Abstract
Despite undergoing castration, most individuals with prostate cancer (PCa) experience progression to castration-resistant PCa (CRPC), in which the androgen receptor (AR) remains an important driver. Concurrent genetic alterations in SPOP and CHD1 define a unique subtype of PCa, but their interactions in tumor progression and therapy response remain unclear. Here, we provide genetic evidence supporting that CHD1 loss accelerates disease progression and confers resistance to castration in males with SPOP-mutated PCa. By leveraging genetic engineering and multiomics, we uncovered a noncanonical function of CHD1 in lipid metabolism reprogramming via repressing the SREBP2 transcriptome. Loss of CHD1 induces cholesterol production, supplies intratumoral androgen biosynthesis and enhances AR activity, leading to castration resistance of SPOP-mutated PCa. Combining anti-androgen therapy with cholesterol-lowering drugs showed synergistic and durable activity against CRPC harboring CHD1 loss and SPOP mutations. These findings advance our understanding of an emerging PCa subtype and offer biomarker-driven combinatorial treatment strategies for men with CRPC.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
CUT&RUN-seq data supporting this study’s findings have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE272802. scRNA-seq data generated in this study have been deposited in the GEO under accession code GSE273102. Metabolism profiling data are available in Supplementary Table 2. The CHD1 RIME dataset generated in this study has been deposited in the PRIDE (ProteomeXchange) database under accession code PXD061854. Previously published scRNA-seq data that were reanalyzed here are available under accession code GSE210358 (ref. 50). The human pan-cancer and PCa genomic and bulk RNA-seq data46 were derived from the TCGA Research Network (http://cancergenome.nih.gov/). The human metastatic PCa genomic and bulk RNA-seq data were derived from the SU2C/PCF metastatic PCa dataset53, which is available at www.cbiportal.org and has been deposited in GitHub at https://github.com/cBioPortal/datahub/tree/master/public/prad_su2c_2019. Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author upon reasonable request
References
Desai, K., McManus, J. M. & Sharifi, N. Hormonal therapy for prostate cancer. Endocr. Rev. 42, 354–373 (2021).
Huggins, C. & Hodges, C. V. Studies on prostatic cancer. I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. Cancer Res. 1, 293–297 (1941).
Stanbrough, M. et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 66, 2815–2825 (2006).
Montgomery, R. B. et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 68, 4447–4454 (2008).
Holzbeierlein, J. et al. Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am. J. Pathol. 164, 217–227 (2004).
Attard, G. et al. Selective inhibition of CYP17 with abiraterone acetate is highly active in the treatment of castration-resistant prostate cancer. J. Clin. Oncol. 27, 3742–3748 (2009).
Potter, G. A., Barrie, S. E., Jarman, M. & Rowlands, M. G. Novel steroidal inhibitors of human cytochrome P45017 α (17 α-hydroxylase-C17,20-lyase): potential agents for the treatment of prostatic cancer. J. Med. Chem. 38, 2463–2471 (1995).
Ryan, C. J. et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med. 368, 138–148 (2013).
de Bono, J. S. et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 364, 1995–2005 (2011).
Ryan, C. J. et al. Abiraterone acetate plus prednisone versus placebo plus prednisone in chemotherapy-naive men with metastatic castration-resistant prostate cancer (COU-AA-302): final overall survival analysis of a randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 16, 152–160 (2015).
James, N. D. et al. Abiraterone for prostate cancer not previously treated with hormone therapy. N. Engl. J. Med. 377, 338–351 (2017).
Danila, D. C. et al. Phase II multicenter study of abiraterone acetate plus prednisone therapy in patients with docetaxel-treated castration-resistant prostate cancer. J. Clin. Oncol. 28, 1496–1501 (2010).
Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015).
Barbieri, C. E. et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 44, 685–689 (2012).
Robinson, D. et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015).
Geng, C. et al. Androgen receptor is the key transcriptional mediator of the tumor suppressor SPOP in prostate cancer. Cancer Res. 74, 5631–5643 (2014).
An, J., Wang, C., Deng, Y., Yu, L. & Huang, H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 6, 657–669 (2014).
Gan, W. et al. SPOP promotes ubiquitination and degradation of the ERG oncoprotein to suppress prostate cancer progression. Mol. Cell 59, 917–930 (2015).
An, J. et al. Truncated ERG oncoproteins from TMPRSS2–ERG fusions are resistant to SPOP-mediated proteasome degradation. Mol. Cell 59, 904–916 (2015).
Groner, A. C. et al. TRIM24 is an oncogenic transcriptional activator in prostate cancer. Cancer Cell 29, 846–858 (2016).
Li, C. et al. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene 30, 4350–4364 (2011).
Shi, L. et al. Mutated SPOP E3 ligase promotes 17βHSD4 protein degradation to drive androgenesis and prostate cancer progression. Cancer Res. 81, 3593–3606 (2021).
Blattner, M. et al. SPOP mutation drives prostate tumorigenesis in vivo through coordinate regulation of PI3K/mTOR and AR signaling. Cancer Cell 31, 436–451 (2017).
Swami, U. et al. Association of SPOP mutations with outcomes in men with de novo metastatic castration-sensitive prostate cancer. Eur. Urol. 78, 652–656 (2020).
Nakazawa, M. et al. Clinical and genomic features of SPOP-mutant prostate cancer. Prostate 82, 260–268 (2022).
Stopsack, K. H. et al. Oncogenic genomic alterations, clinical phenotypes, and outcomes in metastatic castration-sensitive prostate cancer. Clin. Cancer Res. 26, 3230–3238 (2020).
Swami, U. et al. SPOP mutations as a predictive biomarker for androgen receptor axis-targeted therapy in de novo metastatic castration-sensitive prostate cancer. Clin. Cancer Res. 28, 4917–4925 (2022).
Boysen, G. et al. SPOP-mutated/CHD1-deleted lethal prostate cancer and abiraterone sensitivity. Clin. Cancer Res. 24, 5585–5593 (2018).
Stockdale, C., Flaus, A., Ferreira, H. & Owen-Hughes, T. Analysis of nucleosome repositioning by yeast ISWI and CHD1 chromatin remodeling complexes. J. Biol. Chem. 281, 16279–16288 (2006).
Lusser, A., Urwin, D. L. & Kadonaga, J. T. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 12, 160–166 (2005).
Li, H., Gigi, L. & Zhao, D. CHD1, a multifaceted epigenetic remodeler in prostate cancer. Front. Oncol. 13, 1123362 (2023).
Burkhardt, L. et al. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res. 73, 2795–2805 (2013).
Berger, M. F. et al. The genomic complexity of primary human prostate cancer. Nature 470, 214–220 (2011).
Grasso, C. S. et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012).
Huang, S. et al. Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene 31, 4164–4170 (2012).
Liu, W. et al. Identification of novel CHD1-associated collaborative alterations of genomic structure and functional assessment of CHD1 in prostate cancer. Oncogene 31, 3939–3948 (2012).
Baca, S. C. et al. Punctuated evolution of prostate cancer genomes. Cell 153, 666–677 (2013).
Augello, M. A. et al. CHD1 loss alters AR binding at lineage-specific enhancers and modulates distinct transcriptional programs to drive prostate tumorigenesis. Cancer Cell 35, 603–617 (2019).
Zhang, Z. et al. Loss of CHD1 promotes heterogeneous mechanisms of resistance to AR-targeted therapy via chromatin dysregulation. Cancer Cell 37, 584–598 (2020).
Metzger, E. et al. Assembly of methylated KDM1A and CHD1 drives androgen receptor-dependent transcription and translocation. Nat. Struct. Mol. Biol. 23, 132–139 (2016).
Rodrigues, L. U. et al. Coordinate loss of MAP3K7 and CHD1 promotes aggressive prostate cancer. Cancer Res. 75, 1021–1034 (2015).
Jillson, L. K. et al. MAP3K7 loss drives enhanced androgen signaling and independently confers risk of recurrence in prostate cancer with joint loss of CHD1. Mol. Cancer Res. 19, 1123–1136 (2021).
Zhao, D. et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature 542, 484–488 (2017).
Zhao, D. et al. Chromatin regulator CHD1 remodels the immunosuppressive tumor microenvironment in PTEN-deficient prostate cancer. Cancer Discov. 10, 1374–1387 (2020).
Li, H. et al. CHD1 promotes sensitivity to aurora kinase inhibitors by suppressing interaction of AURKA with its coactivator TPX2. Cancer Res. 82, 3088–3101 (2022).
Hoadley, K. A. et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173, 291–304 (2018).
Ellwood-Yen, K. et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 4, 223–238 (2003).
Watson, P. A. et al. Context-dependent hormone-refractory progression revealed through characterization of a novel murine prostate cancer cell line. Cancer Res. 65, 11565–11571 (2005).
Drost, J. et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 11, 347–358 (2016).
Chan, J. M. et al. Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science 377, 1180–1191 (2022).
Horton, J. D., Goldstein, J. L. & Brown, M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 (2002).
Xue, L. et al. Targeting SREBP-2-regulated mevalonate metabolism for cancer therapy. Front. Oncol. 10, 1510 (2020).
Abida, W. et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl Acad. Sci. USA 116, 11428–11436 (2019).
Scacchetti, A. et al. CHRAC/ACF contribute to the repressive ground state of chromatin. Life Sci. Alliance 1, e201800024 (2018).
Baldi, S. et al. Genome-wide rules of nucleosome phasing in Drosophila. Mol. Cell 72, 661–672 (2018).
Wiles, E. T., Mumford, C. C., McNaught, K. J., Tanizawa, H. & Selker, E. U. The ACF chromatin-remodeling complex is essential for Polycomb repression. eLife 11, e77595 (2022).
Kamisuki, S. et al. A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem. Biol. 16, 882–892 (2009).
Luirink, I. K. et al. 20-Year follow-up of statins in children with familial hypercholesterolemia. N. Engl. J. Med. 381, 1547–1556 (2019).
Hamilton, R. J., Goldberg, K. C., Platz, E. A. & Freedland, S. J. The influence of statin medications on prostate-specific antigen levels. J. Natl Cancer Inst. 100, 1511–1518 (2008).
Murtola, T. J., Tammela, T. L., Lahtela, J. & Auvinen, A. Cholesterol-lowering drugs and prostate cancer risk: a population-based case–control study. Cancer Epidemiol. Biomarkers Prev. 16, 2226–2232 (2007).
Harshman, L. C. et al. The impact of statin use on the efficacy of abiraterone acetate in patients with castration-resistant prostate cancer. Prostate 77, 1303–1311 (2017).
Shi, W. et al. Immune checkpoint B7-H3 is a therapeutic vulnerability in prostate cancer harboring PTEN and TP53 deficiencies. Sci. Transl. Med. 15, eadf6724 (2023).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Mohammed, H. et al. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc. 11, 316–326 (2016).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Zhang, Y. et al. Model-based analysis of ChIP–seq (MACS). Genome Biol. 9, R137 (2008).
Amemiya, H. M., Kundaje, A. & Boyle, A. P. The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep. 9, 9354 (2019).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Acknowledgements
We thank R. A. DePinho at The University of Texas MD Anderson for sharing the P and PC mouse models. We thank A. Sutton, who is in Editing Services of the Research Medical Library at MD Anderson, for editing the article. We also acknowledge the MD Anderson Small Animal Imaging Facility (supported by NIH/NCI P30 CA016672 Cancer Center Support Grant), Metabolomics Core Facility (supported by NIH/NCI P30 CA016672 Cancer Center Support Grant) and CCSG Research Histology Core Laboratory (supported by NIH/NCI P30 CA016672 Cancer Center Support Grant). D.E.F. has been supported by NIH R01 CA275138, DOD/PCRP W81XWH-22-1-0686 and DOD/PCRP W81XWH-22-1-0187. B.G. has been supported by RP230072 from the Cancer Prevention & Research Institute of Texas (CPRIT) and R01CA181196, R01CA244144, R01CA247992 and R01CA269646 from the NIH. D.Z. is a CPRIT Scholar in Cancer Research and has been supported by the CPRIT Recruitment of First-Time Tenure-Track Faculty Award RR190021, NIH R00 CA226360, NIH R01 CA275990 and CA278889, Prostate Cancer Foundation Challenge Award FP00016492 and DOD CDMRP IDA award PC230358. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
F.C., H.L. and D.Z. designed the research. F.C. and D.Z. wrote the main text of the manuscript, and H.L. wrote part of the Methods. F.C. performed experiments other than those listed below. H.L. prepared samples for lipidomic profiling and RIME and did initial data analysis, prepared CUT&RUN samples for sequencing and performed co-IP, ChIP–qPCR, luciferase assays and some experiments related to SREBP2 and SNF2H. D.Z. conducted the clinical relevance analysis and scRNA-seq analysis. Y.W. provided mouse husbandry of GEMM mice and assisted in surgical castration and histopathologic staining. X.T. and M.G.R. performed pathological analyses. K.L. and Y.L. performed CUT&RUN-seq data analysis and supported RIME data analysis. Q.L. performed part of the western blotting. C.M. provided SPOP-mutant plasmids and provided technical support. W.S., J.L. and X.L. provided technical support. J.Z. assisted in preparing pathological samples. V.V. aided in conducting MRI scanning. I.M., B.W. and P.L.L. performed lipidomic profiling and data analysis. A.A., D.E.F. and B.G. provided intellectual contributions throughout the project.
Corresponding author
Ethics declarations
Competing interests
D.E.F. has received research funding from GTx and has a familial relationship with March Biosciences, Biocity Biopharmaceuticals, Hummingbird Bioscience and Barricade Therapeutics. The other authors declare no competing interests.
Peer review
Peer review information
Nature Cancer thanks Scott Cramer, David Labbé, Jean-Philippe Theurillat and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Combined CHD1Del and SPOPMut Promote Prostate Tumorigenesis.
a, Co-occurrence of CHD1Del and SPOPMut in prostate adenocarcinoma (Log2 Odds Ratio > 3; p < 0.001). Oncoprint was generated from cbioportal. b, Quantification of MRI of prostate tumors from 10-month-old male PC (n = 8 mice), PSp (n = 8 mice), and PCSp (n = 19 mice). c, Histopathology analysis of prostate tumors from PC, PSp, and PCSp male GEMM mice. AP: anterior prostate lobes; VP: ventral prostate lobes; DLP: dorsal and lateral prostate lobes. d, Histopathology analysis revealed the total area of carcinoma region in prostate tumors from male PC (n = 8 mice), PSp (n = 8 mice), and PCSp (n = 6 mice). e, IHC staining (Ser473-phosphorylated AKT; CHD1; Flag; AR and Ki67) of prostate tumors from PC, PSp, and PCSp male GEMM mice at 6 months of age. Representative images are from AP lobes. Scale bar = 50 μm. Data (b) are presented as the mean ± standard deviation. Experiments were performed at least three times with similar results. One-way ANOVA with Tukey’s post hoc tests (b) was performed using GraphPad Prism v.9.2.0.
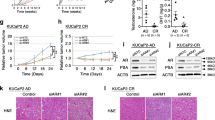
Extended Data Fig. 2 CHD1Del confers resistance to castration in SPOPMut PCa.
a-b, Histologic analysis of Ki67 and Cleaved caspase-3 in PCSp prostate tumors with or without surgical castration (n = 3 tumors per group). Tumors that showed response or resistance to castration were compared. Representative images are from anterior prostate (AP) lobes. Cx: Castration. Scale bar = 50 μm. c, Western blot assays of MyC-Cap cells expressing SPOPMut (W131G) with or without Chd1 knockout (KO). Sp, SPOPW131G; CSp, SPOPW131G/Chd1KO. d, Growth curves of Sp and CSp MyC-CaP cells overexpressing vector control (CSp-Vec) or CHD1 (CSp-CHD1) cultured in medium supplied with regular FBS (Normal) or charcoal-stripped FBS (CS). n = 3 biological replicates. e, MYC IHC staining in Sp and CSp MyC-Cap tumors with or without castration. Scale bar = 50 μm. f, Quantification of Ki67 IHC staining in Sp and CSp MyC-Cap tumors with or without castration (n = 3 tumors per group). g, Western blot assays of LNCaP cells expressing SPOP mutation (W131G) with or without Chd1 knockout (KO). Sp, SPOPW131G; CSp, SPOPW131G/CHD1KO. h, Quantification of Ki67 IHC staining in Sp and CSp LNCaP xenograft tumors with or without castration (n = 3 tumors per group). i, Western blot assays of LAPC4 cells expressing SPOP mutants with or without CHD1 depletion. j, Growth curves of SPOPMut LAPC4 cells with or without CHD1 depletion cultured in medium supplied with regular FBS (Normal) or charcoal-stripped FBS (CS) (n = 3 biological replicates per group). Data in b, d, f, h and j are presented as the mean ± standard deviation. Experiments were performed at least three times with similar results. Two-tailed unpaired Student’s t-test (d and j) or one-way ANOVA with Tukey’s post hoc tests (b, f, and h) was performed using GraphPad Prism v.9.2.0.
Extended Data Fig. 3 CHD1Del retains AR signaling in SPOPMut PCa upon castration.
a, Representative images of GFP+ organoids derived from P and PC GEMMs, cultured with different dosages of DHT for 6 days. Each GFP+ dot indicates an organoids. Scale bar = 2000 μm. b-c, Histopathology analysis of P and PC tumor organoids cultured with or without DHT (n = 3 organoid samples per group). Scale bar = 50 μm. d, Representative images and quantification of AR IHC staining in prostate tumors from PSp and PCSp GEM mice with or without castration (n = 3 tumors). Representative images are from anterior prostate (AP) lobes. Cx: Castration. Scale bar = 50 μm. e, Expression of AR and target genes in P and PC tumor organoids in the absence of DHT, as determined by qPCR (n = 3 biological replicates). f, qPCR analysis of TMPTRSS2 expression in Sp (SPOPW131G) and CSp (SPOPW131G/CHD1KO) MyC-CaP cells cultured in the CS medium (n = 3 biological replicates). g, Western blot assays of LNCaP cells expressing SPOP mutants with or without CHD1 depletion. h, mRNA expression of AR and target genes in LNCaP cells with or without CHD1 knockout, cultured in the CS medium (n = 3 biological replicates). i, V5-tagged CHD1 was overexpressed in CHD1-depleted LNCaP cells, followed by the determination of AR target genes using qPCR (n = 3 biological replicates). j, ChIP-qPCR in control and CHD1 knockout LNCaP cells revealed that CHD1 loss increased AR binding to enhancer regions of PSA and TMPRSS2 genes (n = 3 biological replicates). k, CHD1 expression in 29,861 single epithelial cells from n = 13 mCRPC patients (scRNA-seq dataset, GSE210358). Data (c-f and h-j) presented as the mean ± standard deviation. Experiments were performed at least three times with similar results. Unpaired two-tailed Student’s t-test (e, f, h and j), one-way ANOVA with Tukey’s post hoc test (d and i), or two-way ANOVA (c) with Tukey’s post hoc test were performed using GraphPad Prism v.9.2.0.
Extended Data Fig. 4 Single-cell transcriptomic profiling of PSp and PCSp GEMMs.
a-e, ScRNA-seq was performed in prostate tumors from PSp and PCSp GEMM mice one month after surgical castration (n = 2 mice per group). a, The UMAP views of 22,943 single cells color-coded by nine major clusters (C1-C9). b, The bubble plot presents marker gene expression for each cluster, where dot size and color represent the percentage of marker gene expression and the averaged scaled expression value, respectively. c, The UMAP view of prostate epithelial cells color-coded by two genotype groups. d, The violin plot of CHD1 gene expression for each subcluster in PSp versus PCSp samples. e, UMAP views of prostate epithelial cells, color-coded by the count of signature genes in indicated pathways. Two-tailed Wilcoxon tests (d) were performed using Bioturing.
Extended Data Fig. 5 CHD1Del promotes cholesterol biosynthesis and intratumoral androgen production.
a, Heatmap displaying the abundance of all 1013 lipid metabolites in control versus CHD1-depleted LNCaP cells cultured in CS medium. b, Principal Component Analysis (PCA) scores plot for spectral data of lipid metabolites from control and CHD1-depleted LNCaP cells. c, The abundance of each category of lipid metabolites in control versus CHD1-depleted LNCaP cells cultured in CS medium (n = 3 biological replicates). The full name of each lipid metabolite is listed in Supplemental Table 2. d, g, Control and CHD1-depleted LNCaP cells expressing wildtype SPOP or W131G mutant were cultured in CS medium, followed by the determination of cholesterol (d, n = 3 biological replicates) and testosterone (g, n = 6 biological replicates). e, h, MyC-CaP cells expressing SPOPW131G with or without CHD1 depletion were cultured in CS medium. Cells and culture media were collected to determine total cholesterol (e, n = 3 biological replicates) and testosterone (h, n = 3 biological replicates), respectively. f, Total cholesterol and cholesterol esters in P, PC, PSp, and PCSp organoids cultured in the DHT-free medium (n = 3 biological replicates). i, Enrichment of cholesterol metabolism geneset in SPOPMut prostate tumors with or without CHD1Del, determined by Gene Set Enrichment Analysis (GSEA). The normalized Enrichment Score (NES) and False Discovery Rate (FDR) q-value are shown. Data are presented as the mean ± standard deviation. Experiments were performed at least three times with similar results. Two-tailed unpaired Student’s t-test (c, e, g, and h) or one-way ANOVA (d and f) with Tukey’s post hoc tests were performed using GraphPad Prism v.9.2.0.
Extended Data Fig. 6 CHD1Del promotes cholesterol biosynthesis enzymes by inducing SREBP2 expression.
a-d, qPCR and Western blot analysis of indicated genes in LNCaP cells expressing SPOPMut (W131G or F102C) with or without CHD1 knockout (clone A or C) cultured in normal or charcoal-stripped (CS) medium (n = 3 biological replicates for qPCR assays). e-f, mRNA levels of indicated genes in LAPC4 or Myc-Cap cells expressing SPOPW131G with or without CHD1 depletion (n = 3 biological replicates for qPCR assays). g, mRNA expression of indicated genes in Sp (SPOPW131G) and CSp (SPOPW131G/CHD1KO) LNCaP cells cultured in the normal medium, CS medium, or CS medium supplied with cholesterol (Ch, 30 μM) (n = 3 biological replicates). h, Western blot analysis of SREBP2 in CHD1-depleted LNCap cells upon CHD1 re-expression. i, Expression of CHD1 and SREBF2 in tumors with different CHD1 copy number variation (CNV) (Pan-Cancer TCGA dataset). j, Pearson correlation analysis of CHD1 expression with SREBF2 and its target genes in metastatic PCa containing SPOPMut (n = 13 patients; SU2C/PCF dataset). Data are presented as the mean ± standard deviation. Experiments were performed at least three times with similar results. Two-tailed unpaired Student’s t-test (a, c, e, and f) or one-way ANOVA with Tukey’s post hoc tests (g and i) were performed using GraphPad Prism v.9.2.0.
Extended Data Fig. 7 Non-canonical function of CHD1 in repressing SREBP2 by interacting with SNF2H/ACF1 complex.
a, ChIP-Seq profile showing the binding of CHD1 to the promoter region of the Srebf2 gene in murine PCa cells. Two antibodies against CHD1 were used for ChIP-seq. b, Peptide coverage of the bait protein (CHD1) and its interacting protein SNF2H, as determined by RIME. Regions shown in blue represent peptides detected by mass spectrometry for each protein. c, Co-IP assays were performed in C4-2 and PC3-M cells using CHD1 antibody, followed by Western blot analysis of CHD1 and SNF2H. d, Western blot analysis of SNF2H in control and CHD1-depleted LNCaP cells. e, ChIP was performed using IgG, CHD1, and SNF2H antibodies in LNCaP cells cultured in CS medium. The binding of CHD1 and SNF2H proteins to the promoter region of the SREBF2 gene was determined using qPCR (n = 3 biological replicates). The locations of primers (P1-P6) in the up- or down-stream of SREBF2’s transcription start site (TSS) and Exon 1 (E1) region are shown. f, Expression of HMGCS1 in control and CHD1-depleted LNCaP cells with or without SNF2H (SMARCA5) knockdown (n = 3 biological replicates). g, CUT&RUN tracks illustrating the binding of SNF2H to the promoter region of the SREBF2 gene in SPOPMut LNCaP cells with or without CHD1 knockout. Data are presented as the mean ± standard deviation. Experiments were performed at least three times with similar results. Two-tailed unpaired Student’s t-test (e) or one-way ANOVA with Tukey’s post hoc tests (f) were performed using GraphPad Prism v.9.2.0.
Extended Data Fig. 8 SREBP2 is required for intratumoral cholesterol and androgen synthesis driven by CHD1Del.
a-b, SPOPMut LNCaP cells with SREBF2 overexpression were cultured in CS medium for 48 hours, followed by the determination of SREBF2 gene expression (a) and production of total cholesterol and testosterone (b). c, Growth curves of SPOP-mutated LNCaP cells with SREBF2 overexpression cultured in CS medium for 6 days. Data represent the results of triplicates. d-e, Control and CHD1-depleted SPOPMut LNCaP cells were cultured in the CS medium, followed by treatment with Fatostatin (10 μM) for 24 hours. mRNA expression of SREBF2 target genes (d) and AR target genes (e) was determined by qPCR. f, Control and CHD1-depleted SPOPMut LNCaP cells were cultured in CS medium with or without Fatostatin (10 μM) for 6 days. Cell proliferation was determined, and growth curves were presented. g, PSp and PCSp organoids were cultured in the DHT-free medium, followed by treatment of Fatostatin (10 μM) for 6 days. FKBP5 gene expression was determined by qPCR. n = 3 biological replicates in all figures. Data in all figures are presented as the mean ± standard deviation. Experiments were performed at least three times with similar results. Two-tailed unpaired Student’s t-test (a, b, c, and f) or one-way ANOVA with Tukey’s post hoc tests (d, e and g) was performed using GraphPad Prism v.9.2.0.
Extended Data Fig. 9 Abiraterone synergizes with atorvastatin in suppressing CHD1Del/SPOPMut PCa cells.
a, CHD1-depleted SPOP-mutated LNCaP cells were cultured in the charcoal-stripped (CS) medium, followed by treatment with Abiraterone at different doses for 48 hours. mRNA expression of SREBF2 and its target genes was determined by qPCR (n = 3 biological replicates). b-d, Dose-response matrix and drug interaction landscapes revealing the combination effects of Abiraterone (Abi) and Atorvastatin (Ato) on LNCaP (a,b), MyC-CaP (c), and LAPC4 (d) with or without SPOPW131G and CHD1 deletion. Drug interaction landscapes and synergy scores were generated using Synergyfinder. e, Representative GFP images of P organoids after 6-day treatment of Abiraterone or Atorvastatin, alone or in combination, in the absence of DHT. Scale bar = 2000 μm. f, H&E and IHC staining of indicated markers in P organoids treated with Abiraterone and/or Atorvastatin. Scale bar = 50 μm. g. Quantification of Ki67 (upper), Cleaved caspase-3 (middle), and AR (bottom) IHC staining in P organoids treated with Abiraterone or Atorvastatin, alone or in combination (n = 3 organoid samples per group). Data are presented as the mean ± standard deviation. Experiments were performed at least three times with similar results. One-way ANOVA with Dunnett’s post hoc tests (a) or one-way ANOVA with Tukey’s post hoc tests (g) were performed using GraphPad Prism v.9.2.0.
Extended Data Fig. 10 Abiraterone combined with cholesterol-lowing drugs shows therapeutic potential in CHD1Del/SPOPMut CRPC.
a, Body weights of male PSp (n = 3 mice) and PCSp (n = 4 mice) were documented throughout the treatment period. b-c, MRI imaging and tumor volume quantification of prostate tumors from male PSp (n = 3 mice) before and after treatment with Abiraterone and Atorvastatin. Prostate tumor regions are circled with orange dashed lines. Cx: Castration. d, H&E and AR staining of prostate tumors from castrated PSp male mice, with and without combination treatment. Scale bar = 100 μm. Experiments were performed at least three times with similar results. Two-tailed paired Student’s t-test (c) was performed using GraphPad Prism v.9.2.0.
Supplementary information
Supplementary Information
Supplementary Fig. 1. Gating strategy.
Supplementary Tables 1–5
Supplementary Tables 1–5.
Source data
Source Data Figs. 1–8 and Extended Data Figs. 1–3 and 5–10
Statistical source data.
Source Data Fig. 5 and Extended Data Figs. 2, 3, 6 and 7
Unprocessed western blots and/or gels.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, F., Li, H., Wang, Y. et al. CHD1 loss reprograms SREBP2-driven cholesterol synthesis to fuel androgen-responsive growth and castration resistance in SPOP-mutated prostate tumors. Nat Cancer 6, 854–873 (2025). https://doi.org/10.1038/s43018-025-00952-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43018-025-00952-z
This article is cited by
-
CHD1 status drives divergent metabolic pathways in SPOP-mutant prostate cancer
Nature Cancer (2025)