Abstract
Mitochondrial diseases (MDs) are a heterogeneous group of disorders resulting from mutations in nuclear or mitochondrial DNA genes encoding mitochondrial proteins1,2. MDs cause pathologies with severe tissue damage and ultimately death3,4. There are no cures for MDs and current treatments are only palliative5,6,7. Here we show that tetracyclines improve fitness of cultured MD cells and ameliorate disease in a mouse model of Leigh syndrome. To identify small molecules that prevent cellular damage and death under nutrient stress conditions, we conduct a chemical high-throughput screen with cells carrying human MD mutations and discover a series of antibiotics that maintain survival of various MD cells. We subsequently show that a sub-library of tetracycline analogues, including doxycycline, rescues cell death and inflammatory signatures in mutant cells through partial and selective inhibition of mitochondrial translation, resulting in an ATF4-independent mitohormetic response. Doxycycline treatment strongly promotes fitness and survival of Ndufs4−/− mice, a preclinical Leigh syndrome mouse model8. A proteomic analysis of brain tissue reveals that doxycycline treatment largely prevents neuronal death and the accumulation of neuroimmune and inflammatory proteins in Ndufs4−/− mice, indicating a potential causal role for these proteins in the brain pathology. Our findings suggest that tetracyclines deserve further evaluation as potential drugs for the treatment of MDs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Datasets generated during this study are included as Supplementary Information or are publicly available. Chemical screen data have been deposited in PubChem under accession 1508586 and proteomic data in PRIDE under accession PXD022860. Source data are provided with this paper. Any additional data not included in this manuscript are available from the corresponding author upon request.
References
Area-Gomez, E. & Schon, E. A. Mitochondrial genetics and disease. J. Child Neurol. 29, 1208–1215 (2014).
Wallace, D. C. & Chalkia, D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol. 5, a021220 (2013).
El-Hattab, A. W., Adesina, A. M., Jones, J. & Scaglia, F. MELAS syndrome: clinical manifestations, pathogenesis and treatment options. Mol. Genet. Metab. 116, 4–12 (2015).
Gorman, G. S. et al. Mitochondrial diseases. Nat. Rev. Dis. Primers 2, 16080 (2016).
El-Hattab, A. W., Zarante, A. M., Almannai, M. & Scaglia, F. Therapies for mitochondrial diseases and current clinical trials. Mol. Genet. Metab. 122, 1–9 (2017).
Pfeffer, G., Majamaa, K., Turnbull, D. M., Thorburn, D. & Chinnery, P. F. Treatment for mitochondrial disorders. Cochrane Database Syst. Rev. https://doi.org/10.1002/14651858.CD004426.pub3 (2012).
Russell, O. M., Gorman, G. S., Lightowlers, R. N. & Turnbull, D. M. Mitochondrial diseases: hope for the future. Cell 181, 168–188 (2020).
Kruse, S. E. et al. Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell Metab. 7, 312–320 (2008).
Sproule, D. M. & Kaufmann, P. Mitochondrial encephalopathy, lactic acidosis and stroke-like episodes. Ann. NY Acad. Sci. 1142, 133–158 (2008).
Srivastava, S. et al. PGC-1α/β-induced expression partially compensates for respiratory chain defects in cells from patients with mitochondrial disorders. Hum. Mol. Genet. 18, 1805–1812 (2009).
Pallotti, F. et al. Biochemical analysis of respiratory function in cybrid cell lines harbouring mitochondrial DNA mutations. Biochem. J. 384, 287–293 (2004).
Khan, N. A. et al. mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab. 26, 419–428 (2017).
Johnson, S. C. et al. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science 342, 1524–1528 (2013).
Civiletto, G. et al. Rapamycin rescues mitochondrial myopathy via coordinated activation of autophagy and lysosomal biogenesis. EMBO Mol. Med. 10, e8799 (2018).
Yu, A. K., Datta, S., McMackin, M. Z. & Cortopassi, G. A. Rescue of cell death and inflammation of a mouse model of complex 1-mediated vision loss by repurposed drug molecules. Hum. Mol. Genet. 26, 4929–4936 (2017).
Diaz, F., Enríquez, J. A. & Moraes, C. T. Cells lacking Rieske iron-sulfur protein have a reactive oxygen species-associated decrease in respiratory complexes I and IV. Mol. Cell. Biol. 32, 415–429 (2012).
Barrow, J. J. et al. Bromodomain inhibitors correct bioenergetic deficiency caused by mitochondrial disease complex I mutations. Mol. Cell 64, 163–175 (2016).
Moullan, N. et al. Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep. 10, 1681–1691 (2015).
Sun, T. & Zhang, Y. Pentamidine binds to tRNA through non-specific hydrophobic interactions and inhibits aminoacylation and translation. Nucleic Acids Res. 36, 1654–1664 (2008).
Chukwudi, C. U. rRNA binding sites and the molecular mechanism of action of the tetracyclines. Antimicrob. Agents Chemother. 60, 4433–4441 (2016).
Meydan, S. et al. Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Mol. Cell 74, 481–493 (2019).
Yun, J. & Finkel, T. Mitohormesis. Cell Metab. 19, 757–766 (2014).
Charest, M. G., Lerner, C. D., Brubaker, J. D., Siegel, D. R. & Myers, A. G. A convergent enantioselective route to structurally diverse 6-deoxytetracycline antibiotics. Science 308, 395–398 (2005).
Sun, C. et al. A robust platform for the synthesis of new tetracycline antibiotics. J. Am. Chem. Soc. 130, 17913–17927 (2008).
Wright, P. M. & Myers, A. G. Methodological advances permit the stereocontrolled construction of diverse fully synthetic tetracyclines containing an all-carbon quaternary center at position C5a. Tetrahedron 67, 9853–9869 (2011).
Golub, L. M., McNamara, T. F., D’Angelo, G., Greenwald, R. A. & Ramamurthy, N. S. A non-antibacterial chemically modified tetracycline inhibits mammalian collagenase activity. J. Dent. Res. 66, 1310–1314 (1987).
Leibowitz, R. D. The effect of ethidium bromide on mitochondrial DNA synthesis and mitochondrial DNA structure in HeLa cells. J. Cell Biol. 51, 116–122 (1971).
Quirós, P. M. et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045 (2017).
Alano, C. C., Kauppinen, T. M., Valls, A. V. & Swanson, R. A. Minocycline inhibits poly(ADP-ribose) polymerase-1 at nanomolar concentrations. Proc. Natl Acad. Sci. USA 103, 9685–9690 (2006).
Zhong, W. et al. Doxycycline directly targets PAR1 to suppress tumor progression. Oncotarget 8, 16829–16842 (2017).
Golub, L. M. et al. Tetracyclines inhibit tissue collagenase activity. A new mechanism in the treatment of periodontal disease. J. Periodontal Res. 19, 651–655 (1984).
Jin, Z., Wei, W., Yang, M., Du, Y. & Wan, Y. Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by tipping the balance of macrophage–osteoclast polarization. Cell Metab. 20, 483–498 (2014).
Soustek, M. S. et al. Inhibition of the ER stress IRE1α inflammatory pathway protects against cell death in mitochondrial complex I mutant cells. Cell Death Dis. 9, 658 (2018).
Lorenzl, S., Albers, D. S., Narr, S., Chirichigno, J. & Beal, M. F. Expression of MMP-2, MMP-9 and MMP-1 and their endogenous counterregulators TIMP-1 and TIMP-2 in postmortem brain tissue of Parkinson’s disease. Exp. Neurol. 178, 13–20 (2002).
Montero, R. et al. GDF-15 is clevated in children with mitochondrial diseases and is induced by mitochondrial dysfunction. PLoS One 11, e0148709 (2016).
Sliter, D. A. et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262 (2018).
Balsa, E. et al. Defective NADPH production in mitochondrial disease complex I causes inflammation and cell death. Nat. Commun. 11, 2714 (2020).
Lake, N. J., Compton, A. G., Rahman, S. & Thorburn, D. R. Leigh syndrome: one disorder, more than 75 monogenic causes. Ann. Neurol. 79, 190–203 (2016).
Gengenbacher, M. et al. Tissue distribution of doxycycline in animal models of tuberculosis. Antimicrob. Agents Chemother. 64, e02479-19 (2020).
McAlister, G. C. et al. MultiNotch MS3 enables accurate, sensitive and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 (2014).
Stephan, A. H., Barres, B. A. & Stevens, B. The complement system: an unexpected role in synaptic pruning during development and disease. Annu. Rev. Neurosci. 35, 369–389 (2012).
Torres-Odio, S. et al. Progression of pathology in PINK1-deficient mouse brain from splicing via ubiquitination, ER stress and mitophagy changes to neuroinflammation. J. Neuroinflammation 14, 154 (2017).
Benninger, F., Glat, M. J., Offen, D. & Steiner, I. Glial fibrillary acidic protein as a marker of astrocytic activation in the cerebrospinal fluid of patients with amyotrophic lateral sclerosis. J. Clin. Neurosci. 26, 75–78 (2016).
Liuyu, T. et al. Induction of OTUD4 by viral infection promotes antiviral responses through deubiquitinating and stabilizing MAVS. Cell Res. 29, 67–79 (2019).
Dodd, M. E. et al. The ENTH domain protein Clint1 is required for epidermal homeostasis in zebrafish. Development 136, 2591–2600 (2009).
Zhang, Y. et al. Effect of ApoA4 on SERPINA3 mediated by nuclear receptors NR4A1 and NR1D1 in hepatocytes. Biochem. Biophys. Res. Commun. 487, 327–332 (2017).
Lampropoulou, V. et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 24, 158–166 (2016).
Pegg, A. E. Functions of polyamines in mammals. J. Biol. Chem. 291, 14904–14912 (2016).
Ying, W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid. Redox Signal. 10, 179–206 (2008).
Medina, C. B. et al. Metabolites released from apoptotic cells act as tissue messengers. Nature 580, 130–135 (2020).
Madeo, F., Eisenberg, T., Pietrocola, F. & Kroemer, G. Spermidine in health and disease. Science 359, eaan2788 (2018).
Holmström, K. M. & Finkel, T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 15, 411–421 (2014).
Suomalainen, A. & Battersby, B. J. Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat. Rev. Mol. Cell Biol. 19, 77–92 (2018).
Johnson, S. C. et al. Regional metabolic signatures in the Ndufs4(KO) mouse brain implicate defective glutamate/α-ketoglutarate metabolism in mitochondrial disease. Mol. Genet. Metab. 130, 118–132 (2020).
Cerovic, M., Forloni, G. & Balducci, C. Neuroinflammation and the gut microbiota: possible alternative therapeutic targets to counteract Alzheimer’s disease? Front. Aging Neurosci. 11, 284 (2019).
Sampson, T. R. et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469–1480 (2016).
Ferrari, M. et al. Hypoxia treatment reverses neurodegenerative disease in a mouse model of Leigh syndrome. Proc. Natl Acad. Sci. USA 114, E4241–E4250 (2017).
Teves, S. S. & Henikoff, S. Transcription-generated torsional stress destabilizes nucleosomes. Nat. Struct. Mol. Biol. 21, 88–94 (2014).
Belaghzal, H., Dekker, J. & Gibcus, J. H. Hi-C 2.0: an optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation. Methods 123, 56–65 (2017).
Jha, P., Wang, X. & Auwerx, J. Analysis of mitochondrial respiratory chain supercomplexes using blue native polyacrylamide gel electrophoresis (BN–PAGE). Curr. Protoc. Mouse Biol. 6, 1–14 (2016).
Balsa, E. et al. ER and nutrient stress promote assembly of respiratory chain supercomplexes through the PERK–eIF2α axis. Mol. Cell 74, 877–890 (2019).
Shalem, O. et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Sasarman, F. & Shoubridge, E. A. Radioactive labeling of mitochondrial translation products in cultured cells. Methods Mol. Biol. 837, 207–217 (2012).
Gajda, A., Posyniak, A. & Tomczyk, G. LC-MS/MS analysis of doxycycline residues in chicken tissues after oral administration. Bull. Vet. Inst. Pulawy 58, 573–579 (2014).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators and developers. Protein Sci. https://doi.org/10.1002/pro.3943 (2020).
Rhee, H.-W. et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013).
RCSB PDB. 5AJ4: structure of the 55S mammalian mitoribosome. https://www.rcsb.org/structure/5AJ4.
Acknowledgements
We thank members of the Puigserver Laboratory for helpful discussions regarding this project. We also thank C. Moraes (University of Miami Medical School) and J. Smeitink and R. Vogel (Radboud University Medical Centre) for providing the cell lines used in this study. We acknowledge the Nikon Imaging Center at Harvard Medical School for assistance with brightfield microscopy; the ICCB-Longwood Screening Facility at Harvard Medical School for facilitating our screening efforts; J.M. Asara and M. Yuan at the Beth Israel Deaconess Medical Center Mass Spectrometry Core for providing metabolomics profiling data, R. Bronson and the Rodent Histopathology Core at Harvard Medical School for careful sectioning and analysis of mouse sections; and the Specialized Histopathology Core at Brigham and Women’s Hospital for immunohistochemistry. We thank C. Vidoudez of the Harvard Center for Mass Spectrometry for analysis of doxycycline tissue concentrations. This work was supported by National Institutes of Health (NIH) grants RO1 DK089883-07 NIDDK and RO1 GM121452 NIGMS (to P.P.), F30 DE028206-01A1 NIDCR (to E.A.P.) and F32 GM125243-01A1 NIGMS (to C.F.B.), an EMBO postdoctoral fellowship and MDA Development Grant (to E.B.), and the Human Frontier Science Program (LT-000033/2019-L) to P.L.M.
Author information
Authors and Affiliations
Contributions
Conceptualization: E.A.P., C.F.B. and P.P.; methodology: E.A.P. and C.F.B.; formal analysis: E.A.P. and C.F.B.; investigation: E.A.P., C.F.B., C.L., K.E.O., E.B., P.L.-M., M.J., R.P.L., K.R. and P.M.W.; resources: M.J., S.P.G., A.G.M. and P.P.; writing (original draft): E.A.P., C.F.B. and P.P.; writing (review and editing): E.A.P., C.F.B., P.P., K.E.O., E.A.P., E.B., P.L.-M., C.L., M.J., R.P.L., K.R., P.M.W., A.G.M. and P.P.; visualization: E.A.P. and C.F.B.; supervision: E.A.P., C.F.B. and P.P.; funding acquisition: E.A.P., C.F.B. and P.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Primary Handling Editors: Christoph Schmitt, Pooja Jha. Nature Metabolism thanks Navdeep Chandel, Riekelt Houtkooper and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 MELAS cybrid cells exhibit deficient mitochondrial respiration and undergo apoptosis from nutrient stress.
a, Oxygen Consumption Rates (OCR) measured in control and MELAS cybrid cells. Measurements 4–6 follow the injection of 4 μM oligomycin, measurements 7–9 follow the injection of 4 μM FCCP, and measurements 10–12 follow the injection of 1.5 μM rotenone/4 μM antimycin (n = 5 biologically independent samples). b, 48 hour low-glucose survival assay of MELAS cybrid cells treated with the pan-caspase inhibitor Z-VAD-FMK or doxycycline (Dox) (n = 2 biologically independent samples). c, RFLP mapping of ND1 and MELAS mtDNA mutations in cybrids after 24 hour galactose or low-glucose conditions with 1 μM doxycycline. Expected band sizes are 193/159 bp for ND1 and 117/213 bp for MELAS mutations. d-f, BN-PAGE of isolated mitochondria from MELAS cybrids, ND1 cybrids, and Rieske KO fibroblasts treated with 1 μM doxycycline. MELAS cybrids were propagated 24 hours in low-glucose media, ND1 cybrids were propagated for 48 hours in galactose media, and Rieske KO fibroblasts were propagated for 48 hours in high-glucose media. Data are presented as mean values ± s.e.m. error bars, Student’s t-test with a two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli, with Q = 5%, * q < 0.05.
Extended Data Fig. 2 Doxycycline promotes cell survival through attenuation of mitochondrial translation.
a, Mitochondrial protein synthesis (%DMSO) versus antibiotic concentration based on band quantification relative to DMSO from Fig. 2b (n = 2 experiments). b, 35S-labelled cysteine and methionine pulse in cybrid cells treated with doxycycline at 1 μM or 10 μM with a 48 hour pre-treatment and 1 hour pulse (n = 2 experiments). c, Western blot of MRPL4 in sgMRPL4 ND1 cybrid cells. d, Western blot of GFM2 in sgGFM2 ND1 cybrid cells.
Extended Data Fig. 3 Doxycycline does not promote cell survival in MELAS or ND1 cybrid cells through the integrated protein response (ATF4) or reported protein targets.
a, Western blot of integrated response proteins in MELAS cybrid cells treated with doxycycline (Dox) for 24 hours (n = 2 experiments). b, Western blot of ATF4 in siATF4 MELAS cybrid cells (n = 2 experiments). c, 48 hour low-glucose survival of siATF4 MELAS cybrid cells treated with doxycycline (n = 3 biologically independent samples). d, 48 hour low-glucose survival of MELAS cybrid cells treated with doxycycline with or without ISRIB (Integrated Stress Response Inhibitor) (n = 3 biologically independent samples over n = 2 independent experiments) e, 48 hour low-glucose survival of MELAS cybrid cells treated with pan matrix-metalloprotease (MMP) inhibitor BB-94 (n = 2 biologically independent samples). f, 4 day galactose survival assay of ND1 cybrid cells treated with BB-94 (n = 2 biologically independent samples). g, 48 hour low-glucose survival of MELAS cybrid cells treated with the PARP inhibitor Olaparib (n = 2 biologically independent samples). h, 4 day galactose survival of ND1 cybrid cells treated with Olaparib (n = 2 biologically independent samples). i, 48 hour low-glucose survival of MELAS cybrid cells treated with the PAR1 inhibitor Vorapaxar (n = 2 biologically independent samples). j, 8 day galactose survival of ND1 cybrid cells treated with Vorapaxar (n = 2 biologically independent samples). k, Western blot of PAR1 in siF2R MELAS cybrid cells (n = 2 experiments). l, 48 hour low-glucose survival of MELAS cybrid cells depleted of PAR1 (siF2R) (n = 3 biologically independent samples). Data are presented as mean values ± s.e.m. error bars, Student’s t-test with a two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli, with Q = 5%, * q < 0.05.
Extended Data Fig. 4 Mitochondrial mutant cells have basally elevated cytokines under nutrient stress conditions which are suppressed by doxycycline.
a, Gene expression of inflammatory cytokine panel under high-glucose conditions for control (CON), ND1, and MELAS cybrid cells (n = 3 biologically independent samples). b, Gene expression of inflammatory cytokines in MELAS cybrid cells after 24 hour low-glucose (LG) conditions (n = 6 biologically independent samples). c, Gene expression of inflammatory cytokines in ND1 cybrid cells after 48 hours galactose (GAL) conditions (n = 3 biologically independent samples). Tetracycline analogues that rescue cell survival such as doxycycline, 7015, 7039, and 7066 suppress inflammatory gene expression. Data are presented as mean values ± s.e.m. error bars, Student’s t-test with a two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli, with Q = 5%, * q < 0.05.
Extended Data Fig. 5 Doxycycline suppresses inflammatory gene expression and metabolite levels in a time-dependent manner.
a, Heatmap of metabolites in ND1 cybrid cells that significantly change with doxycycline (Dox) and 7066 compared to DMSO, 7004, 7013, and CMT-3 (n = 3 biologically independent samples, p < 0.05, Student’s t-test with Bonferroni’s correction). b, Time-dependent gene expression of inflammatory markers in ND1 cybrid cells under galactose conditions (n = 3 biologically independent samples). c, Time-dependent metabolite changes in ND1 cybrid cells under galactose conditions (n = 3 biologically independent samples). Data are presented as mean values ± s.e.m. error bars, Student’s t-test with a two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli, with Q = 5%, * q < 0.05.
Extended Data Fig. 6 p38 inhibition partially mimics the doxycycline anti-inflammatory metabolomic signature.
a, Gene expression of inflammatory markers in ND1 cybrid cells treated with doxycycline (Dox) or p38i SB203580 after 48 hours galactose conditions (n = 4 biologically independent samples). b, Metabolite changes in doxycycline signature in ND1 cybrid cells treated with doxycycline or SB203580 after 48 hours galactose conditions (n = 4 biologically independent samples). c-d, Quantitation of GSSG/GSH and NADP+/NADPH ratio in ND1 cybrid cells with doxycycline or SB203580 treatment after 48 hours galactose conditions (n = 4 biologically independent samples). Data are presented as mean values ± s.e.m. error bars, Student’s t-test with a two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli, with Q = 5%, * q < 0.05.
Extended Data Fig. 7 Doxycycline promotes metabolite redox homeostasis independent of complex I protein levels.
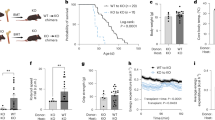
a, Quantitation of doxycycline (Dox) in brains of wild-type and Ndufs4−/− (KO) animals fed 5000 or 8000 ppm doxycycline diets (n = 3 mice per treatment group). b, Quantitation of doxycycline in livers of wild-type and Ndufs4−/− (KO) animals fed 5000 or 8000 ppm doxycycline diets (n = 3 mice per treatment group). c, Heatmap of mitochondrial complex I subunits expressed in the mouse brains quantified through proteomics analysis (n = 3 WT, n = 3 KO, and n = 5 KO Dox mice). d, Heatmap of metabolites altered in Ndufs4−/− (KO) brains (p < 0.15, Student’s t-test, two-sided, unpaired) that are modulated by doxycycline (p < 0.15, Student’s t-test, two-sided, unpaired) (n = 5 mice). e, Heatmap of metabolites altered in ND1 cybrid cells (p < 0.05, Student’s t-test, two-sided, unpaired) that significantly change with doxycycline (p < 0.05, Student’s t-test, two-sided, unpaired) (n = 4 biologically independent samples). f-g, Quantitation of NADP+/NADPH and GSSG/GSH ratios in ND1 cybrid cells or Ndufs4−/− (KO) brains with doxycycline treatment (cybrid cells, n = 4 biologically independent samples; mice, n = 5). Data are presented as mean values ± s.e.m. error bars, Student’s t-test with a two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli, with Q = 5%, * q < 0.05.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots and/or gels.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
About this article
Cite this article
Perry, E.A., Bennett, C.F., Luo, C. et al. Tetracyclines promote survival and fitness in mitochondrial disease models. Nat Metab 3, 33–42 (2021). https://doi.org/10.1038/s42255-020-00334-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42255-020-00334-y
This article is cited by
-
T cell toxicity induced by tigecycline binding to the mitochondrial ribosome
Nature Communications (2025)
-
Selective deficiency of mitochondrial respiratory complex I subunits Ndufs4/6 causes tumor immunogenicity
Nature Cancer (2025)
-
E4F1 coordinates pyruvate metabolism and the activity of the elongator complex to ensure translation fidelity during brain development
Nature Communications (2025)
-
Compensatory activity of the PC-ME1 metabolic axis underlies differential sensitivity to mitochondrial complex I inhibition
Nature Communications (2024)
-
Cannabidiol ameliorates mitochondrial disease via PPARγ activation in preclinical models
Nature Communications (2024)