fluorescence stereomicroscope (Axio Zoom.V16, ZEISS), and embryos exhibiting strong fluorescence were selected and reared to adulthood. F0 fish displaying specific gonadal EGFP signals were crossed with wild-type fish to generate F1 offspring. At 25 dpf, F1 larvae exhibiting specific EGFP expression were selected and raised to adulthood. These F1 adults were then crossed with wild-type fish to generate F2 transgenic offspring. F2 larvae were screened at 25 dpf to exclude non-specific EGFP labeling. Ultimately, the Tg(ddx4:EGFP-UTRddx4) transgenic line was successfully established, and sperm from this line has been cryopreserved at the China Zebrafish Resource Center (CZRC).
Quantification of PGC numbers
Larvae at 6 dpf were anesthetized and mounted horizontally in 2% methylcellulose, positioning the body to align PGCs on one side within a single focal plane for counting (Axio Zoom.V16, ZEISS). After recording PGC numbers on the first side, the body axis and focal plane were adjusted to visualize all PGCs on the opposite side for enumeration. The sum of PGC counts from both sides represented the total PGC number per individual. A total of 56 larvae were analyzed to establish statistical distributions, with individuals categorized into three groups: the top quarter as PGCs-rich, the bottom quarter as PGCs-less, and the middle 50% as PGCs-moderate, based on population-level PGC abundance for subsequent analysis.
CRISPR/Cas9-mediated gene knockout
Mutant lines were generated through CRISPR/Cas9-mediated genome editing following a previous report73. The single-guide RNA (sgRNA) was synthesized using the MEGAscript T7 Transcription Kit (Thermo Fisher Scientific, USA), while Cas9 mRNA was generated via in vitro transcription with the mMESSAGE mMACHINE T3 Kit (Life Technologies, USA). A microinjection cocktail containing 300 ng/μL Cas9 mRNA and 100 ng/μL sgRNA in nuclease-free water (1 nL per embryo) was injected into the animal pole of 1-cell stage embryos to generate the F0 mutant fish. The mutations were identified by Sanger sequencing. F0 fish carrying mutations were raised and outcrossed with wild-type (WT) fish to produce F1 offspring. The F2 progeny were obtained through the intercross of F1 individuals.
Fecundity and fertilization assessment
Female fecundity was assessed by the number of eggs spawned in natural breeding with males. 150 dpf female fish from different groups were paired with 150 dpf wild-type (+/+) males individually for natural spawning. After spawning, the number of eggs released by each female was counted. Fertilization status was examined at 3 hours post-spawning. For each group, three females were subjected to mating every four days, and the mean value of the three mating events was calculated to represent both the fecundity and fertilization rate of each female.
Histological analysis
Juji were anesthetized using MS-222 (Sigma-Aldrich, USA), followed by fixation in Bouin′s fixative for 24 h before further processing. After removal of the head and tail, the samples underwent standard histological processing, followed by embedding in paraffin and sectioning at 2 µm using a HM340E microtome (Thermo Scientific, USA). The sections were then scanned using an Aperio VERSA digital slide scanner (Leica, Germany). Follicle composition was analyzed according our recent report74.
Immunofluorescence
Immunofluorescence staining was performed following previously established protocols21,75. The primary antibody against Ddx4 was generated in a prior study69. Briefly, for whole-mount immunofluorescence staining, Juji gonads were immersion-fixed in 4% paraformaldehyde (PFA) at 4 °C overnight. For sectional immunostaining, Juji gonads were fixed in 3% PFA at room temperature for 2 h, followed by sequential dehydration in sucrose solutions of increasing concentrations (5%, 8%, 12%, 16%, and 20%), remaining 30 min at each concentration. After optimal cutting temperature (OCT) compound embedding, 12 μm sections were obtained using a CM5030 cryostat (Leica, Germany). Sections underwent three 5 min washes in PBSBDT buffer (phosphate-buffered saline containing 2% BSA, 1% DMSO, and 0.1% Triton X-100), followed by 2 h blocking at room temperature using the same buffer. Primary antibodies in PBSBDT were applied overnight at 4 °C, followed by DAPI (Beyotime, China) counterstaining for 2 h at room temperature. Specimens were coverslipped with VECTASHIELD® mounting medium and imaged under a 63× oil-immersion objective on a Leica SP8 laser-scanning confocal microscope (Germany). Images were processed and analyzed using Fiji software76.
Single-molecule fluorescence in situ hybridization (smFISH)
Juji gonads were fixed in 4% PFA overnight at 4 °C and subsequently processed according to our recent study77. Five pairs of probes were designed for each gene detected. After fixation, samples underwent three sequential 5-minute PBST washes (1× PBS with 0.1% Tween-20), followed by tissue permeabilization using 10 µg/mL proteinase K (20 min at room temperature). The samples were then washed with saline-sodium citrate buffer (SSCT) three times for 5 min each, prior to overnight probe hybridization at 37 °C. Unbound probes were removed through four stringent SSCT washes (15 min each). The samples were then incubated with labeled hairpins to initiate hybridization chain reaction (HCR) at room temperature for 4 h under light-protected conditions. Following HCR completion, residual hairpins were eliminated through three SSCT washes (10 min each). Nuclear counterstaining was achieved via 30 min incubation with 1 µg/mL DAPI (Beyotime, China), after which samples were mounted in VECTASHIELD® antifade medium (Vector Laboratories, USA). Confocal images were acquired using a laser-scanning confocal inverted microscope (Leica SP8, Germany) with a 63× oil-immersed objective and processed with Fiji software76.
Quantification of fluorescence intensity
Larvae at various developmental stages were anesthetized and examined under a stereomicroscope (Axio Zoom.V16, ZEISS). Using a consistent magnification, the focal plane was adjusted to maximize the fluorescent area of one gonad while maintaining identical exposure time. The fluorescence intensity of the gonadal region was measured using Fiji software, and the average intensity of bilateral gonads from each fish was recorded as the representative value for that individual. Similarly, cellular fluorescence intensity from immunofluorescence and single-molecule in situ hybridization was measured under a laser-scanning confocal inverted microscope (Leica SP8, Germany) using the same magnification and laser intensity settings. A minimum of five replicates per group were analyzed for statistical evaluation.
Quantitative PCR (qPCR)
Juji were first sex-typed based on EGFP fluorescence intensity at 20 dpf, then dissected to remove heads, tails, and visceral masses, retaining only trunk tissues (including gonads). Each trunk was processed as a single sample for RNA extraction. Total RNA extraction was performed using the FlaPure Animal Tissue Total RNA Extraction Kit (Genesand Biotech, RE715, China). Reverse transcription was carried out using HiScript III All-in-One RT SuperMix (Vazyme, R333-01, China) to generate cDNA. Quantitative PCR was conducted on a CFX96 Touch Real-Time PCR Detection System (BioRad, USA) with Taq Pro Universal SYBR qPCR Master Mix (Vazyme, Q712-02, China). The ef1a gene served as a reference gene due to its stable expression patterns in both female and male gonadal tissues. The expression levels of foxl2l, amh, dmrt1, and foxl2 were analyzed. The primer pairs were: foxl2l: foxl2l-F (5′-CTTCACCGACTCGTACTGCATG-3′) and foxl2l-R (5′-GGTAGCCGGAGATGTGCGTC-3′); amh: amh-F (5′-CCGACACCTCTGGATGATGC-3′) and amh-R (5′-CACCAGTCCTGACAGAAGTGTC-3′); dmrt1: dmrt1-F (5′-GAACGAGGTCATGGGTGATGTG-3′) and dmrt1-R (5′-CATGGAACGGCTCTCTAGGTTG-3′); foxl2: foxl2-F (5′-GGATGGCAGAACAGCATCCG-3′) and foxl2-R (5′-GGTCCAGGGTCCAGTAGTTGC-3′); and ef1a: ef1a-F (5′-GGTATTGGAACTGTGCCCGTG-3′) and ef1a-R (5′-GTGGTGCATCTCAACGGACTTG-3′).
Testicular germline stem cell transplantation (tGSCT)
To generate germ cell-depleted hosts, we collected fertilized eggs from crosses between dmrt1+/- males and dmrt1-/- females. At the 1-cell stage, embryos were microinjected with a dnd morpholino (MO) (5′-GCTGGGCATCCATGTCTCCGACCAT-3′, 50 μM) to ablate endogenous germ cells. These embryos were reared to 6 dpf and used as sterile hosts for transplantation. For donor cell preparation, testes were dissected from two sexually mature Tg(ddx4:EGFP-UTRddx4) males. Testicular tissues were enzymatically digested using our previously published protocol to isolate spermatogonial stem cells20. The donor tGSCs were transplanted into the gonadal ridge of hosts at 6 dpf. After transplantation, hosts were screened with EGPF signal at 6 dpf, 10 dpf, 30 dpf, and 160 dpf using a stereomicroscope (Axio Zoom.V16, ZEISS).
RNA sequencing and analysis
Juji were first sex-typed based on EGFP fluorescence intensity at 20 dpf, then dissected to remove heads, tails, and visceral masses, retaining only trunk tissues (including gonads). Each trunk was processed as an individual sample for total RNA extraction and purification using the FastPure Complex Tissue/Cell Total RNA Isolation Kit (Vazyme, China). After passing quality control, 50 ng of RNA from each sample was used to construct VAHTS RNA-seq libraries with the Universal V6 RNA-seq Library Prep Kit for Illumina (Vazyme, China). Sequencing was performed on the Illumina NextSeq 500 platform (USA), generating 150 bp paired-end (PE) reads at the Analysis and Testing Center, Institute of Hydrobiology, Chinese Academy of Sciences, China. Clean reads were mapped to Juji transcripts derived from the ASM2302916v1 genome assembly (NCBI release)19. Differential expression analysis was conducted using DESeq278, with genes meeting the criteria of |log2 fold change(female/male)|≥1 and adjusted P < 0.05 classified as differentially expressed genes (DEGs). The identified DEGs were subsequently subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. Enrichment analysis and visualization were performed using RStudio.
Statistics and Reproducibility
All results are presented as mean ± standard error of the mean (SEM) from independent biological replicates. Statistical analyses were performed using Prism software (GraphPad, USA) with appropriate parametric tests: unpaired two-tailed Student′s t-test for comparisons between two groups, or one-way analysis of variance (ANOVA) for multi-group comparisons. Significance thresholds were defined as *P < 0.05, **P < 0.01, and ***P < 0.001, ****: P < 0.0001. All experiments were performed at least twice.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are available within the Article and Supplementary Files, or obtained from the corresponding authors on reasonable request. The sequence data that support the findings of this study have been deposited in the Genome Sequence Archive of the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA027239 https://ngdc.cncb.ac.cn/gsa). The numerical source data for all graphs and charts are provided in Supplementary Data 1.
References
Nagahama, Y., Chakraborty, T., Paul-Prasanth, B., Ohta, K. & Nakamura, M. Sex determination, gonadal sex differentiation, and plasticity in vertebrate species. Physiol. Rev. 101, 1237–1308 (2021).
Li, J. & Ge, W. Zebrafish as a model for studying ovarian development: Recent advances from targeted gene knockout studies. Mol. Cell Endocrinol. 507, 110778 (2020).
Ren, Z. et al. foxl2l is a germ cell-intrinsic gatekeeper of oogenesis in zebrafish. Zool. Res. 45, 1116–1130 (2024).
Kossack, M. E. & Draper, B. W. Genetic regulation of sex determination and maintenance in zebrafish (Danio rerio). Curr. Top. Dev. Biol. 134, 119–149 (2019).
Wang, X., Bártfai, R., Sleptsova-Freidrich, I. & Orbán, L. The timing and extent of ‘juvenile ovary’ phase are highly variable during zebrafish testis differentiation. J. Fish. Biol. 70, 33–44 (2007).
Zhang, Q. et al. Zebrafish cyp11c1 Knockout Reveals the Roles of 11-ketotestosterone and Cortisol in Sexual Development and Reproduction. Endocrinology 161, https://doi.org/10.1210/endocr/bqaa048 (2020).
Aharon, D. & Marlow, F. L. Sexual determination in zebrafish. Cell Mol. Life Sci 79, 8 (2021).
Wilson, C. A. et al. Wild sex in zebrafish: loss of the natural sex determinant in domesticated strains. Genetics 198, 1291–1308 (2014).
Siegfried, K. R. & Nüsslein-Volhard, C. Germ line control of female sex determination in zebrafish. Dev. Biol. 324, 277–287 (2008).
Ye, D. et al. Abundance of Early Embryonic Primordial Germ Cells Promotes Zebrafish Female Differentiation as Revealed by Lifetime Labeling of Germline. Mar. Biotechnol. 21, 217–228 (2019).
Dranow, D. B., Tucker, R. P. & Draper, B. W. Germ cells are required to maintain a stable sexual phenotype in adult zebrafish. Dev. Biol. 376, 43–50 (2013).
Tzung, K. W. et al. Early depletion of primordial germ cells in zebrafish promotes testis formation. Stem Cell Rep 4, 61–73 (2015).
Houwing, S. et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129, 69–82 (2007).
Zhang, R. et al. A germline-specific regulator of mitochondrial fusion is required for maintenance and differentiation of germline stem and progenitor cells. Adv. Sci. 9, e2203631 (2022).
Wu, K. et al. Genetic evidence for differential functions of figla and nobox in zebrafish ovarian differentiation and folliculogenesis. Commun. Biol. 6, 1185 (2023).
Xiong, X., Luo, S., Wu, B. & Wang, J. Comparative Developmental Toxicity and Stress Protein Responses of Dimethyl Sulfoxide to Rare Minnow and Zebrafish Embryos/Larvae. Zebrafish 14, 60–68 (2017).
Bai, Y. et al. Species and Life-Stage Sensitivity of Chinese Rare Minnow (Gobiocypris rarus) to Chemical Exposure: A Critical Review. Environ. Toxicol. Chem. 40, 2680–2692 (2021).
Liang, X. & Zha, J. Toxicogenomic applications of Chinese rare minnow (Gobiocypris rarus) in aquatic toxicology. Comp. Biochem Physiol. Part D. Genomics Proteom. 19, 174–180 (2016).
Hu, X. et al. Genomic deciphering of sex determination and unique immune system of a potential model species rare minnow (Gobiocypris rarus). Sci. Adv. 8, eabl7253 (2022).
Zhang, F. et al. Surrogate production of genome-edited sperm from a different subfamily by spermatogonial stem cell transplantation. Sci. China Life Sci 65, 969–987 (2022).
Wang, X. et al. Induced formation of primordial germ cells from zebrafish blastomeres by germplasm factors. Nat. Commun. 14, 7918 (2023).
Xu, C., Cao, Y. & Bao, J. Building RNA-protein germ granules: insights from the multifaceted functions of DEAD-box helicase Vasa/Ddx4 in germline development. Cell Mol. Life Sci 79, 4 (2021).
Hay, B., Jan, L. Y. & Jan, Y. N. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell 55, 577–587 (1988).
Raz, E. The function and regulation of vasa-like genes in germ-cell development. Genome Biol. 1, REVIEWS1017 (2000).
Knaut, H., Pelegri, F., Bohmann, K., Schwarz, H. & Nüsslein-Volhard, C. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J. Cell Biol. 149, 875–888 (2000).
Krøvel, A. V. & Olsen, L. C. Expression of a vas::EGFP transgene in primordial germ cells of the zebrafish. Mech. Dev. 116, 141–150 (2002).
Tanaka, M., Kinoshita, M., Kobayashi, D. & Nagahama, Y. Establishment of medaka (Oryzias latipes) transgenic lines with the expression of green fluorescent protein fluorescence exclusively in germ cells: a useful model to monitor germ cells in a live vertebrate. Proc. Natl. Acad. Sci. USA 98, 2544–2549 (2001).
Yoshizaki, G., Takeuchi, Y., Sakatani, S. & Takeuchi, T. Germ cell-specific expression of green fluorescent protein in transgenic rainbow trout under control of the rainbow trout vasa-like gene promoter. Int. J. Dev. Biol. 44, 323–326 (2000).
Krøvel, A. V. & Olsen, L. C. Sexual dimorphic expression pattern of a splice variant of zebrafish vasa during gonadal development. Dev. Biol. 271, 190–197 (2004).
Nishimura, T. et al. Sex determination. foxl3 is a germ cell-intrinsic factor involved in sperm-egg fate decision in medaka. Science 349, 328–331 (2015).
Yan, Y.-L. et al. A Hormone That Lost Its Receptor: Anti-Müllerian Hormone (AMH) in Zebrafish Gonad Development and Sex Determination. Genetics 213, 529–553 (2019).
Zhang, Z., Wu, K., Ren, Z. & Ge, W. Genetic evidence for Amh modulation of gonadotropin actions to control gonadal homeostasis and gametogenesis in zebrafish and its noncanonical signaling through Bmpr2a receptor. Development 147, https://doi.org/10.1242/dev.189811 (2020).
Lin, Q. et al. Distinct and Cooperative Roles of amh and dmrt1 in Self-Renewal and Differentiation of Male Germ Cells in Zebrafish. Genetics 207, 1007–1022 (2017).
Nanda, I. et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 99, 11778–11783 (2002).
Webster, K. A. et al. Dmrt1 is necessary for male sexual development in zebrafish. Dev. Biol. 422, 33–46 (2017).
Herpin, A. & Schartl, M. Dmrt1 genes at the crossroads: a widespread and central class of sexual development factors in fish. FEBS J. 278, 1010–1019 (2011).
Rey, R. A. & Grinspon, R. P. Anti-Müllerian hormone, testicular descent and cryptorchidism. Front Endocrinol. (Lausanne) 15, 1361032 (2024).
Shang, G. et al. Steroidogenic acute regulatory protein and luteinizing hormone are required for normal ovarian steroidogenesis and oocyte maturation in zebrafish. Biol. Reprod. 101, 760–770 (2019).
Zirkin, B. R. & Papadopoulos, V. Leydig cells: formation, function, and regulation. Biol. Reprod. 99, 101–111 (2018).
Zhai, G. et al. Characterization of Sexual Trait Development in cyp17a1-Deficient Zebrafish. Endocrinology 159, 3549–3562 (2018).
Tanaka, M. Germline stem cells are critical for sexual fate decision of germ cells. Bioessays 38, 1227–1233 (2016).
Gross-Thebing, T. & Raz, E. Dead end and Detour: The function of the RNA-binding protein Dnd in posttranscriptional regulation in the germline. Curr. Top. Dev. Biol. 140, 181–208 (2020).
Wu, Y.-K. & Fan, H.-Y. Revisiting ZAR proteins: the understudied regulator of female fertility and beyond. Cell Mol. Life Sci 79, 92 (2022).
Shi, D.-L. Interplay of RNA-binding proteins controls germ cell development in zebrafish. J. Genet Genomics 51, 889–899 (2024).
Yang, Y.-J., Wang, Y., Li, Z., Zhou, L. & Gui, J.-F. Sequential, Divergent, and Cooperative Requirements of Foxl2a and Foxl2b in Ovary Development and Maintenance of Zebrafish. Genetics 205, 1551–1572 (2017).
Costa, Y. & Cooke, H. J. Dissecting the mammalian synaptonemal complex using targeted mutations. Chromosome Res 15, 579–589 (2007).
Biswas, L. et al. Meiosis interrupted: the genetics of female infertility via meiotic failure. Reproduction 161, R13–R35 (2021).
Qin, Y., Jiao, X., Simpson, J. L. & Chen, Z.-J. Genetics of primary ovarian insufficiency: new developments and opportunities. Hum. Reprod. Update 21, 787–808 (2015).
de Boer, E. & Heyting, C. The diverse roles of transverse filaments of synaptonemal complexes in meiosis. Chromosoma 115, 220–234 (2006).
Strunnikov, A. Cohesin complexes with a potential to link mammalian meiosis to cancer. Cell Regen 2, 4 (2013).
Ito, M., Fujita, Y. & Shinohara, A. Positive and negative regulators of RAD51/DMC1 in homologous recombination and DNA replication. DNA Repair 134, 103613 (2024).
San Filippo, J., Sung, P. & Klein, H. Mechanism of eukaryotic homologous recombination. Annu Rev. Biochem 77, 229–257 (2008).
Li, X.-Y., Mei, J., Ge, C.-T., Liu, X.-L. & Gui, J.-F. Sex determination mechanisms and sex control approaches in aquaculture animals. Sci. China Life Sci 65, 1091–1122 (2022).
Kikuchi, M. et al. Novel components of germline sex determination acting downstream of foxl3 in medaka. Dev. Biol. 445, 80–89 (2019).
Kikuchi, M., Nishimura, T., Ishishita, S., Matsuda, Y. & Tanaka, M. foxl3, a sexual switch in germ cells, initiates two independent molecular pathways for commitment to oogenesis in medaka. Proc. Natl. Acad. Sci. USA 117, 12174–12181 (2020).
Dai, S. et al. Germline sexual fate is determined by the antagonistic action of dmrt1 and foxl3/foxl2 in tilapia. Development 148, https://doi.org/10.1242/dev.199380 (2021).
Josso, N. & di Clemente, N. TGF-beta Family Members and Gonadal Development. Trends Endocrinol. Metab. 10, 216–222 (1999).
Grinspon, R. P., Bergadá, I. & Rey, R. A. Male Hypogonadism and Disorders of Sex. Development. Front Endocrinol. (Lausanne) 11, 211 (2020).
Li, M. et al. A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia, Oreochromis niloticus. PLoS Genet 11, e1005678 (2015).
Zhang, Z., Zhu, B., Chen, W. & Ge, W. Anti-Müllerian hormone (Amh/amh) plays dual roles in maintaining gonadal homeostasis and gametogenesis in zebrafish. Mol. Cell Endocrinol. 517, 110963 (2020).
Morinaga, C. et al. The hotei mutation of medaka in the anti-Mullerian hormone receptor causes the dysregulation of germ cell and sexual development. Proc. Natl. Acad. Sci. USA 104, 9691–9696 (2007).
Nakamura, S. et al. Hyperproliferation of mitotically active germ cells due to defective anti-Müllerian hormone signaling mediates sex reversal in medaka. Development 139, 2283–2287 (2012).
Yi, W., Ross, J. M. & Zarkower, D. Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development 127, 4469–4480 (2000).
Matsuda, M. et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417, 559–563 (2002).
Masuyama, H. et al. Dmrt1 mutation causes a male-to-female sex reversal after the sex determination by Dmy in the medaka. Chromosome Res. 20, 163–176 (2012).
Lewis, Z. R., McClellan, M. C., Postlethwait, J. H., Cresko, W. A. & Kaplan, R. H. Female-specific increase in primordial germ cells marks sex differentiation in threespine stickleback (Gasterosteus aculeatus). J. Morphol. 269, 909–921 (2008).
Li, Q., Fujii, W., Naito, K. & Yoshizaki, G. Application of dead end-knockout zebrafish as recipients of germ cell transplantation. Mol. Reprod. Dev. 84, 1100–1111 (2017).
Saito, D. et al. Proliferation of germ cells during gonadal sex differentiation in medaka: Insights from germ cell-depleted mutant zenzai. Dev. Biol. 310, 280–290 (2007).
Ye, D. et al. Identification of fish spermatogenic cells through high-throughput immunofluorescence against testis with an antibody set. Front Endocrinol 14, 1044318 (2023).
Luzio, A., Santos, D., Monteiro, S. M. & Coimbra, A. M. Zebrafish male differentiation: Do all testes go through a “juvenile ovary” stage? Tissue Cell 72, 101545 (2021).
Lau, E. S.-W., Zhang, Z., Qin, M. & Ge, W. Knockout of Zebrafish Ovarian Aromatase Gene (cyp19a1a) by TALEN and CRISPR/Cas9 Leads to All-male Offspring Due to Failed Ovarian Differentiation. Sci. Rep. 6, 37357 (2016).
Nakamoto, M. et al. Ovarian aromatase loss-of-function mutant medaka undergo ovary degeneration and partial female-to-male sex reversal after puberty. Mol. Cell Endocrinol. 460, 104–122 (2018).
Zhang, F. et al. Efficient generation of zebrafish maternal-zygotic mutants through transplantation of ectopically induced and Cas9/gRNA targeted primordial germ cells. J. Genet Genomics 47, 37–47 (2020).
Li, Y. et al. Endogenous biosynthesis of docosahexaenoic acid (DHA) regulates fish oocyte maturation by promoting pregnenolone production. Zool. Res. 45, 176–188 (2024).
Chen, Z. et al. Intestinal DHA-PA-PG axis promotes digestive organ expansion by mediating usage of maternally deposited yolk lipids. Nat. Commun. 15, 9769 (2024).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Wang, Y. et al. Cyp11a2 Is Essential for Oocyte Development and Spermatogonial Stem Cell Differentiation in Zebrafish. Endocrinology 163, https://doi.org/10.1210/endocr/bqab258 (2022).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014).
Acknowledgements
The authors express sincere gratitude to Kuoyu Li and Linglu Li from CZRC for experimental fish care and sperm cryopreservation. Special thanks are extended to Zhixian Qiao and Xiaocui Chai for technical support with high-throughput sequencing, and to Fang Zhou and Guangxin Wang for confocal microscopy assistance (Analysis and Testing Center of the Institute of Hydrobiology). We acknowledge Qin Wang and Menghan Wu from Prof. Jianwei Wang′s Lab of the Institute of Hydrobiology for their help on Juji breeding. This study was supported by the National Natural Science Foundation of China (32025037), Ministry of Science and Technology of China (2023YFD2401603), Ministry of Agriculture and Rural Affairs of China (NK2022010207), Hubei Provincial Natural Science Foundation of China (2025AFA053), Natural Science Foundation of Wuhan, Science and Technology Special Fund of Hainan Province (ZDYF2024XDNY256), and the State Key Laboratory of Breeding Biotechnology and Sustainable Aquaculture (2024BBSA01).
Author information
Authors and Affiliations
Contributions
Y.S. conceived and designed the study, wrote the manuscript, and revised the manuscript as the corresponding author. N.S. performed experiments, analyzed data, wrote the manuscript, and revised the manuscript. Z.R. analyzed data and wrote the manuscript. H.W., C.W., and Y.H. provided assistance with experiments. D.Y. and M.H. analyzed data. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Mengtan Xing. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Su, N., Ren, Z., Wang, H. et al. Cyprinid Juji (Gobiocypris rarus) as a model fish to study germ cell development and gonadal differentiation. Commun Biol 9, 106 (2026). https://doi.org/10.1038/s42003-025-09378-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09378-w