Abstract
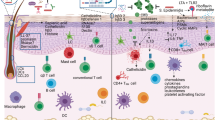
Older adults represent a vulnerable population with elevated risk for numerous morbidities. To explore the association of the microbiome with aging and age-related susceptibilities, including frailty and infectious disease risk, we conducted a longitudinal study of the skin, oral, and gut microbiota in 47 community- or skilled nursing facility-dwelling older adults versus younger adults. We found that microbiome changes were not associated with chronological age so much as frailty; we identified prominent changes in microbiome features associated with susceptibility to pathogen colonization and disease risk, including diversity, stability, heterogeneity and biogeographic determinism, which were moreover associated with a loss of Cutibacterium acnes in the skin microbiome. Strikingly, the skin microbiota were also the primary reservoir for antimicrobial resistance, clinically important pathobionts and nosocomial strains, suggesting a potential role particularly for the skin microbiome in disease risk and dissemination of multidrug resistant pathogens.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Metagenomic sequence files from participants who consented to making their deidentified metagenomic data available in public access data can be accessed in National Center for Biotechnology Information BioProject PRJNA699281. HMP data can be accessed from https://www.hmpdacc.org/hmp/; SRS IDs of the samples used in this study are detailed in Supplementary Table 2. Oh et al.29,30 data can be accessed via National Center for Biotechnology Information BioProject PRJNA46333.
References
Franceschi, C. et al. The continuum of aging and age-related diseases: Common mechanisms but different rates. Front. Med. 5, 61 (2018).
Inouye, S. K., Studenski, S., Tinetti, M. E. & Kuchel, G. A. Geriatric syndromes: Clinical, research and policy implications of a core geriatric concept. J. Am. Geriatrics Soc. 55, 780 (2007).
Tang, W. H. W. et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368, 1575 (2013).
Koren, O. et al. Colloquium Paper: Human oral, gut, and plaque microbiota in patients with atherosclerosis. PNAS 108, 4592 (2011).
Pickard, J. M., Zeng, M. Y., Caruso, R. & Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses and inflammatory disease. Immunol. Rev. 279, 70–89 (2017).
Byrd, A. L., Belkaid, Y. & Segre, J. A. The human skin microbiome. Nat. Rev. Microbiol. 16, 143 (2018).
Xue, Q. The frailty syndrome: Definition and natural history. Clin. Geriatr. Med 27, 1–15 (2011).
Meehan, C. J., Langille, M. G. I. & Beiko, R. G. Frailty and the microbiome. Top. Gerontol. Geriatr. 41, 54–65 (2015).
Rockwood, K. et al. A frailty index based on deficit accumulation quantifies mortality risk in humans and in mice. Sci. Rep. 7, 43068 (2017).
Mitnitski, A., Howlett, S. E. & Rockwood, K. Heterogeneity of human aging and its assessment. J. Gerontol. A Biol. Sci. Med Sci. 72, 877–884 (2017).
Ferrucci, L. & Kuchel, G. A. Heterogeneity of aging: individual risk factors, mechanisms, patient priorities, and outcomes.J. Am. Geriatr. Soc. 69, 610–612 (2021).
Nguyen, Q. D. Health heterogeneity in older adults: Exploration in the Canadian longitudinal study on aging.J. Am. Geriatr. Soc. 69, 678–687 (2021).
Kuchel, G. A. Inclusion of older adults in research: ensuring relevance, feasibility, and rigor. J. Am. Geriatrics Soc. 67, 203–204 (2019).
Haran, J. P., Bucci, V., Dutta, P., Ward, D. & McCormick, B. The nursing home elder microbiome stability and associations with age, frailty, nutrition and physical location. J. Med. Microbiol. 67, 40–51 (2018).
Claesson, M. J. et al. Gut microbiota composition correlates with diet and health in the elderly. Nat. (Lond.) 488, 178–184 (2012).
Gómez-Zorrilla, S. et al. Prospective observational study of prior rectal colonization status as a predictor for subsequent development of Pseudomonas aeruginosa clinical infections. Antimicrob. Agents Chemother. 59, 5213–5219 (2015).
Barbier, F. et al. Infection-related ventilator-associated complications in ICU patients colonised with extended-spectrum β-lactamase-producing Enterobacteriaceae. Intensive Care Med. 44, 616–626 (2018).
Grasselli, G. et al. Gastrointestinal colonization with multidrug-resistant Gram-negative bacteria during extracorporeal membrane oxygenation: effect on the risk of subsequent infections and impact on patient outcome.Ann. Intensive Care 18, 141 (2019).
Nelson, R. E. et al. Methicillin-resistant Staphylococcus aureus colonization and pre- and post-hospital discharge infection risk. Clin. Infect. Dis. 68, 545–553 (2019).
Gmehlin, C. G. & Silvia Munoz-Price, L. Coronavirus disease 2019 (COVID-19) in long-term care facilities: A review of epidemiology, clinical presentations, and containment interventions.Infect. Control Hosp. Epidemiol. 43, 504–509 (2022).
Aleman, F. D. D. & Valenzano, D. R. Microbiome evolution during host aging. PLoS Pathog. 15, e1007727 (2019).
Xu, C., Zhu, H. & Qiu, P. Aging progression of human gut microbiota. BMC Microbiol. 19, 236 (2019).
Dréno, B. et al. Microbiome in healthy skin, update for dermatologists. J. Eur. Acad. Dermatol. Venereol. 30, 2038 (2016).
Prescott, S. L. et al. The skin microbiome: impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. World Allergy Organ. J. 10, 29 (2017).
Maguire, M. & Maguire, G. The role of microbiota, and probiotics and prebiotics in skin health. Arch. Dermatol Res 309, 411–421 (2017).
Willis, J. R. & Gabaldón, T. The human oral microbiome in health and disease: From sequences to ecosystems. Microorganisms 8, 308 (2020).
Huang, S. et al. Human skin, oral, and gut microbiomes predict chronological age. mSystems 5, e00630–19 (2020).
Zhou, W. et al. Host-specific evolutionary and transmission dynamics shape the functional diversification of Staphylococcus epidermidis in human skin. Cell 180, 454–470 (2020).
Oh, J. et al. Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64 (2014).
Oh, J., Byrd, A. L., Park, M., Kong, H. H. & Segre, J. A. Temporal stability of the human skin microbiome. Cell 165, 854–866 (2016).
Li, Z. et al. New insights into the skin microbial communities and skin aging. Front. Microbiol. 11, 565549 (2020).
Shibagaki, N. et al. Aging-related changes in the diversity of women’s skin microbiomes associated with oral bacteria.Sci. Rep. 7, 10567 (2017).
Roghmann, M. et al. Comparison of the microbiota of older adults living in nursing homes and the community. mSphere 2, 210 (2017).
Nagase, S. et al. Distinct skin microbiome and skin physiological functions between bedridden older patients and healthy people: A single-center study in Japan.Front. Med. 7, 101 (2020).
Ranjan, R., Rani, A., Metwally, A., McGee, H. S. & Perkins, D. L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 469, 967–977 (2016).
Brumfield, K. D., Huq, A., Colwell, R. R., Olds, J. L. & Leddy, M. B. Microbial resolution of whole genome shotgun and 16S amplicon metagenomic sequencing using publicly available NEON data. PLoS One 15, e0228899 (2020).
Fried, L. P. et al. Frailty in older adults: evidence for a phenotype. J. Gerontol. A Biol. Sci. Med. Sci. 56, 146 (2001).
Washburn, R. A., Smith, K. W., Jette, A. M. & Janney, C. A. The physical activity scale for the elderly (PASE): Development and evaluation. J. Clin. Epidemiol. 46, 153–162 (1993).
Hills, R. D. Gut microbiome: Profound implications for diet and disease.Nutrients 11, 1613 (2019).
Turnbaugh, P. J. et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1, 6ra14 (2009).
Peterson, J. et al. The NIH Human microbiome project. Genome Res. 19, 2317–2323 (2009).
Human Microbiome Project Consortium A framework for human microbiome research. Nature 486, 215–221 (2012).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Dickson, I. Stability and individuality of adult microbiota. Nature Milestones June, S11 (2019).
Oh, J. et al. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res. 23, 2103–2114 (2013).
Nagpal, R. et al. Gut microbiome and aging: Physiological and mechanistic insights. Nutr. healthy aging 4, 267–285 (2018).
Whelan, F. J. et al. The loss of topography in the microbial communities of the upper respiratory tract in the elderly. Ann. ATS 11, 513–521 (2014).
Kerns, M. L., et al. Fitzpatrick’s Dermatology. (McGraw-Hill Education, 2019).
Dréno, B. et al. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: a brief look at the latest updates. J. Eur. Acad. Dermatol. Venereol. 32, 5–14 (2018).
Claesen, J. et al. A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci. Transl. Med. 12, eaay5445 (2020).
Bolla, B. S. et al. Cutibacterium acnes regulates the epidermal barrier properties of HPV-KER human immortalized keratinocyte cultures. Sci. Rep. 10, 1–13 (2020).
Jasson, F. et al. Different strains of Propionibacterium acnes modulate differently the cutaneous innate immunity. Exp. Dermatol. 22, 587–592 (2013).
Otto, M. Staphylococcus epidermidis: the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7, 555 (2009).
Magne, F. The Firmicutes/Bacteroidetes ratio: A relevant marker of gut dysbiosis in obese patients?. Nutrients 12, 1474 (2020).
Castaner, O. et al. The gut microbiome profile in obesity: A systematic review. Int. J. Endocrinol. 2018, 4095789 (2018).
Wu, G. D. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011).
Park, S., Chung, H. & Lee, M. Clinical and microbiological characteristics of six staphylococcus pettenkoferi isolates from blood samples. Ann. Lab. Med. 35, 250 (2015).
Aubin, G. G. et al. Propionibacterium namnetense sp. nov., isolated from a human bone infection. Int J. Syst. Evol. Microbiol 66, 3393–3399 (2016).
Yan, Y., Nguyen, L. H., Franzosa, E. A. & Huttenhower, C. Strain-level epidemiology of microbial communities and the human microbiome. Genome Med. 12, 71 (2020).
Sonnenborn, U. Escherichia coli strain Nissle 1917—from bench to bedside and back: history of a special Escherichia coli strain with probiotic properties. FEMS Microbiol. Lett. 363, fnw212 (2016).
Lim, J. Y., Yoon, J. W. & Hovde, C. J. A brief overview of Escherichia coli O157:H7 and its plasmid O157. J. Microbiol. Biotechnol. 20, 5 (2010).
Conlan, S. et al. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 13, R64 (2012).
Mitchell, J. Streptococcus mitis: Walking the line between commensalism and pathogenesis. Mol. Oral. Microbiol. 26, 89–98 (2011).
Brinkac, L., Voorhies, A., Gomez, A. & Nelson, K. E. The threat of antimicrobial resistance on the human microbiome. Microb. Ecol. 74, 1001 (2017).
Keller, R., Pedroso, M. Z., Ritchmann, R. & Silva, R. M. Occurrence of virulence-associated properties in Enterobacter cloacae. Infect. Immun. 66, 645–649 (1998).
Yamamoto, S. & Shinoda, S. [Iron uptake mechanisms of pathogenic bacteria].Nihon Saikingaku Zasshi 51, 523–547 (1996).
Saffrey, M. & Saffrey, M. Aging of the mammalian gastrointestinal tract: a complex organ system. AGE 36, 1019–1032 (2014).
Sovran, B. et al. Age-associated impairment of the mucus barrier function is associated with profound changes in microbiota and immunity. Sci. Rep. 9, 1–13 (2019).
Muller, C. et al. The intraperitoneal transcriptome of the opportunistic pathogen Enterococcus faecalis in Mice. PLoS One 10, e0126143 (2015).
Vrancianu, C. O., Popa, L. I., Bleotu, C. & Chifiriuc, M. C. Targeting plasmids to limit acquisition and transmission of antimicrobial resistance. Front Microbiol 11, 761 (2020).
Orlek, A. et al. Plasmid classification in an era of whole-genome sequencing: Application in studies of antibiotic resistance epidemiology. Front. Microbiol. 8, 182 (2017).
Kao, K. et al. Risk factors of methicillin-resistant Staphylococcus aureus infection and correlation with nasal colonization based on molecular genotyping in medical intensive care units: A prospective observational study. Medicine (Baltimore) 94, e1100 (2015).
Sender, R., Fuchs, S. & Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016).
Flowers, L. & Grice, E. A. The skin microbiota: Balancing risk and reward. Cell Host Microbe 28, 190–200 (2020).
Plewig, G. & Kligman, A. M. Proliferative activity of the sebaceous glands of the aged.J. Invest. Dermatol. 70, 314–317 (1978).
Luna, P. C. Skin microbiome as years go by. Am. J. Clin. Dermatol. 21, 12–17 (2020).
Rockwood, K., Theou, O. & Mitnitski, A. What are frailty instruments for? Age Ageing 44, 545–547 (2015).
Wade, W. G. The oral microbiome in health and disease. Pharmacol. Res. 69, 137–143 (2013).
McInnes, P. & Cutting, M. Human Microbiome Project: Core Microbiome Sampling Protocol A HMP Protocol # 07-001 (2010). https://www.hmpdacc.org/hmp/doc/HMP_MOP_Version12_0_072910.pdf
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357 (2012).
Zhou, W. et al. ReprDB and panDB: minimalist databases with maximal microbial representation. Microbiome 6, 15 (2018).
Segata, N. et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 9, 811–814 (2012).
Kostic, A. D. et al. PathSeq: A comprehensive computational tool for the identification or discovery of microorganisms by deep sequencing of human tissue. Nat. Biotechnol. 29, 393–396 (2011).
Morris, E. K. et al. Choosing and using diversity indices: Insights for ecological applications from the German Biodiversity Exploratories. Ecol. Evol. 4, 3514–3524 (2014).
Yue, J. C. & Clayton, M. K. A similarity measure based on species proportions. Commun. Stat. Theory Methods 34, 2123–2131 (2005).
Liaw, A. & Weiner, M. Classification and Regression by randomForest.R News 2, 18–22 (2002).
Emiola, A., Zhou, W. & Oh, J. Metagenomic growth rate inferences of strains in situ. Sci. Adv. 6, eaaz2299 (2020).
O’Leary, N. A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, 733 (2016).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016).
Treangen, T. J., Ondov, B. D., Koren, S. & Phillippy, A. M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15, 524 (2014).
Hong, C. et al. PathoScope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome 2, 33 (2014).
Chen, L. et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, 325 (2005).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2014).
Franzosa, E. A. et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 15, 962–968 (2018).
Li, D., Liu, C., Luo, R., Sadakane, K. & Lam, T. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Krawczyk, P. S., Lipinski, L. & Dziembowski, A. PlasFlow: predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acids Res. 46, e35 (2018).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 11, 119 (2010).
Arango-Argoty, G. et al. DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 6, 1–15 (2018).
R Core Team. R: A language and environment for statistical computing. 3.4.0 (2017). https://www.R-project.org/
Acknowledgements
Funding for this project were provided by internal UConn support via the UConn Research Excellence Program and the UConn Microbiome Research Seed Grant. Investigator salaries were additionally supported by the National Institute on Aging (R56 AG060746 and P30 AG067988) and Claude D. Pepper Older Americans Independence Center at UConn. JO is additionally supported by the National Institutes of Health (DP2 GM126893-01, K22 AI119231-01, 1U54NS105539, 1 U19 AI142733 and 1 R21 AR075174), the National Science Foundation (1853071), the American Cancer Society, the Leo Foundation and the Mackenzie Foundation.
Author information
Authors and Affiliations
Contributions
Conceptualization: P.J.L., G.A.K., J.T.R. and J.O. Methodology: J.O., J.T.R. and G.A.K. Software: P.J.L., W.Z. and J.O. Validation: P.J.L., W.Z. and J.O. Formal analysis: P.J.L. and W.Z. Investigation: P.J.L., A.S., S.D., A.Y.V., E.F. and W.Z. Resources: G.A.K., J.T.R., J.O. Data Curation: J.T.R., A.S., S.D., W.Z. and J.O. Writing – Original Draft: P.J.L. and W.Z. Writing – Review & Editing: J.O., GK, J.T.R., A.S., S.D., A.Y.V. and W.Z. Visualization: P.J.L. and W.Z. Supervision: G.A.K., J.T.R. and J.O. Project Administration: J.T.R. and J.O. Funding acquisition: G.A.K., J.T.R., JG, O.K.C. and J.O.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Falk Hildebrand and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information:
Supplementary figure and table legends and supplementary Figs.1–23.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Larson, P.J., Zhou, W., Santiago, A. et al. Associations of the skin, oral and gut microbiome with aging, frailty and infection risk reservoirs in older adults. Nat Aging 2, 941–955 (2022). https://doi.org/10.1038/s43587-022-00287-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43587-022-00287-9
This article is cited by
-
Smart Textiles with Living Interfaces: Microbiome–Electronics Integration for Advanced Skin Health Management
Advanced Fiber Materials (2026)
-
Chronological age estimation from human microbiomes with transformer-based Robust Principal Component Analysis
Communications Biology (2025)
-
Exposure to ileal feces with frailty-associated dysbiosis elevates gastrointestinal complication risk after intracorporeal urinary diversion
Scientific Reports (2025)
-
Succinate modulates oral dysbiosis and inflammation through a succinate receptor 1 dependent mechanism in aged mice
International Journal of Oral Science (2025)
-
The human skin microbiome: from metagenomes to therapeutics
Nature Reviews Microbiology (2025)