Abstract
Background
As COVID-19 becomes endemic and vaccines are annually adapted, exposure intervals and immune imprinting become critical considerations for vaccination strategy. Imprinting by the ancestral spike protein affected bivalent Wuhan-Hu-1/BA.4-5 vaccine responses. We assess the persistence of imprinting in antibody responses to the more recent XBB.1.5 monovalent formulation.
Methods
We quantified live virus-neutralizing antibodies by focus reduction neutralization test and ancestral spike receptor-binding isotype titers by immunosorbent assay in individuals before and after XBB.1.5 vaccination. We compared responses between those who previously received three to four doses of Wuhan-Hu-1 vaccine and one dose of bivalent Wuhan-Hu-1/BA.4-5 (bivalent recipients) and those who received three to four doses of Wuhan-Hu-1 (bivalent non-recipients).
Results
We report that before XBB.1.5 vaccination, bivalent non-recipients have decreased breadth and potency of neutralization. At post-vaccination, non-recipients exhibit greater boosting of neutralizing antibodies against XBB.1.5 (18.4X versus 6.2X), EG.5.1 (30.9X versus 7.0X), and JN.1 (9.2X versus 3.7X) variants with comparable breadth and trends toward greater potency. Greater boosting in non-recipients is similarly observed for spike-binding IgA and total IgG/A/M but not IgG nor IgM. Bivalent non-recipients had longer intervals between vaccination, which may enhance antibody responses; however, bivalent receipt and interval are tightly linked, preventing isolation of individual contributions to boosting. Nonetheless, back-boosting of ancestral SARS-CoV-2 titers in both participant groups provides interval-independent evidence that imprinting persists.
Conclusions
Our findings indicate that immune imprinting continues to affect humoral immunity elicited by the XBB.1.5 vaccine. Both imprinting and exposure intervals are important phenomena underlying immunogenicity of future variant-adapted COVID-19 vaccines.
Plain language summary
COVID-19 vaccines help us produce antibodies, which are protective proteins that can recognize and neutralize viruses. However, the immune system tends to strongly remember the first version of the virus it saw, which can limit how well it responds to new variants—a phenomenon called “immune imprinting.” We studied how people responded to a COVID-19 vaccine that targeted a newer variant, XBB.1.5. We found that it still boosted antibodies against the original virus, showing that imprinting remains a concern. People with fewer previous vaccines that targeted older variants responded better, possibly because of less imprinting or longer time since their last dose. Our findings suggest that vaccine history and timing between doses affect how well updated COVID-19 vaccines work.
Similar content being viewed by others
Introduction
The World Health Organization (WHO) declared the end of COVID-19 as a public health emergency in May 20231, but SARS-CoV-2 variants continue to emerge and spread2. To maintain population immunity, the XBB.1.5 monovalent vaccine was subsequently approved in 2023 and was shown to successfully induce cellular responses and neutralizing antibodies against circulating variants3,4,5,6. The JN.1-adapted vaccines were further formulated to match prevalent strains in 20247. However, the diversity and potency of new immune responses generated through updated COVID-19 vaccines may be influenced by immune imprinting following exposure to historic spike proteins during prior vaccination or natural infection8.
Immune imprinting has been extensively explored for influenza and is characterized by reduced induction of neutralizing antibodies against novel variants of a viral antigen upon exposure. Back-boosting is also highly characteristic of imprinting and describes boosting of titers against ancestral antigen upon exposure to a novel variant8. Recognition of immune imprinting in the context of COVID-19 initially arose with observations that infection resulted in back-boosting of binding antibodies against seasonal coronaviruses OC43 and HKU19. Ancestral Wuhan-Hu-1 infection prior to Omicron BA.1 infection resulted in poorer Omicron-targeting antibody responses compared to infection with BA.1 alone10. Similarly, vaccination with monovalent Wuhan-Hu-1 prior to infection with Alpha or Delta variants generated weaker variant-specific responses than in unvaccinated, infected individuals11. As the first widely administered variant-adapted vaccine, the bivalent Wuhan-Hu-1/BA.4-5 vaccine failed to significantly enhance Omicron-neutralizing antibodies compared to monovalent Wuhan-Hu-1 boosting, likely due to imprinting by the ancestral spike protein12,13,14. Consistent with this observation, minimal additional protection was provided against COVID-19 infection and hospitalization in bivalent versus Wuhan-Hu-1 monovalent recipients15. More recently, studies have demonstrated that targeting immune responses to ancestral SARS-CoV-2 by means of additional booster doses increases susceptibility to reinfection with emerging Omicron variants16. Though these reinfection study designs may be confounded by selection bias17, reports assessing XBB.1.5 vaccine-induced pseudovirus neutralizing antibodies indeed observe the back-boosting of responses against historic variants that is characteristic of immune imprinting5,8.
To further evaluate the impact of immune imprinting on XBB.1.5 vaccine immunogenicity, we identified a group of individuals who did not previously receive the bivalent vaccine in 2022 but did receive the XBB.1.5 monovalent vaccine in 2023 (bivalent non-recipients). Between bivalent non-recipients and participants who received the bivalent vaccine prior to XBB.1.5 vaccination (bivalent recipients), we compared serum neutralizing antibody titers that effectively block cellular infection by live clinical isolates of SARS-CoV-2 representing ancestral (WA1), vaccine-matched (XBB.1.5), and emergent variants (EG.5.1 and JN.1) and measured IgG, IgA, IgM, and total IgG/A/M isotypes targeting the ancestral spike receptor-binding domain (RBD).
Across both groups, the XBB.1.5 vaccine successfully boosts neutralizing antibodies against XBB.1.5, EG.5.1, and JN.1. As evidence of persistent immune imprinting, XBB.1.5 vaccination back-boosts antibodies against WA1 as well as IgG, IgA, and total IgG/A/M isotypes targeting the ancestral spike RBD. Also indicative of imprinting, we observe greater boosting of antibodies against XBB.1.5, EG.5.1, and JN.1 as well as IgA and total IgG/A/M in bivalent non-recipients. Non-recipients had a longer rest interval since prior vaccination, which has been shown to further enhance antibody responses18,19,20; however, collinearity between bivalent receipt and interval prevents the isolation of individual contributions to greater non-recipient boosting in multiple predictors regression modeling. Thus, we present evidence that immune imprinting persists, and that imprinting and exposure interval together are likely critical variables determining antibody responses to the XBB.1.5 vaccine and future variant-adapted formulations.
Materials and methods
Study design
The purpose of this study was to compare humoral immune responses to the XBB.1.5 monovalent vaccine in individuals who received all United States Center for Disease Control and Prevention (CDC)-recommended COVID-19 vaccines since the beginning of the pandemic versus those who were missing just the bivalent Wuhan-Hu-1/BA.4-5 vaccine in 2022. Serum samples were collected from participants before and after XBB.1.5 vaccination, which were assessed using ELISAs and FRNTs. Bivalent non-recipients were selected for inclusion based on having received all prior CDC-recommended doses except for the bivalent vaccine. Bivalent recipient controls were selected on the basis of prior vaccine history, sex, age, and days since receiving the bivalent vaccine in order to closely match demographics in the non-recipient group. The number of prior infections, suspected and confirmed, were similar across both groups. All participant samples were tested for anti-nucleocapsid antibodies by ELISA to screen for recent infection and excluded from analysis if deemed positive.
Cohort selection and serum collection
Healthcare workers at Oregon Health and Science University (OHSU) were recruited and enrolled in the study. Written informed consent was obtained at the time of enrollment. Study approval was obtained from the OHSU Institutional Review Board (IRB). This study was carried out in compliance with all relevant ethical regulations as outlined by the IRB. Bivalent recipients were fully vaccinated, defined as having received 3 or 4 doses of ancestral Wuhan-Hu-1 vaccine and 1 dose of bivalent Wuhan-Hu-1/BA.4-5 vaccine. Bivalent non-recipients were defined as missing the bivalent vaccine but otherwise fully vaccinated. At the time of enrollment, each participant contributed 4–6 mL of whole blood, which was then centrifuged at 1000 x g, subjected to heat inactivation at 56 °C for 30 minutes, and subsequently stored at −20 °C to serve as a baseline (pre-boost) serum sample. Roughly three weeks following the administration of the XBB.1.5-adapted monovalent vaccine (Moderna Therapeutics), additional whole blood samples were collected and processed in the same manner to obtain post-boost serum samples.
Cell Culture
Vero E6 monkey kidney epithelial cells (ATCC, CRL-1586) were authenticated by the manufacturer and maintained in tissue culture-treated plates in Dulbecco’s modified Eagle’s medium (DMEM), 10% fetal bovine serum (FBS), 1% nonessential amino acids (NEAAs), and 1% penicillin-streptomycin (PS) at 37 C, 5% CO2, 100% relative humidity. Mycoplasma testing was performed routinely using PCR-based detection kit (Applied Biological Materials Inc., G238).
SARS-CoV-2 Growth and Titration
SARS-CoV-2 clinical isolates were obtained from BEI Resources: WA1, USAWA1/2020 (NR-52281); XBB.1.5, USA/MD-HP40900/2022 (NR-59104); EG.5.1, USA/MD-HP47946/2023 (NR-59503); and JN.1, USA/New York/PV96109/2023 (NR-59693). To generate Passage 1 viruses, ~70% confluent Vero E6 cells were inoculated and cultured until cytopathic effect appeared—72 hours for WA1 and 96 hours for XBB.1.5, EG.5.1, and JN.1. The culture supernatants were collected, centrifuged at 1000 x g for 10 minutes, and then stored in aliquots at −80 °C. Passage 2 viruses were produced in a similar manner using Passage 1 viruses for inoculation.
Virus titers were established via a focus-forming assay with 10-fold dilutions in dilution medium (2% FBS, 1% NEAA, and 1% P/S in Opti-MEM). Confluent Vero E6 cells in 96-well plates were incubated with virus dilutions for one hour at 37 °C in 5% CO2. Overlay medium (1% methylcellulose in dilution medium) was then added, and plates were incubated for 24 hours for WA1 and 48 hours for the other strains. Following this, the overlay medium was removed, and cells were fixed using 4% formaldehyde in PBS for one hour at RT. To stain infection foci, cells were permeabilized in perm buffer (0.1% BSA and 0.1% saponin in PBS) for 30 minutes at RT and then incubated with polyclonal anti-SARS-CoV-2 alpaca serum diluted at 1:5000 for WA1 and 1:500 for XBB.1.5, EG.5.1, and JN.1 overnight at 4 °C. After washing three times with wash buffer (0.01% Tween-20 in PBS) cells were treated with HRP-conjugated anti-alpaca IgG (Novus #NB7242) diluted at 1:20,000 for WA1 and 1:10,000 for the other strains for two hours at RT. Following another three washes, 50 µL of KPL TrueBlue substrate (Seracare #5510-0030) was added per well. After 20 minutes of incubation at room temperature, plates were imaged using a CTL Immunospot Analyzer. Foci were then counted with the Viridot package for R 4.3.121.
Enzyme-linked immunosorbent assay (ELISA)
ELISAs for each Ig isotype were conducted in technical duplicates in 96-well plates (Maxisorp #423501). Each plate was coated overnight at 4 °C with 100 µL per well of purified wild-type SARS-CoV-2 RBD at 1 µg/mL in PBS, with gentle rocking. The plates were washed three times with wash buffer (0.05% Tween-20 in PBS), then incubated with 150 µL per well of blocking buffer (5% nonfat dry milk powder and 0.05% Tween-20 in PBS) for 1 hour at RT, rocking. Participant serum samples were diluted in blocking buffer to create five 4-fold serial dilutions (from 1:25 to 1:6,400). Plates were incubated with serum dilutions for one hour at RT, rocking, followed by three washes. HRP-conjugated secondary antibodies against human IgG (BD Biosciences #555788), IgA (BioLegend #411002), and IgM (Bethyl Laboratories #A80-100P) were used at a 1:3,000 dilution in blocking buffer, and 100 µL was added per well. For total antibody detection, HRP-conjugated goat anti-human IgG/A/M (Invitrogen, #A18847) was used at a 1:10,000 dilution. After 1 hour at RT, rocking, and three washes, o-phenylenediamine dihydrochloride (ThermoFisher Scientific, #34005) was added as per the manufacturer’s instructions. The reaction was halted after 25 minutes with an equal volume of 1 M HCl, and the optical density (OD) was measured at 492 nm on a CLARIOstar plate reader. OD492 values were normalized by subtracting the mean of the negative control wells. The protocol for detecting anti-nucleocapsid antibodies was identical, except the plates were coated with 1 µg/mL nucleocapsid protein (SinoBio #40588-V08B) and detected using HRP goat anti-human IgG/A/M. Serum was deemed positive for anti-nucleocapsid antibodies if the calculated EC50 exceeded the lower limit of detection of 25.
SARS-CoV-2 Focus Reduction Neutralization Test (FRNT)
Participant serum samples were prepared in technical duplicate, with five 10-fold serial dilutions for WA1 (1:10 to 1:100,000) and five 5-fold serial dilutions for XBB.1.5, EG.5.1, and JN.1 (from 1:10 to 1:6250)22. These dilutions were mixed with an equal volume of 2X concentrated virus and incubated for 1 hour at RT. The mixtures were then added to Vero E6 cells in 96-well plates and incubated at 37 °C with 5% CO2 for 1 hour, followed by a further incubation with overlay media for 24 hours for WA1 and 48 hours for XBB.1.5, EG.5.1, and JN.1. Cells were then fixed with 4% formaldehyde in PBS for one hour at room temperature. All subsequent staining, developing, imaging, and counting followed methods used in prior titration experiments. For FRNT, approximately 30 focus-forming units (FFU) of virus per well were used, and for each set of five test wells, a sixth control well without serum was included. Percent neutralization for each test well was calculated by comparing the focus count to the average in the control wells.
ELISA EC50 and FRNT50 calculations
ELISA OD492 readings and FRNT percent neutralization metrics were analyzed using Python (v3.7.6) with the data analysis libraries numpy (v1.18.1), scipy (v1.4.1), Matplotlib (v3.1.3), and pandas (v1.0.1) to compute ELISA EC50 and FRNT50 values. Data from technical replicates were combined and fit using a three-parameter logistic model. Replicate curves for both ELISA EC50 and FRNT50 were generated separately for quality control. If the final ELISA EC50 values were below the quantification limit of 25, they were adjusted to 24; values exceeding the upper limit of 6400 were adjusted to 6401. Similarly, final FRNT50 values below the quantification threshold of 20 were adjusted to 19, and values surpassing the upper limits of 200,000 for WA1 and 12,500 for XBB.1.5, EG.5.1, and JN.1 were set to 200,001 and 12,501, respectively.
Antigenic Cartography
Using the Racmacs package (v1.2.9, https://acorg.github.io/Racmacs/) for R (v.4.3.1), antigenic distances between serum samples and SARS-CoV-2 strains were calculated from FRNT50 neutralizing titer values and visualized23. For 2-dimensional mapping of serum and variants for each study group (bivalent recipients or non-recipients) and at each timepoint (pre- and post-vaccination), optimization was set to 2000 steps, and “minimum column basis” was set to “none.” Manual seeds were set for each map to orient WA1 to the left side of each plot. Map distances between WA1 and each variant were extracted with the dist(ptCoords()) function.
Statistics and reproducibility
For repeated measures comparisons involving antibody isotype titers (EC50), a restricted effect maximum likelihood (REML) model with Holm-Sidak multiple comparisons correction was performed on the log-transformed data. For similar comparisons involving neutralizing antibody titers (FRNT50), a repeated measures one-way ANOVA with Holm-Sidak multiple comparisons correction was performed on log-transformed data. All other multi-group statistics were assessed by standard one-way ANOVA with follow-up “tests for linear trend” (a.k.a. “test for linear contrast,” “post-test for trend”), which assesses systematic increases or decreases in column means in left-to-right column order. Single pairwise comparisons of absolute titers or fold changes between groups were the result of two-tailed, unpaired t tests. Multiple linear regression modeling was performed on log-transformed fold-change (post/pre-vaccination titer) data with bivalent receipt and vaccine interval as explanatory variables. Outliers were identified and removed by Robust Regression and Outlier Removal (ROUT) with Q = 0.1%. P-values less than 0.05 were considered statistically significant. All statistical analyses were performed in GraphPad Prism v10. Each serum sample was assessed in technical duplicate serial dilutions for each isotype or against each variant. These data were combined to generate single EC50 or FRNT50 values, as described in detail above. Statistical results (e.g., geometric means, confidence intervals, p-values) were then derived from biological replicates of antibody titer (EC50s or FRNT50s) within each participant group: n = 30 individual participant/serum samples for the bivalent recipient group and n = 10 for the non-recipient group.
Results
Cohort and study design
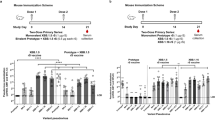
Forty-nine participants were initially recruited; however, serum samples were analyzed for anti-nucleocapsid seropositivity by enzyme-linked immunosorbent assay (ELISA) to screen for recent prior infection. Nine seropositive participants were excluded from the final cohort. Within the final cohort of 40 participants (Table 1), 30 received three or four doses of monovalent Wuhan-Hu-1 followed by one dose of bivalent Wuhan-Hu-1/BA.4-5 vaccine (bivalent recipients), and 10 received three or four doses of monovalent Wuhan-Hu-1 vaccine (bivalent non-recipients) (Fig. 1a). At time of recruitment, all participants received the XBB.1.5 monovalent mRNA vaccine manufactured by Moderna Therapeutics. Suspected and rapid antigen-confirmed infection history was collected from participants. Serum samples were collected from each participant prior to XBB.1.5 vaccination and again at an average of 22 days post-vaccination. These were tested for 50% effective antibody IgG, IgA, IgM, and total IgG/A/M isotype concentrations (EC50) by ELISA against the ancestral spike RBD as well as 50% live SARS-CoV-2 neutralizing titers by focus reduction neutralization tests (FRNT50) against the ancestral WA1, vaccine-matched XBB.1.5, and emergent EG.5.1 and JN.1 strains.
a Pre- and post-XBB.1.5 vaccination sera were collected from healthcare workers who previously received the bivalent vaccine or not. Live SARS-CoV-2 neutralization by serum antibodies was assessed by focus reduction neutralization test (FRNT) and reported as FRNT50s for bivalent recipients (b) and non-recipients (c). Serum antibody isotype titers against ancestral spike RBD were determined by enzyme-linked immunosorbent assay (ELISA) and reported as EC50 for bivalent recipients (d) and non-recipients (e). The dotted lines indicate assay lower limits of detection. Geometric mean titers (GMT) for each bar were calculated in GraphPad Prism. Fold changes were calculated by dividing the post-XBB.1.5 vaccination titer by pre-vaccination titer. Reported p-values are the result of a one-way repeated measures ANOVA (b, c) or restricted maximum likelihood model (d, e) with Holm-Sidak multiple comparisons correction for pairwise comparisons between variant-specific and isotype titers within the bivalent recipient (n = 30) or non-recipient (n = 10) groups.
XBB.1.5 monovalent vaccination boosts antibody titers
In both bivalent recipient and non-recipient groups, the XBB.1.5 monovalent vaccine successfully boosts neutralizing titers against XBB.1.5 (recipient p < 0.0001, non-recipient p = 0.0008), EG.5.1 (recipient p < 0.0001, non-recipient p < 0.0001), and JN.1 (recipient p < 0.0001, non-recipient p = 0.0013) and back-boosts titers against the ancestral WA1 strain (recipient p < 0.0001, non-recipient p = 0.0490). Of note, the JN.1 variant, from which currently dominant strains emerged, demonstrates significant escape from vaccinated sera with neutralizing titers that are 4-fold lower than against the vaccine-matched XBB.1.5, regardless of vaccine history (recipient p = 0.0002, non-recipient p = 0.0490) (Fig. 1b, c). In both groups, the XBB.1.5 vaccine similarly back-boosts IgG (recipient p = 0.0003, non-recipient p = 0.0199), IgA (recipient p = 0.0056, non-recipient p = 0.0286), and total IgG/A/M (recipient p = 0.0127, non-recipient p = 0.0123) isotype antibodies targeting the ancestral spike RBD, but does not significantly boost RBD-specific IgM (recipient p = 0.1457, non-recipient p = 0.0731) (Figs. 1d, e).
Bivalent non-recipients exhibit diminished neutralizing breadth and potency prior to XBB.1.5 vaccination
To visually assess breadth of neutralizing antibody responses, neutralizing titers against the XBB.1.5, EG.5.1, and JN.1 variants were plotted against ancestral WA1 titers. 95% confidence ellipses were drawn to delineate the distribution of each participant group. Antibodies in both groups demonstrate a strong preference toward neutralizing WA1 over variants at pre-vaccination, but these cross-neutralizing responses are improved by XBB.1.5 vaccination as denoted by the upward shift toward increased variant-specific titers (Fig. 2a–f).
For each participant, pre- and post-vaccination neutralizing titers against live, clinical isolates of XBB.1.5 (a, d), EG.5.1 (b, e), or JN.1 (c, f) variants were plotted against neutralizing titers against WA1. The black dotted line represents equal neutralization, and the dotted ellipses represent 95% confidence ellipses. Relative neutralization ratios were also calculated by dividing neutralizing titers against each variant by those against WA1. g Antigenic cartographs were generated using the Racmacs packages in R v4.3.1 for bivalent recipients and non-recipients at each timepoint. Arrows and corresponding values represent quantified map distances between WA1 and variants. Each square-length represents ~2-fold changes in FRNT50 neutralizing titer. Gray triangles represent positions for each serum sample. h Neutralizing potency index (NPI) for pre-XBB.1.5 vaccination (pre) serum samples were calculated by dividing the FRNT50 against WA1, XBB.1.5, EG.5.1, or JN.1 by total IgG/A/M EC50 for each participant. For all bar plots, the plotted geometric mean values are displayed. Error bars are geometric means with 95% confidence intervals. Reported p-values are the result of unpaired t tests between bivalent recipients (n = 30) and non-recipients (n = 10).
In comparing groups at the pre-vaccination timepoint, bivalent non-recipients have a slight skew toward WA1 neutralization relative to recipients. To quantitate these visual observations, relative neutralization ratios were calculated by dividing the neutralizing titer against a given variant by WA1 titers. At pre-vaccination, bivalent non-recipients have significantly lower relative neutralization ratios for XBB.1.5 (p = 0.0305) and EG.5.1 (p = 0.0111) compared to recipients, possibly due to absence of prior vaccine exposure to the BA.4/5 spike variant which would enhance Omicron cross-neutralizing responses. The trend for JN.1 is similar but not statistically significant (p = 0.1935) (Figs. 2a–c). However, after XBB.1.5 vaccination, the bivalent non-recipient group clusters to the upper, right distribution of the total cohort, suggesting a trend toward improved cross-neutralization in addition to overall potency of neutralization. However, this enhancement in cross-neutralization is not significant upon comparing relative neutralization ratios (Figs. 2d–f).
Antigenic cartography was performed to further evaluate variant cross-neutralization. Here, neutralizing titer values against each SARS-CoV-2 strain for each participant serum sample were used to generate antigenic maps. One map was generated for each group (bivalent recipient and non-recipient) and at each time point (pre- and post-vaccination). One square-length map distance represents a ~2-fold change in FRNT50 neutralizing titer and is a measure of antigenic difference. Across all maps, XBB.1.5 and EG.5.1 are positioned closest together and appear most antigenically similar from the perspective of serum titers; this is consistent with the fact that EG.5.1 is a closely related descendent of the XBB.1.5 lineage while WA1 and JN.1 are phylogenetically more distant24. In both groups, map distances between WA1 and variants are shorter at post-vaccination compared to pre-vaccination, demonstrating successful boosting of cross-neutralizing responses by the XBB.1.5 monovalent vaccine (Fig. 2g).
Unsurprisingly, bivalent non-recipients exhibit longer map distances than recipients at pre-vaccination between WA1 and XBB.1.5 (6.4 versus 5.2), EG.5.1 (7.2 versus 5.6), and JN.1 (6.8 versus 5.8), suggesting diminished cross-neutralization from lack of prior vaccination with the BA.4/5 spike variant. At post-vaccination, however, bivalent non-recipients have slightly shorter but overall similar map distances between WA1 and XBB.1.5 (3.6 versus 3.7), EG.5.1 (3.6 versus 3.9), and JN.1 (4.9 versus 5.0) (Fig. 2g). These analyses suggest that despite reduced pre-vaccination cross-neutralization due to lack of the bivalent vaccine, non-recipients recover comparable cross-neutralization to bivalent recipients upon XBB.1.5 vaccination.
Finally, neutralizing potency of antibody responses at pre-vaccination was measured by assessing the neutralizing potency index (NPI), which is calculated by dividing the serum neutralizing titer for a given variant by the total IgG/A/M titer. Biologically, each variant-specific NPI quantifies the subset of spike RBD-binding antibodies that lead to neutralization of infection by the assessed variant. The neutralizing potency of serum antibodies are significantly lower against XBB.1.5 (p = 0.0432), EG.5.1 (p = 0.0195), and JN.1 (p = 0.0371) but not WA1 (p = 0.4560) in bivalent non-recipients prior to XBB.1.5 vaccination. As with significantly diminished variant cross-neutralization, reduced neutralizing potency is likely the result of lacking exposure to the BA.4/5 spike variant and highlights the importance of bivalent vaccination in enhancing Omicron-targeting responses (Fig. 2h).
XBB.1.5 vaccine elicits greater boosting of variant-neutralizing antibodies in bivalent non-recipients
Pre-vaccination neutralizing titers against the contemporary variants trend lower in bivalent non-recipients against XBB.1.5 (1.6-fold; p = 0.3107), EG.5.1 (2-fold; p = 0.1040), and JN.1 (1.4-fold; p = 0.3792), consistent with greater waning of antibody responses in this group due to the missing bivalent vaccine and longer time interval since prior vaccination. Strikingly, however, after final vaccination, trends toward higher absolute titers are observed in the bivalent non-recipient group against WA1 (1.7-fold; p = 0.1158), XBB.1.5 (1.9-fold; p = 0.3188), EG.5.1 (2.2-fold; p = 0.1773), JN.1 (1.8-fold; p = 0.1500), suggesting an anchoring effect due to immune imprinting in bivalent recipients (Figs. 1b, 1c). Although these differences in post-vaccination absolute titers between bivalent recipients and non-recipients do not reach statistical significance, boosting of variant-specific neutralizing antibodies (i.e. fold-change after vaccination) is significantly greater in non-recipients against XBB.1.5 (18.4X versus 6.2X, p = 0.0134), EG.5.1 (30.9X versus 7.0X, p = 0.0027), and JN.1 (9.2X versus 3.7X, p = 0.0389). However, no significant difference is observed against WA1 (2.7X versus 2.2X, p = 0.5910) (Fig. 3a–d).
Live SARS-CoV-2 neutralizing antibodies were quantified by focus reduction neutralization test (FRNT) and reported as FRNT50s. Fold changes were calculated by dividing the post-XBB.1.5 vaccination titer by pre-vaccination titer for each participant against WA1 (a), XBB.1.5 (b), EG.5.1 (c), and JN.1 (d). Ancestral spike RBD-binding antibodies were determined by enzyme-linked immunosorbent assay (ELISA) and reported as EC50s. Fold-changes were calculated for IgG (e), IgA (f), IgM (g), and total IgG/A/M (h). The dotted lines indicate no change in titer from pre- to post-vaccination (fold change = 1). Error bars show geometric means with 95% confidence intervals. Reported p-values are the result of unpaired t tests between bivalent recipients (n = 30) and non-recipients (n = 10).
The bivalent recipient group was further stratified based upon having received either three or four doses of ancestral monovalent vaccine prior to the bivalent vaccine. Using an ANOVA follow-up test called “test for linear trend” (a.k.a. “test for linear contrast,” “post-test for trend”), which statistically assesses systematic decreases in left-to-right column order, these recipient subgroups were then compared to the bivalent non-recipient group in order to assess the effect of ancestral spike vaccinations on XBB.1.5 vaccine immunogenicity. This reveals a generally inverse relationship between number of ancestral spike vaccinations and absolute post-titer that approaches significance for XBB.1.5 (p = 0.0803) and EG.5.1 (p = 0.0504) while boosting has similar and statistically significant relationships for XBB.1.5 (p = 0.0124) and EG.5.1 (p = 0.0030). Within the bivalent recipient group, however, having 3 or 4 doses of ancestral spike results in similar responses against JN.1 and thus lack of linear trend in terms of absolute post-titer (p = 0.1875) and boosting (p = 0.1219), suggesting that vaccine immunogenicity against JN.1 and strains emerging from this distinct lineage may suffer less from ancestral spike imprinting. While the number of ancestral vaccinations has an impact on absolute post-titers against WA1 (p = 0.0323), no difference is observed in boosting (p = 0.6491), likely due to relatively high baseline neutralizing antibody titers from repetitive vaccine- and infection-mediated back-boosting across all participant groups (Supplementary Fig. 1).
XBB.1.5 vaccine elicits greater boosting and absolute ancestral spike-binding isotype titers in bivalent non-recipients
Pre-vaccination titers trend higher for bivalent non-recipients despite longer interval since prior vaccination for IgG (1.7-fold; p = 0.0926), IgA (1.7-fold; p = 0.1283), and total IgG/A/M (2.2-fold; p = 0.0236) (Fig. 1d, e). Our assay was optimized to detect antibody isotypes targeting the ancestral spike RBD and, though unexpected, these higher pre-vaccination titers in the non-recipient group are validated by similarly elevated neutralizing titers against the ancestral WA1 strain (Fig. 1b, c, Supplementary Fig. 1a). These increased pre-vaccination responses against ancestral antigen in bivalent non-recipients may be explained by natural infection histories skewed toward older strains. In other words, non-recipients opted out of the bivalent vaccine and were thus more susceptible to infection by early or pre-Omicron lineages that circulated in 2022. Infection by these lineages would more likely maintain responses against older spike variants, whereas an infection history more dominated by post-XBB Omicron would more drastically draw responses away from ancestral antigen. However, it is important to note that both bivalent recipient and non-recipient groups have similar overall numbers of prior infection.
At post-vaccination, absolute titers are significantly higher in the bivalent non-recipient group for IgG (2.7-fold; p = 0.0098), IgA (3.6-fold; p = .0089), and total IgG/A/M (5.1-fold; p = 0.0004) (Fig. 1d, e). Furthermore, more profound boosting of isotypes is observed in bivalent non-recipients than in recipients for IgA (3.1X versus 1.5X, p = 0.0131) and total IgG/A/M (3.5X versus 1.6X, p = 0.0130), though weaker and not statistically significant for IgG (3.2X versus 2.0X, p = 0.1422) and indifferent for IgM (1.5X versus 1.4X, p = 0.8288) (Fig. 3e–h).
When stratifying study participants by number of ancestral Wuhan-Hu-1 doses received prior to bivalent vaccination and performing ANOVA follow-up column “tests for linear trend,” significant inverse relationships are found with post-vaccination titers for IgG (p = 0.0222), IgA (p = 0.0003), and total IgG/A/M (p = 0.0014) as well as for boosting of IgA (p = 0.0154). This relationship approaches statistical significance for boosting of total IgG/A/M (p = 0.0799). No relationship is observed for absolute IgM post-titer (p = 0.4371) nor boosting of IgG (p = 0.6057) and IgM (p = 0.9400) (Supplementary Fig. 2).
Bivalent receipt and vaccination interval are tightly linked variables
It has been previously demonstrated that longer intervals between exposures, whether through natural infection or vaccination, may result in enhanced potency and breadth of neutralizing antibody responses18,19,20. Compared to bivalent recipients, the non-recipient group have a greater interval between the XBB.1.5 monovalent vaccine and prior vaccination (mean of 361.2 ± 48.3 days versus mean of 706.9 ± 58.9 days). It is therefore of interest to evaluate the contribution of these longer intervals to the increased boosting observed in bivalent non-recipients. However, despite being statistically significant, representative multiple predictors linear regression modeling on EG.5.1 titer fold-change reveals that bivalent receipt and vaccine interval are highly collinear variables within our cohort as evident in variance inflation factors >10 and R2 > 0.9 between variables (Supplementary Fig. 3). In other words, those who opted to not receive the bivalent vaccine reliably had longer rest intervals between vaccines. This is an expected problem due to the inherently intertwined nature of these variables in real-world vaccinated communities and indicates that regression modeling within this cohort cannot be used to fully isolate the independent effect of vaccine intervals on antibody boosting. However, it is important to emphasize that ancestral titers are independently back-boosted within each participant group, providing evidence that immune imprinting persists in the context of XBB.1.5 monovalent vaccination regardless of the differences, including vaccine intervals, between the two groups and is likely remnant of original ancestral monovalent vaccines (Fig. 1b–e).
Discussion
We examine variant-specific humoral responses to XBB.1.5 monovalent vaccination in individuals who did and did not receive prior boosting with the bivalent Wuhan-Hu-1/BA.4-5 vaccine. In each group, XBB.1.5 vaccination results in significant boosting of humoral immunity as measured by RBD-specific IgG, IgA, and total IgG/A/M isotypes and neutralizing titers against XBB.1.5, EG.5.1, and JN.1. Importantly, both groups exhibit back-boosting of neutralizing titers against the ancestral WA1 strain and isotype titers binding the ancestral spike RBD, which is highly characteristic of persistent immune imprinting8. While EG.5.1 is neutralized by vaccinated sera to the same degree as XBB.1.5, JN.1 demonstrates marked escape. These data indicate that while the XBB.1.5 vaccine is highly immunogenic, additional updates will be required to counter continued viral evolution. Indeed, the JN.1 lineage gave rise to currently circulating variants, including KP.2, KP.3, LP.8.1, and the dominant XEC strains25. At post-XBB.1.5 vaccination, neutralizing titers against KP.2 appear to fall 3.1-fold compared to JN.1, with KP.3.1.1 demonstrating similar evasion of neutralization2, 26. Thus, vaccines have been formulated to target this emerging lineage of variants and are being rolled out at present; however, our findings suggest that imprinting and vaccination intervals will be important considerations for these and future updates. In particular, the effectiveness of JN.1 formulations should be evaluated in terms of immune imprinting from prior vaccines, including the 2023 XBB.1.5 monovalent vaccine assessed herein.
Beyond back-boosting, we demonstrate that immune imprinting also affects responses to the updated XBB.1.5 vaccine by direct comparisons to a group of participants missing the prior bivalent vaccine. Experiencing less imprinting upon XBB.1.5 vaccination, these bivalent non-recipients have greater boosting of neutralizing antibodies with trends toward higher absolute titers and comparable cross-neutralization. Within the bivalent recipient group, those who had one less ancestral monovalent vaccine also appear to exhibit greater boosting of XBB.1.5 and EG.5.1 neutralization after XBB.1.5 vaccination. This is in line with previous reports that repetitive vaccination with the ancestral spike protein causes profound immune imprinting27. Indeed, despite the exclusion of ancestral Wuhan-Hu-1 spike protein in the XBB.1.5 monovalent vaccine, XBB.1.5 vaccination largely results in recall of ancestral spike-reactive memory B cells instead of generating novel responses28. Fortunately, additional Omicron exposures through natural infection or vaccination may break immune imprinting by ancestral spike protein29. Therefore, vaccines that exclude historic variant spike proteins will be preferable to minimize imprinting, though multivalent formulations may still be effective if they combine circulating variants30. Encouragingly, responses against JN.1 are similar whether bivalent recipients received 3 or 4 ancestral vaccinations, suggesting that the grip of ancestral imprinting on vaccine immunogenicity against this and more contemporary lineages may be weakening.
We further demonstrate that vaccine intervals and bivalent receipt are highly collinear variables in our cohort and likely in the vaccinated community, prohibiting the isolation of each factor’s independent contribution to enhanced bivalent non-recipient boosting. However, longer exposure intervals have been extensively shown to enhance humoral immunity against SARS-CoV-2 variants and likely contribute to enhanced non-recipient boosting18,19. Our previous study similarly demonstrated this interval effect, but with participants who had up to 400 days between exposures20. This suggests that a fruitful strategy for COVID-19 vaccines will be to optimize recommended intervals while minimizing disease risk from waning immunity. Of particular interest will be comprehensive studies characterizing efficacy and risk when extending the current trend of annual recommendations to longer, perhaps biennial intervals.
While we observe that bivalent non-recipients experience greater boosting of neutralizing antibodies, it is important to emphasize that this group has lower pre-vaccination titers against variants, cross-neutralization, and neutralizing potency due to waning immunity and lack of Omicron boosting by the BA.4/5 spike variant. Though both groups have similar infection histories, these diminished responses underline a possible increase in infection susceptibility in bivalent non-recipients before XBB.1.5 vaccination. Indeed, prior to being excluded from the final cohort based on anti-nucleocapsid positivity, 2/12 (16.7%) participants in the initial bivalent non-recipient group compared to 3/37 (8.1%) in the initial recipient group were infected in between serum collection timepoints before the XBB.1.5 vaccination could sufficiently boost adaptive immunity. These trends highlight the risk we must avoid when optimizing the current vaccine strategy. Importantly, current annual vaccines are highly effective at reducing disease burden with estimates of XBB.1.5 vaccine effectiveness against COVID-19 hospitalization ranging from 43% to 76.1%31,32. Collectively, the foregoing observations support public health recommendations for remaining current with updated vaccinations to protect against disease caused by novel, emerging variants.
The primary limitation of this study is a small sample size for the bivalent non-recipient group, which is comprised of relatively rare individuals missing only the 2022 bivalent vaccine and opting for the 2023 formulation. However, our assessment of immune imprinting on XBB.1.5 vaccine immunogenicity is corroborated by orthogonal studies with larger cohorts5,28. Furthermore, vaccine interval and bivalent receipt are highly collinear within our cohort, limiting interval-independent observations of imprinting to ancestral back-boosting, meanwhile extended intervals may contribute to increased non-recipient boosting. The isolation of imprinting influence on this particular phenotype will thus require granular cohorts with more overlapping vaccine intervals. We evaluate only antibody responses, and other immune facets such as T cell activity, memory B cell response, and non-neutralizing Fc effector functions will need to be assessed in the context of immune imprinting. Despite our study limitations, we demonstrate immune imprinting through back-boosting of ancestral titers and, most importantly, direct comparisons of groups with fewer imprinting vaccines. Of greatest need will be studies connecting these diminished immune responses to clinical outcomes while controlling for selection bias.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data for all figures (FRTN50 and EC50 values) are publicly available on Zenodo at https://zenodo.org/records/1398898533. All other data may be acquired from the corresponding authors with reasonable request.
Code availability
The FRNT50/EC50 calculation code used in this study is available on Zenodo at https://zenodo.org/records/515865534.
References
Harris, E. WHO Declares End of COVID-19 Global Health Emergency. JAMA 329, 1817 (2023).
Kaku, Y. et al. Virological characteristics of the SARS-CoV-2 KP.2 variant. Lancet Infect. Dis. 24, e416 (2024).
Stankov, M. V. et al. Humoral and cellular immune responses following BNT162b2 XBB.1.5 vaccination. Lancet Infect. Dis. 24, e1–e3 (2024).
Patel, N. et al. XBB.1.5 spike protein COVID-19 vaccine induces broadly neutralizing and cellular immune responses against EG.5.1 and emerging XBB variants. Sci. Rep. 13, 19176 (2023).
Wang, Q. et al. XBB.1.5 monovalent mRNA vaccine booster elicits robust neutralizing antibodies against XBB subvariants and JN.1. Cell Host Microbe 32, 315–321.e3 (2024).
Nguyenla, X. H. et al. Evaluating Humoral Immunity Elicited by XBB.1.5 Monovalent COVID-19 Vaccine - Volume 30, Number 6—June 2024 - Emerging Infectious Diseases journal - CDC. https://doi.org/10.3201/eid3006.240051.
Research, C. for B. E. and. Updated COVID-19 Vaccines for Use in the United States Beginning in Fall 2024. FDA (2024).
Koutsakos, M. & Ellebedy, A. H. Immunological imprinting: Understanding COVID-19. Immunity 56, 909–913 (2023).
Aydillo, T. et al. Immunological imprinting of the antibody response in COVID-19 patients. Nat. Commun. 12, 3781 (2021).
Reynolds, C. J. et al. Immune boosting by B.1.1.529 (Omicron) depends on previous SARS-CoV-2 exposure. Science 377, eabq1841 (2022).
Röltgen, K. et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell 185, 1025–1040.e14 (2022).
Collier Ai-ris, Y. et al. Immunogenicity of BA.5 bivalent mrna vaccine boosters. N. Engl. J. Med. 388, 565–567 (2023).
Qian, Wang et al. Antibody response to Omicron BA.4–BA.5 bivalent booster. N. Engl. J. Med. 388, 567–569 (2023).
Wang, Q. et al. SARS-CoV-2 neutralising antibodies after bivalent versus monovalent booster. Lancet Infect. Dis. 23, 527–528 (2023).
Cerqueira-Silva, T., Boaventura, V. S. & Barral-Netto, M. Effectiveness of monovalent and bivalent COVID-19 vaccines. Lancet Infect. Dis. 23, 1208–1209 (2023).
Chemaitelly, H. et al. History of primary-series and booster vaccination and protection against Omicron reinfection. Sci. Adv. 9, eadh0761 (2023).
Hansen, C. H. Evidence of immune imprinting or the effect of selection bias? Sci. Adv. 9, eadk5668 (2023).
Grunau, B. et al. Immunogenicity of extended mRNA SARS-CoV-2 vaccine dosing intervals. JAMA 327, 279–281 (2022).
Hall, V. G. et al. Delayed-interval BNT162b2 mRNA COVID-19 vaccination enhances humoral immunity and induces robust T cell responses. Nat. Immunol. 23, 380–385 (2022).
Bates, T. A. et al. An extended interval between vaccination and infection enhances hybrid immunity against SARS-CoV-2 variants. JCI Insight 8, e165265 (2023).
Katzelnick, L. C. et al. Viridot: An automated virus plaque (immunofocus) counter for the measurement of serological neutralizing responses with application to dengue virus. PLoS Negl. Trop. Dis. 12, e0006862 (2018).
Bates, T. A. et al. Neutralization of SARS-CoV-2 variants by convalescent and BNT162b2 vaccinated serum. Nat. Commun. 12, 5135 (2021).
Smith, D. J. et al. Mapping the Antigenic and Genetic Evolution of Influenza Virus. Science 305, 371–376 (2004).
Planas, D. et al. Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion. Nat. Commun. 15, 2254 (2024).
COVID-19 variants | WHO COVID-19 dashboard. datadot https://data.who.int/dashboards/covid19/variants.
Kaku, Y., Uriu, K., Okumura, K., Ito, J. & Sato, K. Virological characteristics of the SARS-CoV-2 KP.3.1.1 variant. Lancet Infect. Dis. 24, e609 (2024).
Wang, Q. et al. Deep immunological imprinting due to the ancestral spike in the current bivalent COVID-19 vaccine. Cell Rep. Med. 4, 101258 (2023).
Tortorici, M. A. et al. Persistent immune imprinting occurs after vaccination with the COVID-19 XBB.1.5 mRNA booster in humans. Immunity 57, 904–911.e4 (2024).
Yisimayi, A. et al. Repeated Omicron exposures override ancestral SARS-CoV-2 immune imprinting. Nature 625, 148–156 (2024).
Chang, S. et al. Strategy to develop broadly effective multivalent COVID-19 vaccines against emerging variants based on Ad5/35 platform. Proc. Natl Acad. Sci. 121, e2313681121 (2024).
Hansen, C. H. et al. Short-term effectiveness of the XBB.1.5 updated COVID-19 vaccine against hospitalisation in Denmark: a national cohort study. Lancet Infect. Dis. 24, e73–e74 (2024).
DeCuir, J. Interim Effectiveness of Updated 2023–2024 (Monovalent XBB.1.5) COVID-19 Vaccines Against COVID-19–Associated Emergency Department and Urgent Care Encounters and Hospitalization Among Immunocompetent Adults Aged ≥18 Years — VISION and IVY Networks, September 2023–January 2024. MMWR Morb. Mortal. Wkly. Rep. 73, (2024).
Wrynla, X. H. Source data for ‘Immune imprinting and vaccination interval determine antibody responses to XBB.1.5 vaccination’. Zenodo https://doi.org/10.5281/zenodo.13988985 (2025).
tbates0. tbates0/FRNT50_calculator: First release of FRNT50 calculator. Zenodo https://doi.org/10.5281/zenodo.5158655 (2021).
Acknowledgements
This work was funded in part by the National Institutes of Health (grant no. R01AI141549 to F.G.T.). The authors thank the many participants in this study for their generous contribution. We gratefully acknowledge the efforts of the entire OHSU COVID-19 serology study team. We also express gratitude to Lauren Rodda for helpful discussions.
Author information
Authors and Affiliations
Contributions
Conceptualization: X.W., M.C., F.T. Methodology: X.W., T.B., M.T., M.W., A.H., M.C., F.T. Investigation: X.W., T.B., M.T., M.W., A.H. Visualization: X.W. Funding acquisition: M.C., F.T. Project administration: X.W., M.T., M.W., A.H. Supervision: M.C., F.T. Writing – original draft: X.W. Writing – review & editing: X.W., T.B., M.T., M.W., A.H., M.C., F.T.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Wilfredo Garcia-Beltran and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wrynla, X.H., Bates, T.A., Trank-Greene, M. et al. Immune imprinting and vaccine interval determine antibody responses to monovalent XBB.1.5 COVID-19 vaccination. Commun Med 5, 182 (2025). https://doi.org/10.1038/s43856-025-00898-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43856-025-00898-4