Abstract
Glucose deficiency promotes the secretion of cytokines and inflammatory factors to rewire the immune compartment and restore blood flow. Here we show that cancer cells subjected to glucose deprivation or hypoxia, but not to other metabolic stressors, secrete LIF, an interleukin-6 family cytokine implicated in the development of solid tumours. We find that mannose supplementation prevents LIF release by sustaining multiple metabolic pathways in the absence of glucose. Mechanistically, LIF release is associated with impairment of N-glycosylation and activation of PERK and MEK MAP kinases. In mouse models of non-small-cell lung cancer, reduction of LIF impairs angiogenesis and tumour growth, rewires the immune system toward an antitumour phenotype and inhibits tumour implantation in the lung. In individuals with non-small-cell lung cancer, LIF levels correlate with markers of hypoxia, glucose deprivation and angiogenesis. Overall, these findings identify LIF as a metabolic stress-induced cytokine that could be targeted to disrupt adaptive responses in cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data that support the findings of this study are available as Extended Data figures or are provided in the Supplementary Information in this article. Source data are provided with this paper.
Code availability
The code packages used in this article are publicly available as described in Methods.
References
Longchamp, A. et al. Amino acid restriction triggers angiogenesis via GCN2/ATF4 regulation of VEGF and H2S production. Cell 173, 117–29.e14 (2018).
Kerbel, R. S. Tumor angiogenesis. N. Engl. J. Med. 358, 2039–2049 (2025).
Sullivan, M. R. et al. Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. eLife 8, e44235 (2019).
Shaw, R. J. et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl Acad. Sci. USA 101, 3329–3335 (2004).
Dibble, C. C. & Manning, B. D. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 15, 555–564 (2013).
Shanware, N. P. et al. Glutamine deprivation stimulates mTOR–JNK-dependent chemokine secretion. Nat. Commun. 5, 4900 (2014).
Gameiro, P. A. & Struhl, K. Nutrient deprivation elicits a transcriptional and translational inflammatory response coupled to decreased protein synthesis. Cell Rep. 24, 1415–1424 (2018).
Püschel, F. et al. Starvation and antimetabolic therapy promote cytokine release and recruitment of immune cells. Proc. Natl Acad. Sci. USA 117, 9932–9941 (2020).
Cruz-Rodríguez, M., Chevet, E. & Muñoz-Pinedo, C. Glucose sensing and the unfolded protein response. FEBS J. 292, 3581–3595 (2025).
Chen, Y. et al. Gprc5a deletion enhances the transformed phenotype in normal and malignant lung epithelial cells by eliciting persistent STAT3 signaling induced by autocrine leukemia inhibitory factor. Cancer Res. 70, 8917–8926 (2010).
Albrengues, J. et al. LIF mediates proinvasive activation of stromal fibroblasts in cancer. Cell Rep. 7, 1664–1678 (2014).
Ohata, Y. et al. Leukemia inhibitory factor produced by fibroblasts within tumor stroma participates in invasion of oral squamous cell carcinoma. PLoS ONE 13, e0191865 (2018).
Wang, M. T. et al. Blockade of leukemia inhibitory factor as a therapeutic approach to KRAS driven pancreatic cancer. Nat. Commun. 10, 3055 (2019).
Shi, Y. et al. Targeting LIF-mediated paracrine interaction for pancreatic cancer therapy and monitoring. Nature 569, 131–135 (2019).
Kristensen, D. M., Kalisz, M. & Nielsen, J. H. Cytokine signalling in embryonic stem cells. APMIS 113, 756–772 (2005).
Li, Z. et al. LIF in embryo culture medium is a predictive marker for clinical pregnancy following IVF-ET of patients with fallopian tube obstruction. J. Reprod. Immunol. 141, 103164 (2020).
Duluc, D. et al. Tumor-associated leukemia inhibitory factor and IL-6 skew monocyte differentiation into tumor-associated macrophage-like cells. Blood 110, 4319–4330 (2007).
Abe, T. et al. Macrophage differentiation inducing factor from human monocytic cells is equivalent to murine leukemia inhibitory factor. J. Biol. Chem. 264, 8941–8945 (1989).
Pascual-García, M. et al. LIF regulates CXCL9 in tumor-associated macrophages and prevents CD8+ T cell tumor-infiltration impairing anti-PD1 therapy. Nat. Commun. 10, 2416 (2019).
Sierra Gonzalez, P. et al. Mannose impairs tumour growth and enhances chemotherapy. Nature 563, 719–723 (2018).
Zhang, C. S. et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 548, 112–116 (2017).
Datema, R. & Schwarz, R. T. Formation of 2-deoxyglucose-containing lipid-linked oligosaccharides: interference with glycosylation of glycoproteins. Eur. J. Biochem. 90, 505–516 (1978).
Yoo, J. et al. GlcNAc-1-P-transferase–tunicamycin complex structure reveals basis for inhibition of N-glycosylation. Nat. Struct. Mol. Biol. 25, 217–224 (2018).
Lopez-Sambrooks, C. et al. Oligosaccharyltransferase inhibition induces senescence in RTK-driven tumor cells. Nat. Chem. Biol. 12, 1023–1030 (2016).
Szaruga, M. et al. Activation of the integrated stress response by inhibitors of its kinases. Nat. Commun. 14, 5535 (2023).
Shin, S. et al. ERK2 mediates metabolic stress response to regulate cell fate. Mol. Cell 59, 382–398 (2015).
Favaro, F. et al. TRAIL receptors promote constitutive and inducible IL-8 secretion in non-small cell lung carcinoma. Cell Death Dis. 13, 1046 (2022).
Yoon, S. et al. NF-κB and STAT3 cooperatively induce IL6 in starved cancer cells. Oncogene 31, 3467–3481 (2012).
Pecharromán, I. et al. IκB kinase-α coordinates BRD4 and JAK/STAT signaling to subvert DNA damage-based anticancer therapy. EMBO J. 42, e114719 (2023).
Zhang, C., Liu, J., Wang, J., Hu, W. & Feng, Z. The emerging role of leukemia inhibitory factor in cancer and therapy. Pharmacol. Ther. 221, 107754 (2021).
Lambrechts, D. et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277–1289 (2018).
Kubota, Y., Hirashima, M., Kishi, K., Stewart, C. L. & Suda, T. Leukemia inhibitory factor regulates microvessel density by modulating oxygen-dependent VEGF expression in mice. J. Clin. Invest. 118, 2393–2403 (2008).
Wu, H. X. et al. LIFR promotes tumor angiogenesis by up-regulating IL-8 levels in colorectal cancer. Biochim. Biophys. Acta 1864, 2769–2784 (2018).
Li, P. et al. LIF, a mitogen for choroidal endothelial cells, protects the choriocapillaris: implications for prevention of geographic atrophy. EMBO Mol. Med. 14, e14511 (2022).
Huang, Y. et al. Pulmonary vascular destabilization in the premetastatic phase facilitates lung metastasis. Cancer Res. 69, 7529–7537 (2009).
Rashidfarrokhi, A. et al. Tumor-intrinsic LKB1–LIF signaling axis establishes a myeloid niche to promote immune evasion and tumor growth. Preprint at bioRxiv https://doi.org/10.1101/2023.07.15.549147 (2023).
Taftaf, R. et al. ICAM1 initiates CTC cluster formation and trans-endothelial migration in lung metastasis of breast cancer. Nat. Commun. 12, 4867 (2021).
Beretta, E., Dhillon, H., Kalra, P. S. & Kalra, S. P. Central LIF gene therapy suppresses food intake, body weight, serum leptin and insulin for extended periods. Peptides 23, 975–984 (2002).
Mori, M., Yamaguchi, K. & Abe, K. Purification of a lipoprotein lipase-inhibiting protein produced by a melanoma cell line associated with cancer cachexia. Biochem. Biophys. Res. Commun. 160, 1085–1092 (1989).
Yang, X. et al. Leukemia inhibitory factor suppresses hepatic de novo lipogenesis and induces cachexia in mice. Nat. Commun. 15, 627 (2024).
Kwon, Y.-Y. & Hui, S. IL-6 promotes tumor growth through immune evasion but is dispensable for cachexia. EMBO Rep. 25, 2592–2609 (2024).
Santos, G. C. et al. Leukemia inhibitory factor (LIF) overexpression increases the angiogenic potential of bone marrow mesenchymal stem/stromal cells. Front. Cell Dev. Biol. 8, 778 (2020).
Santio, N. M. et al. Endothelial PIM3 kinase protects the vascular barrier during lung metastasis. Nat. Commun. 15, 10514 (2024).
Orozco, J. M. et al. Dihydroxyacetone phosphate signals glucose availability to mTORC1. Nat. Metab. 2, 893–901 (2020).
Kalender, A. et al. Metformin, independent of AMPK, inhibits mTORC1 in a Rag GTPase-dependent manner. Cell Metab. 11, 390–401 (2010).
Liu, X. et al. Cystine transporter regulation of pentose phosphate pathway dependency and disulfide stress exposes a targetable metabolic vulnerability in cancer. Nat. Cell Biol. 22, 476–486 (2020).
Nikonorova, I. A. et al. Time-resolved analysis of amino acid stress identifies eIF2 phosphorylation as necessary to inhibit mTORC1 activity in liver. J. Biol. Chem. 293, 5005–5015 (2018).
Iurlaro, R. et al. Glucose deprivation induces ATF4-mediated apoptosis through TRAIL death receptors. Mol. Cell. Biol. 37, e00479-16 (2017).
León-Annicchiarico, C. L. et al. ATF4 mediates necrosis induced by glucose deprivation and apoptosis induced by 2-deoxyglucose in the same cells. FEBS J. 282, 3647–3658 (2015).
Hung, C.-H. et al. Defective N-glycosylation of IL6 induces metastasis and tyrosine kinase inhibitor resistance in lung cancer. Nat. Commun. 15, 7885 (2024).
Steelman, L. S. et al. Roles of the Raf/MEK/ERK and PI3K/PTEN/AKT/mTOR pathways in controlling growth and sensitivity to therapy—implications for cancer and aging. Aging 3, 192–222 (2011).
Ramírez-Peinado, S. et al. 2-Deoxyglucose induces Noxa-dependent apoptosis in alveolar rhabdomyosarcoma. Cancer Res. 71, 6796–6806 (2011).
Wellen, K. E. et al. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 24, 2784–2799 (2010).
Huber, A. L. et al. p58IPK-mediated attenuation of the proapoptotic PERK–CHOP pathway allows malignant progression upon low glucose. Mol. Cell 49, 1049–1059 (2013).
Hunter, S. A. et al. An engineered ligand trap inhibits leukemia inhibitory factor as pancreatic cancer treatment strategy. Commun. Biol. 4, 452 (2021).
Hyman, D. M. et al. A phase 1 study of MSC-1, a humanized anti-LIF monoclonal antibody, in patients with advanced solid tumors. J. Clin. Oncol. 36, TPS2602 (2018).
Borazanci, E. et al. Phase I dose escalation of MSC-1, a humanized anti-LIF monoclonal antibody, in patients (pts) with advanced solid tumours: updated results. Ann. Oncol. 30, v487–v488 (2019).
Andersen, C. L., Jensen, J. L. & Ørntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250 (2004).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013).
Hijazo-Pechero, S. et al. Transcriptional analysis of landmark molecular pathways in lung adenocarcinoma results in a clinically relevant classification with potential therapeutic implications. Mol. Oncol. 18, 453–470 (2024).
Vincent, E. E. et al. Mitochondrial phosphoenolpyruvate carboxykinase regulates metabolic adaptation and enables glucose-independent tumor growth. Mol. Cell 60, 195–207 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Korotkevich, G. et al. Fast gene set enrichment analysis. Preprint at bioRxiv https://doi.org/10.1101/060012 (2021).
Hammerman, P. S. et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012).
Collisson, E. A. et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Méndez-Lucas, A. et al. Identifying strategies to target the metabolic flexibility of tumours. Nat. Metab. 2, 335–350 (2020).
Acknowledgements
We thank CERCA Programme/Generalitat de Catalunya for institutional support. This study was funded by Ministerio de Ciencia e Innovación y Universidades (MICINN/MCIU), which is part of Agencia Estatal de Investigación, through the Generación de Conocimiento grant numbers PID2022-140457OB-I00, PID2019-107213GB-I00 (https://doi.org/10.13039/501100011033) and BFU2016-78154-R (cofunded by the European Regional Development Fund, ‘A way to build Europe’), the ‘Personal Tecnico de Apoyo’ fellowship numbers PTA2018-016371-I and PRE2020-096391 (cofunded by European Social Fund, ESF investing in your future), European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie Grant Agreements 675448 (TRAINERS) and 766214 (META-CAN) and the 201929-30 project, funded by La Marató de TV3. Funding was also provided by Instituto de Salud Carlos III (PI18/00920, PI21/00789 and PI24/00702) to E.N. (cofunded by the European Regional Development Fund/FEDER) and F.L.-M. (CD20/00191; cofunded by European Social Fund, ESF investing in your future); AGAUR (cofunded by European Social Fund, ESF investing in your future; 2021FI_B 00485) to M.C.-R.; Fundación Científica Asociación Española Contra el Cáncer (grant number LABAE20040GENT) and Ministerio de Ciencia e Innovación (grant number PID2021-122125NB-I00) to A.G.; and Programa de Formación del Profesorado Universitario grant agreement FPU21/06884, from the Spanish Ministry of Universities, to S.P.-M. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. We thank A. R. Mitjana, L. Pijuan, J. Repúlles, S. Mendizuri and P. Monaco for experimental help and E. Chevet, M. À. Pujana, E. Blommaert, H. Kalkavan, M. Mazzone, J.-E. Ricci and the META-CAN network for suggestions and tools. We thank the Bioinformatics Unit at IDIBELL for their contributions and the Histology Platform of IDIBELL Scientific and Technical Services for assistance with experiments.
Author information
Authors and Affiliations
Contributions
Investigation: M.H.-M., F.P., F.L.-M., L.C.-R., F.F., V.V.-A., S.H.-P., N.F.-D., F.J.-H., J.R.-P., P.F.V., J.M.-C., M.C.-R., D.P.-G., S.P.-M., L.d.B.-G., A.F. and A.M.-L. Conceptualization: C.M.-P., M.H.-M., F.L.-M., A.G., A.M.-L., J.C.P., F.V. and X.S. Formal analysis: M.H.-M., S.H.-P., F.L.-M. and J.M.-C. Supervision: E.N. and C.M.-P.
Corresponding author
Ethics declarations
Competing interests
E.N. declares research funding from Roche, Pfizer, Merck Serono and Bristol Myers Squibb; advisory board and consulting roles at Amgen, Apollomics, AstraZeneca, BeiGene, Bristol Myers Squibb, Boehringer Ingelheim, Daiichi Sankyo, Genmab, Johnson and Johnson, Lilly, Merck Sharp and Dohme (MSD), Merck Serono, Pfizer, Pierre Fabre, Qiagen, Regeneron, Roche, Sanofi and Takeda; honoraria for lectures from Amgen, AstraZeneca, BeiGene, Bristol Myers Squibb, Boehringer Ingelheim, Daiichi Sankyo, Illumina, Johnson and Johnson, Lilly, MSD, Merck Serono, Pfizer, Pierre Fabre, Qiagen, Regeneron, Roche, Sanofi and Takeda; and travel support from Roche, Takeda, Johnson and Johnson and MSD. C.M.-P. declares honorarium for lecture fees from Bristol Myers Squibb. The other authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Alfredo Gimenez-Cassina and Christoph Schmitt, in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of cell lines and antibodies.
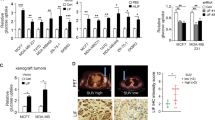
A. Mutational landscape of human and murine cell lines used. B. LIF protein secretion measured using ELISA in supernatants from human breast cancer (MCF7) cells after 24 h incubation in presence or absence of glucose. Expressed as pg/mL (left, n = 3) and normalized by protein content (right, n = 3). C. LIF protein measured using ELISA in supernatants from alveolar rhabdomyosarcoma Rh4 cells, Rh28 cells and Rms13 cells and embryonal rhabdomyosarcoma RD cells after 24 h incubation (or also after 48 h in the case of RD cells) in DMEM in presence or absence of glucose (n = 1). D. LIF protein secretion in supernatants from cell line CT26 using Mouse LIF R&D Duoset kit after 24 h incubation in DMEM in presence or absence of glucose (n = 4). E. LIF protein secretion in non-transformed cell lines HBEC and BEAS-2B using ELISA after incubation for 24 h with decreasing glucose concentrations (n = 3). F. Protein content per well of HBEC and BEAS-2B at 24 h (panel Extended Data Fig. 1E, n = 3) G. Demonstration of low sensitivity of anti-LIF antibodies for detection by WB. 100, 10, 1 and 0.1 ng of recombinant human LIF protein from R&D (generated in E. coli) were loaded and subjected to SDS PAGE and WB with indicated antibody (R&D AF-250-SP). H. LIF Antibody testing by WB using different recombinant LIFs (from R&D systems or Sino Biological, check Materials section) loading 200 and 100 ng. I. Measurement using hLIF R&D Duoset ELISA (discontinued, ref: DY7734-05) of two commercial recombinant LIF isoforms: glycosylated generated in HEK293, and non-glycosylated generated in E. coli (n = 3). J. Measurement, using hLIF R&D Quantikine ELISA (ref: DLF00B), of two commercial recombinant LIF isoforms: glycosylated, generated in HEK, and non-glycosylated generated in E. coli (n = 3). K. Measurement of murine LIF using R&D Duoset ELISA (ref: DY449) of glycosylated form of recombinant murine LIF. Standard used is non-glycosylated murine LIF generated in E. coli (provided in the kit) (n = 1). Graphs show average values and individual replicates, and error bars represent the standard error of the mean (SEM); analyzed by two-tailed paired t-test, except E (HBEC) that has been analyzed by one-way ANOVA with multiple comparisons.
Extended Data Fig. 2 Kinetics of secretion of glycosylated and unglycosylated LIF.
A. LIF secretion kinetics measured using R&D hLIF Quantikine ELISA (total LIF, upper panels n ≥ 3, check figure for exact n values) and R&D hLIF Duoset ELISA (for non-glycosylated LIF, lower panels, n = 3). Glc + in grey and Glc 0 mM in red. B. LIF mRNA kinetics in cellular lysates from A549 cells incubated in Plasmax measured by qPCR (n = 4 (0-24 h), 3(48 h), 2(72 h)). C. Translation efficiency of LIF, calculated as the ratio between ribosome protected fragments and RNA expression in an immortalized breast epithelial cell line (MCF10A) untransformed (EtOH) and transformed (TAM) to tumorigenic breast epithelium, and subject to glucose or glutamine deprivation for 30 min and 4 h. Calculated from public data, n = 2 (Gameiro & Struhl, 2018)7. D. LIF protein secretion, not normalized to protein content, in A549 cells after incubation for 24 h in 0 or 25 mM Glc (grey or red) in presence or absence of actinomycin D (ActD) (n = 3). E. LIF protein secretion in SW900 cells as in D, after incubation for 16 h (n = 4 (total LIF), n = 3 (non-glyc LIF)). F. LIF protein measured using R&D hLIF Quantikine ELISA (total LIF) in supernatants from A549 cells after 24 h incubation in presence or absence of glucose and increasing concentrations of H2O2. Results normalized by protein content (n = 3). G. Secreted LIF protein from A549 cells after 24 h treatment with the inhibitor of glucose-6-phosphate dehydrogenase, dehydroepiandrosterone (DHEA), at indicated concentrations, measured using R&D hLIF Quantikine ELISA (total LIF). Results normalized to protein content (n = 2). H. Protein content per well after incubating cells for 24 h under hypoxic conditions (0.1 or 1% O2) in DMEM + 10% dFBS (A549 (n = 3), H1299 (n = 4) and LLC (n = 5)) or RPMI + 10% dFBS (SW900, n = 3). I. LIF protein measured using ELISA in supernatants from A549 cells after 24 h incubation in presence or absence of glucose and increasing concentrations of CoCl2. Results were normalized by protein content (n = 3). J. LIF mRNA expression measured by qPCR and expressed as relative expression versus normoxia conditions (21% O2) after incubating cells for 24 h under hypoxic conditions (1% O2) in DMEM + 10% dFBS (H1299 and LLC, n = 3) or RPMI + 10% dFBS (SW900, n = 3). Graphs show average values and individual replicates, and error bars represent the standard error of the mean (SEM); statistical significance was determined by two-way ANOVA with multiple comparisons test (A-B), one-way ANOVA with multiple comparisons test (D-G and I) or two-tailed paired t-test (H and J).
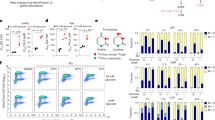
Extended Data Fig. 3 Mannose attenuates LIF release but does not substitute for glucose in restoring all metabolite pools.
A. Cell death measured by PI-positivity in A549 cells after incubation for 24 h at different glucose concentrations (n = 3). B. Protein content per well in A549 cells after 24 h incubation in glucose deprived conditions with re-addition of several glucose derived metabolites (n = 6, 4 for Man-ol condition). Glc=glucose; Man= mannose; Fru=fructose; Gal=galactose; GlcNAc= N-acetyl-glucosamine; Lac=lactate; Pyr=pyruvate, Man-ol=mannitol. C. LIF protein secretion (top) and protein content per well (bottom) in H1299 cells measured using ELISA after 24 h incubation in glucose deprived conditions with re-addition of several glucose derived metabolites (n = 3, 4 for Glc, Man and Man-ol condition). Glc=glucose; Man= mannose; Fru=fructose; Gal=galactose; GlcNAc= N-acetyl-glucosamine; Lac=lactate; Pyr=pyruvate, Man-ol=mannitol. D. LIF protein secretion (top) and protein content per well (bottom) in SW900 cells measured using ELISA after 24 h incubation in glucose deprived conditions with re-addition of several glucose derived metabolites (n = 3, and 5 for Glc and GlcNAc conditions). Glc=glucose; Man= mannose; Fru=fructose; Gal=galactose; GlcNAc= N-acetyl-glucosamine; Lac=lactate; Pyr=pyruvate, Man-ol=mannitol. E. Diagram depicting the fate of labelled mannose when glucose is present (left) or absent (right). Font size reduction indicated reduced pools, color in arrows and fonts indicate labelling. F. Total content of the indicated metabolites in A549 cells from the experiment shown in Fig. 3 (3 h - GC-MS, n = 3). G. LIF secreted from H1299 or SW900 cells after 24 h treatment with the NADPH replenisher N-acetylcysteine (NAC) measured using ELISA. Results normalized by protein content (n = 3). H. LIF secreted from H1299 cells after 24 h treatment with the inhibitor of glycolysis (3PO), measured using ELISA (n = 3). I. Representative WB of A549 cells at the times and conditions indicated. β-actin (“β-Act”) as loading control (n = 2). J. Protein content per well measured by BCA of panel Fig. 3i (n = 3). K. LIF secreted by H1299 cells after 24 h treatment with activator of AMPK, A769662 (A76) measured using ELISA. Results normalized by protein content (n = 2). L. Representative WB of A549 cells at 24 h and indicated conditions. Ponceau was used as loading control (n = 2). Graphs show average values and individual replicates, and error bars represent the standard error of the mean (SEM); statistical significance was determined by one-way ANOVA with multiple comparisons test except for panel F, which was analyzed by one-way ANOVA plus Tukey’s Test for Post-hoc Analysis.
Extended Data Fig. 4 Effects of glycosylation inhibitors on LIF release and cellular protein content.
A. Glycosylation as fold change vs. Glc 25 mM in A549. Densitometric quantification of the concanavalin A WB in Fig. 4b (n = 4). B. LIF secretion (non-glycosylated) from A549 treated for 24 h (n = 3). C-E. A549 cells were treated for 24 h with indicated drugs or DMSO (labeled as “D”). LIF secretion was measured at 24 h using R&D hLIF Quantikine ELISA (total LIF) (C, D), and protein content is indicated in panel E). n indicated in figure. F-H. SW900 and H1299 cells were incubated for 24 h with indicated drugs or DMSO (labeled as “D”). Total LIF release is shown in F and G. Protein content is shown in H. n indicated in figure. Graphs show average values and individual replicates, and error bars represent the standard error of the mean (SEM); statistical significance was determined by one-way ANOVA with multiple comparisons test.
Extended Data Fig. 5 Effects of the UPR and ISR on LIF release.
A. Relative expression of indicated mRNA, measured by qPCR, after 24 h of treatment (n = 3). B. Relative expression of indicated mRNA, measured by qPCR, after silencing (48 h) and 24 h of further treatment (n = 3). C. Relative expression of ATF6, measured by qPCR, after silencing (48 h) and 16 h of further treatment (n = 3). D. Western blot of ATF6 after silencing (48 h) and subsequent treatment (16 h). E. LIF secretion in the cell lines indicated after siRNA silencing for 48 h with ATF6. Incubated with glucose-free media for 16 h (n = 3 for A549, 5 for H1299 and SW900). F-H. Protein content per well measured by BCA of panel Fig. 5e-f (F,H n = 3, G n = 4). I. LIF in supernatants of A549 cells subject to PERK knockdown (24 h) and subsequent glucose deprivation (24 h), normalized to protein content (n = 4). J. Protein content per well measured by BCA of panel I (n = 4). K. Western blot of PERK after silencing (24 h) and subsequent treatment (24 h) in A549 cells. L. Western blot of LLC cells treated with glucose deprivation and the ISR inhibitor ISRIB (6 h) in media with 10% dFBS. M. Secreted LIF in supernatants of LLC cells under glucose deprivation (6-16 h) and treated with ISRIB, normalized to protein content (n = 3). N. Protein content per well (panel 5H, n = 3). O. Western blot of ATF4 after silencing (48 h) and subsequent treatment (16 h). P. As in E, with siRNA against ATF4 (n = 6 for A549, 4 for H1299 and SW900). Q. Protein content per well (Fig. 5i and Extended Data Fig. 5P, H1299, n = 3) R. Cells treated with non-targeting siRNA (“c”) or siRNA against QRICH were treated for 16 h and subjected to WB (up) and their supernatant to LIF ELISA (bottom, n = 3). S. Protein content per well (panel 5 K, n = 3) T. Cells treated with thapsigargin at the indicated concentrations for 24 h and subjected to WB (up), and their supernatants to LIF ELISA (bottom, n = 3). U-V. LIF secretion (non-glycosylated in U, n = 4, or total in V, n = 1) from A549 24 h after silencing p65 for 48 h. W. Protein content per well (panel 5 M, n = 3). X. Densitometric analysis of phospho ERK1/2 normalized to 1 h glc+ (Fig. 5o, n indicated in figure). Graphs show average values and individual replicates, and error bars represent the standard error of the mean (SEM); statistical significance was determined by one-way ANOVA with multiple comparisons test (A, B, C and Q), two-way ANOVA (F, G, M, N, S, W, X) or two-tailed paired t-test (E, H, I, J, P, R, U, X).
Extended Data Fig. 6 Characterization of LIF-deficient LLC.
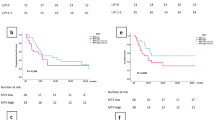
A. Distribution of LIF expression in tumours in the TCGA NSCLC LUAD and LUSC datasets grouped per tumour stage, as indicated, or grouped as early stage (I-II) and late stage (III-IV). Statistical tests used were two-sided Kruskal-Wallis or Wilcoxon, as indicated. The centre line is the median (P50); box bounds are the first and third quartiles (P25 and P75); whiskers extend to the most extreme observations within 1.5×IQR of the quartiles (IQR = P75–P25). Minima and maxima are defined as the lower and upper whisker ends (that is, the smallest and largest observations within 1.5×IQR), not the absolute extremes when outliers are present. Individual observations are shown as dots on the right; ‘n’ is the number of tumours with both expression and stage available. B. LIF mRNA levels and LIF secretion measured by ELISA in scramble, LIF-KO pool, and LIF-KO C3 groups measured in 25 mM Glucose and in 0 mM Glucose after 24 h (n = 3). C. Cell proliferation after 7 days, measured by cell counting (n = 3). D. Cell death analysis after 7 days, quantified by counting of trypan blue-positive cells (n = 3). E. Images of lungs from LLC injected mice separated into cohorts from LIF-KO pool LLC and Scramble vector LLC injected mice. Arrows indicate visible macroscopic lesions. F. Representative image of Ki67HIGH (green channel) and DAPI (blue channel) staining of Scramble and LIF-KO pool tumours, used to quantify tumoral areas (scale bars 1 mm). G. Tumour area quantification of each injected mice (n = 9 per group), each point representing a single tumour. Graphs B-D, G show average values, and error bars represent the standard error of the mean (SEM). Statistical significance was determined using one-way ANOVA with multiple comparisons.
Extended Data Fig. 7 Effects of LIF on HUVEC.
A. Representative histological images of subcutaneous tumours (scale bar 50 µm). B. Quantification of LIF positive areas in tumours using images from Fig. 7h. Results are expressed as the percentage of the LIF-positive area relative to the field area (n = 6, Scrambled 3 males and 3 females, LIF-KO C3 4 males and 2 females). C. Single-cell RNA-Seq average expression for LIFR in samples from previously untreated patients with NSCLC (n = 5) stratified by cell-type. Average expression is shown in each cell type without category aggregation. D. HUVEC cell proliferation measured by crystal violet assay after 24 h treatment with or without LIF (n = 3). E. Migration assay (6 h) quantification in HUVECs treated with increasing concentrations of glycosylated and non-glycosylated LIF, with increasing concentrations of FGF2 and VEGF as positive controls. Data is represented as fold change relative to control (media containing 0.5% of FBS only, n = 3). F. Migration assay (6 h) quantification in HUVECs treated with glycosylated and non-glycosylated LIF, with FGF2 and VEGF as positive controls. Data is represented as fold change relative to control (media only, n = 3 and n = 4 for 1% FBS). G-H. Representative images (G) and quantification (H) of tube formation assays in HUVECs treated with glycosylated and non-glycosylated LIF at 16 h. FGF2 and VEGF were included as positive controls. Unsupplemented HUVEC media was used as control. 20 ng/ml of glycosylated LIF were used instead of 10 to compensate for its double molecular weight. Quantification is expressed as fold change relative to control (media only, n = 3 for 0% FBS and n = 4 for 1% FBS). Dialyzed serum was used. I-J. Representative western blot (J) and quantification (I, n = 2) of pSTAT3 levels in HUVECs treated with glycosylated and non-glycosylated LIF at different concentrations (0, 1, 2.5, and 10 ng/mL). K. mRNA expression levels of genes related to VEGF in subcutaneous tumours from scramble and LIF-KO groups (n = 6). Graphs show average values and error bars represent the standard error of the mean (SEM). Statistical significance was determined by two-tailed Mann-Whitney unpaired test (B) or one-way ANOVA with multiple comparisons (I).
Supplementary information
Supplementary Information
Supplementary Tables 1–4.
Supplementary Data 1
Meta-analysis correlations LIF hypoxia and angiogenesis.
Supplementary Data 2
Hypoxia Hallmarks in-house glucose deprivation signature (48 h).
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Figs. 4 and 5 and Extended Data Figs. 1, 3, 5 and 7
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Luciano-Mateo, F., Moreno-Caceres, J., Hernández-Madrigal, M. et al. Glucose deprivation drives LIF-dependent lung cancer. Nat Metab (2026). https://doi.org/10.1038/s42255-025-01437-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42255-025-01437-0