Abstract
Background
Prenatal exposure genocide-related to trauma has been previously associated with increased morbidity. Whether the prenatal exposure to genocide and rape also impacts various aspects of biological regulation, including patterns of DNA methylation, remains unknown. The purpose of this study was to evaluate whether prenatal exposure to genocide-related trauma, including conception through rape, is associated with accelerated epigenetic aging, a molecular indicator of biological aging.
Methods
We used a cross-sectional dataset to evaluate whether prenatal exposure to genocide or genocidal rape, among individuals conceived during the 1994 genocide against Tutsi in Rwanda were associated with differences in age acceleration in a range of clocks (first generation: Horvath and Hannum; second generation: PhenoAge; third generation: GrimAge and DunedinPace; and fourth generation: YingDamAge and YingAdaptAge), while taking into account exposure to adverse childhood experiences (ACEs). Participants were 24 years old during the time of data collection, and we enrolled 46 female and 45 male participants. Control participants were those of Rwandan descent who did not live in Rwanda during the genocide.
Results
We show that there are no differences in age acceleration observed with first- or second-generation age clocks. However, age acceleration was associated with prenatal exposure to extreme stress for all other clocks, with the greatest acceleration observed in the genocidal rape conception group. For the YingDamAge clock, acceleration effects were strengthened after the inclusion of ACEs.
Conclusions
Our findings suggest that prenatal trauma exposure is associated with epigenetic age acceleration. Third and fourth-generation clocks may more accurately capture these relationships.
Plain language summary
Early life adverse experiences, including during prenatal development, are thought to reduce one’s lifespan. We evaluated whether mothers who experienced genocide related trauma during the 1994 genocide against Tutsi in Rwanda had children with faster biological aging. We measured biological aging using multiple epigenetic “clocks”, which calculates biological age, as opposed to chronological age, by looking at DNA changes across many genes. Our results showed that prenatal exposure to genocide-related trauma was associated with signs of faster aging for several epigenetic clocks. These findings suggest that supportive care and structural assistance for trauma-affected pregnant women and their children should be prioritized during human rights crises to mitigate these effects.
Similar content being viewed by others
Introduction
Genocide has profound and long-lasting effects on humanity. In Rwanda, more than 1,000,000 lives were lost in a period of 100 days in the 1994 genocide against the Tutsi1. The genocide not only impacted the lives of those who directly experienced it but also those who were indirectly exposed while in utero. Our previous studies demonstrated that young adults conceived during the genocide against the Tutsi had worse mental and physical function and higher post-traumatic stress disorder (PTSD) scores, anxiety, depression, pain intensity, and sleep disturbance compared to age- and sex-matched young adults who were not prenatally exposed to the genocide2. Other studies conducted in Rwanda also reported significantly higher scores of PTSD, depression and lower cortisol levels among Rwandans prenatally exposed to genocide when compared with those whose parents were living outside the country during the time of genocide3,4.
Rape was used as a systematic weapon during the 1994 genocide and affected approximately 350,000 women, of whom only one in six survived2,5. While the exact number of children born because of genocidal rape will never be known, the total is estimated between 2000–10,0005,6. Individuals conceived during the genocide, including those conceived through rape, were exposed to this trauma during the first trimester, which is a critical stage of development. For those conceived via genocidal rape, stress related to their birth origins extends beyond the acute period of genocide and continues throughout childhood7,8. For example, studies conducted in Rwanda and the former Yugoslavia reported that children born of genocidal rape face physical and emotional abuse from family and community members and endure poverty and other socioeconomic hardships7,9. This can manifest in a significantly higher likelihood of experiencing adverse childhood experiences2.
While research has demonstrated associations between prenatal exposure to genocide and mental and physical health outcomes2,10, it is possible that prenatal exposure to genocide and rape also impacted various aspects of biological regulation, including patterns of DNA methylation. We previously reported that individuals who were born of genocidal rape, relative to controls, had DNA methylation that varied at CpGs in BDNF and SLC6A4, and methylation in these sites was associated with adult mental health outcomes11. While suggestive, site-specific DNA methylation has not proven to be a useful prognosticator of health; by contrast, DNA methylation-based aging estimators have been associated with mental health treatment outcomes12, cancer prognosis, and chronic disease mortality13. There are different machine-learning algorithms to estimate biological aging. First-generation epigenetic age estimators were created by training neural-net models to predict age by comparing the methylation status at CpG sites present on arrays with chronological age (e.g., Horvath14, Hannum15); age acceleration was conceptualized as the positive difference (either raw or residualized) between estimated and actual age. Such estimators have been critiqued for low test-retest reliability16, a lack of generalizability beyond their training data17, and a limited ability to capture biological processes and/or epigenetic patterns related to healthy aging and longevity. In response, so-called “second generation” epigenetic clocks have included phenotypic measures known to associate with biological aging (e.g. PhenoAge) as well as “third generation” clocks that are generated using longitudinal mortality and longevity data (GrimAge, DunedinPACE). Most recently, fourth-generation clocks have been developed using causally-constrained epigenetic markers of adaptive aging or longevity (YingAdaptAge) and/or decreased lifespan or damage-related aging (YingDamAge)18 (see Supplementary Data 1). Though effect sizes vary depending on the clock used, accelerated epigenetic age has been associated with decreased lifespan and healthspan19. Notably, early life adversity (ELA, e.g. both prenatally and in childhood) has been associated with accelerated epigenetic age, although these associations frequently vary depending on the epigenetic clock used and the way adversity is measured20. For example, in a systematic review of ELA and epigenetic aging in the Horvath and Hannum clocks, experiences of threat (vs. deprivation) were most related to biological aging21 in children. In a separate analysis among Congolese newborns, prenatal exposure to general trauma and war trauma was found to be associated with accelerated epigenetic age in the Hannum extrinsic age (but not PhenoAge or GrimAge) clocks22. Lower birth weight has also been associated with accelerated epigenetic aging in the Hannum, DNAmPhenoAge, DunedinPoAm, and DNAmTL (but not GrimAge) in young male, but not female, adults in the Philippines23. Here, we use DNA methylation array data from our previously described cohort of individuals conceived during the 1994 genocide against the Tutsi11 to evaluate epigenetic aging using all seven published epigenetic age estimators.
The primary aim of this study is to evaluate whether different patterns of prenatal exposure to maternal stress are associated with epigenetic age. We evaluate three groups, (1) single-exposed—maternal stress related to genocide; (2) double-exposed—maternal stress related to genocide and genocidal rape; (3) control—not directly exposed to genocide or genocidal rape. In contrast to previous work on similar samples, here we evaluate whether any association between prenatal genocide exposure and epigenetic age remains after adjustment for adverse childhood experiences (ACEs). For our analysis, we hypothesize that adverse early life experiences would be associated with accelerated epigenetic age. For the prenatal adversity groups, we predict that single-exposed individuals will have a significantly accelerated epigenetic age compared to control individuals, and double-exposed individuals will have a significantly accelerated epigenetic age relative to single-exposed and control individuals. We explore this relationship using first-generation (Horvath and Hannum), second-generation (PhenoAge), third-generation (GrimAge and DunedinPACE) and fourth-generation (YingDamAge and YingAdaptAge) epigenetic age estimators. We find that prenatal exposure to genocide is associated with accelerated aging for third- and fourth-generation clocks, with those individuals conceived through genocidal rape during the genocide being the most affected.
Methods
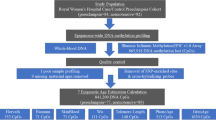
This analysis is part of a comparative and associational cross-sectional study that explored the health impacts of prenatal exposure to genocide among Rwandan young adults conceived during the 100 days of genocide against the Tutsi in Rwanda in 19942,10. Study approvals were obtained from the Institutional Review Boards of the University of Illinois at Chicago (UIC: 2018-1497), the University of Rwanda (UR No 063/CMHS IRB/2019), and Dartmouth College (STUDY0003231). All participants were given an information letter about the study and signed a consent form before data collection2,10. Rwandans aged 24 years old during the time of data collection were enrolled in the study and categorized into three groups accordion to their level of exposure: group 1: single-exposed—maternal stress-related genocide group 2: double-exposed—maternal stress-related genocide and genocidal rape; and group 3: control—not directly exposed to genocide or genocidal rape. The target sample size (n = 90; 30 per group) was generated a priori with power analyses calculated using G*power 3.124. The study had 80% power to detect mean differences between groups with a minimum effect size of r = 0.4, and a p < 0.052, which was a reasonable expected effect size given previous studies4,22. The first participants in both exposed groups were recruited from the Solidarity for the Development of Widows and Orphans to Promote Self-Sufficiency and Livelihoods “SEVOTA” and Association of Genocide Widows Agahozo “AVEGA Agahozo”, non-profit organizations that support genocide survivors2. Participants in the control group were descendants of Rwandans who were living outside the country during the time of the genocide and had no direct experience of the 1994 genocide. Each participant was invited to recommend age- and sex-matched Rwandans who belonged to any of the three groups.
Data were collected by the first author, who is a Rwandan mental health nurse, in a private room. Interviews were conducted in Kinyarwanda. A total of 91 participants completed demographic and health-related surveys in Research Electronic Data Capture (REDCap).
Prenatal exposure to genocide
To determine the level of exposure, we asked each participant if they were conceived by a genocide survivor and whether they were conceived via genocidal rape. Most of the participants in the exposed groups were referred to the study by an organization that supports survivors of genocide and their offspring; these organizations hence knew and shared with us in which category their referred potential participants belonged to. We verified these exposures with participants during their interviews. We conducted screening interviews with participants in the control group to determine if individuals were born to Tutsi women who were living outside the country during the time of the genocide. We excluded participants if their parents left the country due to the genocide or other political unrest in the months leading up to the genocide. We backdated participants’ dates of birth to estimate if they were conceived during the time of the genocide: April 07–July 4, 1994. An equal number of female and male participants were enrolled in each group (Table 1).
Early Life Adversity
Adverse childhood experiences before age 18 were assessed using the Adverse Childhood Experiences International Questionnaire (ACEs IQ)25. This measure includes 13 items—emotional abuse; physical abuse; sexual abuse; violence against household members; living with household members who were substance abusers; living with household members who were mentally ill or suicidal; living with household members who were imprisoned; one or no parents, parental separation or divorce; emotional neglect; physical neglect; bullying; community violence; and collective violence, resulting in an ACEs score of 0-13. This measure has been validated in another African setting26 and had acceptable internal consistency within our sample (α = 0.70).
Dried Blood Spot Collection
Dried blood spots (DBS) were collected for later DNA methylation analysis following the interview. DBS is a minimally invasive and frequently used method that is convenient and cost-effective in research in remote settings that requires long-distance transportation of samples27. The use of DBS has been validated in studies exploring DNAm28. Whole blood drops were collected from a finger stick by sterile lancet on Flinders Technology Associates cards (FTA), with four sample areas of 125 μL each per card. Samples were collected from March 07 to April 06, 2019. The drops were air-dried for at least four hours before placing each card in an airtight envelope with silica-based desiccant and stored at room temperature in Rwanda. Samples were then shipped to the University of Illinois on April 7, 2019, and later to Dartmouth College on August 26, 2021, where they were stored at −80 °C prior to sample processing and DNA methylation analysis in April 2022.
DNA Methylation Sample Processing
DNA was extracted from dried blood spots (DBS) using QIAamp DNA Investigator Kit (Qiagen, Catalog #56504). The manufacturer’s protocol was optimized to improve DNA yield. For each sample (N = 91), two 6 mm hole punches were processed in individual 1.5 microcentrifuge tubes (Eppendorf) and QIAamp MinElute columns (Qiagen). The elution buffer ATE (Qiagen) was heated to 70 °C to improve the release of DNA from the silica membrane. 60 µL of ATE were pipetted onto the silica membrane of the MinElute column and incubated at room temperature (15–25 °C) for 10 min. before centrifugation. The eluate was re-eluted onto the silica membrane and incubated at room temperature (15–25 °C) for 3 min. Following final centrifugation, the eluates were combined into one 1.5 microcentrifuge tube and carefully pipetted up and down to ensure sufficient mixture. Purified DNA was quantified using Invitrogen Qubit 3.0 Fluorometer broad range assay (median = 259.6 ng of DNA). Infinium FFPE QC and DNA Restoration kit (Illumina Inc., WG-321-1001, WG-321-1002) was used to evaluate sample quality and restore degraded DNA, prior to bisulfite treatment (Zymo EZDNA Methylation Kit, Zymo Research, Irvine, CA, USA). Sample quality was evaluated at multiple stages of the DNA methylation data collection process. The Illumina FFPE QC assay, a qPCR-based test, was used to assess sample quality and indicated very high-quality samples. Following the restoration protocol and bisulfite conversion, additional quality control was performed on the converted DNA from 5% of the samples. This DNA methylation data was generated in a single batch using the Illumina MethylationEPIC BeadChip v1.0, interrogating ~850,000 CpG sites. Samples were randomized by the prenatal exposure group across chips.
Data Preprocessing and Normalization
DNA methylation microarray idat files were imported into R (version 4.2.1) and processed using the minfi package (version 1.41.0)29. Quality control included estimating sex and calculating mean detection p-value for CpGs across all samples to evaluate signal reliability. Beta and M values were calculated using the normal-exponential out-of-band (Noob) method, recommended for the 12-immune-cell-type extended deconvolution, which includes normalization and background correction30. Further preprocessing took place before any epigenomic analysis, including filtering CpGs with low detection p-value across samples (20,170), filtering probes on X and Y chromosomes (Y = 135, X = 18,588), and filtering SNPs/cross-hybridizing probes (77,510). DNAm β-values and M-values were extracted and used in subsequent analysis.
Epigenetic age acceleration
All participants were 24 years of age during data collection. Epigenetic age was calculated from the preprocessed methylation beta values across all samples using the methyAge function in package ENmix (version 1.32.0), which includes Horvath, Hannum, and PhenoAge clocks. DunedinPACE, GrimAgeAccel, YingAdaptage, and YingDamAge were similarly calculated using the Biolearn library developed and maintained by the Biomarkers of Aging Consortium31. While several of these clocks were developed using the Illumina 450 K array, each has been validated for use with the EPIC microarray used in the present analysis32. After calculating the predicted epigenetic age, we regressed chronological age on the predicted age (except for GrimAgeAccel, which produces its own residualized score), and the age-corrected residual difference was used in all analyses as our epigenetic age acceleration measure.
Immune cell type deconvolution
Immune cell type proportions were estimated across all samples using a DNA methylation-derived cell type deconvolution method. The package FlowSorted.BloodExtended.EPIC (version 2.0.0) infers proportions of 12 immune cell types; neutrophils (Neu), monocytes (Mono), basophils (Bas), eosinophils (Eos), CD4T naïve cells (CD4nv), CD4T memory cells (CD4mem), B naïve cells (Bnv), B memory cells (Bmem), CD8T naive cells (CD8nv), CD8T memory cells (CD8mem), T regulatory cells (Treg), and natural killer cells (NK)30. To summarize immunophenotype, a principal component analysis was conducted on all cell types; the first principal component explained 68.2% of the variance and was used to control for cell-type composition. The first immune cell type principal component (PC) was included as a covariate in the epigenetic age analysis. We also conducted sensitivity analyses with the first four PCs, accounting for 91% of the variance in sample cell-type composition, which resulted in slightly stronger effects of prenatal exposure, but without substantial differences to the overall direction and magnitude of our findings (see Supplementary Tables 29-42).
Statistics and Reproducibility
All statistical analyses were conducted in R version 4.3.1. Code to reproduce these findings is available at https://github.com/luisamariariverarivera/Rwanda_Aging_Public33. Descriptive statistics were run across the exposure groups before analysis, and epigenetic aging measures were plotted by the exposure group and visually inspected (see Supplementary Fig. 1a & 1b). Because overall rates of smoking in Rwandan youth are low (8%)34, this study did not specifically ask about smoking behavior. As an additional cautionary measure, we estimated smoking status from the DNA methylation data using the package EpiSmokEr and found only two participants (one in the control and one in the single-exposed group) were classified as potential smokers35; no smoking related covariates were thus included in the models. Similarly, we conducted an ANOVA to determine whether body mass index varied significantly across exposure groups and found that there were no significant group differences (F(2,88) = 0.954, p = 0.389).
All participants had complete data and were therefore included in the models. For each epigenetic clock, we conducted a two-step multiple linear regression to determine the relationship between the exposure group and epigenetic age acceleration, with the control group serving as the reference category. In the first step, we adjusted for sex and the first immune cell-type PC; in the second, we added the ACE total score as an additional predictor alongside the exposure group to evaluate whether or how it changed the main effect of exposure group on epigenetic age acceleration. We also produced overlapping density plots and within-individual Pearson correlations between the clocks to assess the consistency of the predictions (Supplementary Fig. 2 & 3). To compare effects across different epigenetic clocks, we calculated the standard mean difference of the regression coefficients for single or double-exposed group status vs. control contrast for models and plotted them together in a forest plot (see Fig. 1).
Comparison of effect sizes (standardized mean difference in age acceleration) and 95% confidence intervals across epigenetic age estimators and exposure groups (double exposed, n = 30, single exposed n = 31, control, n = 30). A Model 1 showing forest plots for single exposed and double exposed individuals, adjusted for sex and cell type. B Model 2 showing forest plots for single exposed and double exposed individuals, adjusted for sex, cell type, and ACEs.
Sensitivity analyses
We repeated the above procedure covarying for the first four cell-type principal components, which together accounted for 91% of the variance in sample cell type composition (see Supplementary Tables 29-42). Adjusting for all four cell type PCs slightly strengthened the association between prenatal exposure and epigenetic aging in several of the clocks (e.g., GrimAge and YingDamAge)—likely by reducing bias from cell type heterogeneity—however, we prioritized parsimony in primary models due to limited degrees of freedom. To explore whether the effects were sex-specific, we also evaluated sex-specific interactions with genocide exposure. There were no substantial differences in our findings (see Supplementary Tables 15-28). That said, we may have been underpowered for this analysis, since there are only 15 individuals of each sex in each exposure group. Likely due to the decrease in power, some effects appear attenuated in these models. The only significant interaction was in the PhenoAge clock models, such that single-exposed males (β = 5.714, p = 0.029, SE = 2.6) are specifically associated with age acceleration, with a directionally similar to the marginal association found for double-exposed males (β = 4.634, p = 0.075, SE = 2.5). PhenoAge was constructed using a panel of peripheral-blood based biomarkers associated with aging, and it is possible that these measures may themselves exhibit sex-dependent biases that render PhenoAge more sensitive to male vs. female aging phenotypes. However, given the multiple tests conducted and low power, we are hesitant to over-interpret this finding.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Sample descriptive statistics are presented in Table 1. As noted earlier, all participants were 24 years of chronological age (mean= 24.1, sd = 0.10) and fairly evenly split by sex (n = 46 female, n = 45 male). The majority (73.7%) had some college education. Reported adverse childhood events ranged from 1 to 11, with a mean of 5.00, and as previously reported, were highest in the doubly exposed group2.
Epigenetic age
Predicted epigenetic age ranges varied between clocks (see Supplementary Data 2). For example, the mean PhenoAge predicted age for the sample was 12.8 years (sd = 5.61). We found no evidence of an association between epigenetic age acceleration and prenatal genocide exposure (single or double) and/or ACEs in the Hannum, Horvath, or PhenoAge clocks (Table 2; Supplementary Tables 1-6).
By contrast, epigenetic age acceleration calculated from YingDamAge, YingAdaptAge, DundeinPACE, and GrimAgeAccel clocks were all associated with prenatal genocide exposure with stronger effects for the double-exposed group seen in the YingDamAge (βsingle = 3.601, p = 0.048, SE = 1.80; βdouble = 6.375, p = 0.0007, SE = 1.81) and YingAdaptAge clocks (βsingle = −6.482, p = 0.0007, SE = 1.84; βdouble = −7.725, p = 0.0001, SE = 1.86) (Table 2). The relationship between prenatal genocide exposure and epigenetic age acceleration calculated from YingAdaptAge remained significant and increased in both exposed groups after controlling for ACEs (βsingle = −7.450, p = 0.0002, SE = 1.93; βdouble = −9.436, p < 0.0001, SE = 2.15). For YingDamAge the double-exposed group, but not the single exposed group, had significant epigenetic age acceleration after adjusting for ACEs (YingDamAge: βdouble = 6.349, p = 0.0036, SE = 2.12). The prenatal exposure group coefficients attenuated after adjustment for the GrimAge measure in particular (βdouble in model 1 = 1.448, p = 0.037, SE = 0.69; βdouble in model 2 = 0.93, p = 0.25, SE = 0.80). Visual examination of regression diagnostic plots in the performance package demonstrated good model performance for each clock.
We calculated the standardized mean difference to compare effect sizes across each of the models tested (Fig. 1). The confidence intervals for each of the Horvath, Hannum, and PhenoAge models contained zero; in DunedinPACE and GrimAge, the single-exposed contrast, but not the double-exposed, also contained zero. Effect sizes were progressively greater (and greater for the double-exposed) in the third and fourth-generation clocks tested. The largest effect size was for reduced adaptive aging in the double-exposed (YingAdaptAge SMD = −0.98, (95% CI = −1.54, −0.52) in Model 1 (unadjusted for ACEs) and Model 2 (YingAdaptAge SMD = − 1.19, 95% CI = −1.73, −0.66).
Discussion
Previous research has demonstrated the sustained impacts of exposure to genocide and war-related trauma on the epigenome of individuals4,22,36,37. Here, we reported the results of an analysis of epigenetic age among two groups of individuals conceived in Rwanda during the 1994 genocide and one group of individuals conceived outside of Rwanda during that time. We found a pattern of increased epigenetic aging between both prenatal exposure groups among nearly all epigenetic aging clocks, with stronger effects for the more severely exposed group and in later (third and fourth) generation clocks. Adjustment for postnatal adversity only slightly attenuated these effects, confirming the importance of prenatal developmental conditions. Our findings are provocative because they indicate that recent efforts to improve epigenetic clock construction by capturing physiological and/or causal variation in aging related DNA methylation may also better capture the impact of prenatal insults on healthspan. This finding contributes valuable knowledge about how sensitive periods of development influence health throughout life, particularly highlighting the independent impact of adverse prenatal exposure to extreme conditions on long-term health outcomes. Effect sizes were largest for the double-exposed group, and the associations remained significant for all initially significant models even with the inclusion of the ACEs measurement. The fourth-generation YingAdaptAge and YingDamAge clocks showed the largest effects, and notably, the latter association was strengthened once ACEs were added. While all later generation clocks, except for PhenoAge, were significantly different for the single-exposed group when adjusting for sex and immune cell composition, YingAdaptAge was the only measure that was significantly different once ACEs were added to the model. This suggests that postnatal environmental experiences may be particularly important moderators or mediators of the relationship between prenatal exposure and epigenetic age within the single-exposed group. Conversely, the YingAdaptAge may be particularly sensitive to prenatal influences, which could also explain the strength of its association in the double-exposed group.
Links between prenatal conditions and earlier onset of age-related disease and decreased lifespan have been theorized within the Developmental Origins of Health and Disease (DOHaD) framework38,39. Low birthweight, prenatal exposure to stress and famine, and other indicators of poor developmental conditions have been associated with increased risk of chronic disease, decreased healthspan, and premature death in a well-established literature40,41. Research exploring epigenetic age acceleration in young adults and associations with early life or prenatal conditions, however, is an emerging field; due to an individual’s limited ability to report on their own prenatal exposures, most studies in this field have investigated aging in children with parental reports of exposure, or childhood adversity as self-reported by adults. Epigenetic age acceleration has been found in adults with in-utero exposure to the Great Depression42, the Dutch Hunger Winter43, and other documented adverse developmental conditions. Our study adds to this literature by demonstrating the impact of prenatal exposure to genocide and genocidal rape in relatively healthy young adults. Whether these responses represent constraint, adaptation, or pathology is currently unclear, and should be a focus of study in future research44.
Our study also contributes to understanding some of the challenges and limitations of epigenetic clocks45, discerning causal relationships between epigenetic aging and healthspan, and the generalizability of epigenetic aging algorithms to non-Western populations17. We note, especially for the first-generation clocks, the substantial variation in estimated age in these measures. The Hannum and PhenoAge clocks predicted age ranges between 2.40−32.1 years and 0.445−30.1 years, respectively. According to these predictions, some individuals had epigenetic age deceleration on the order of 23 years. It is possible that these deviations result from errors in the epigenetic clocks due to our relatively small sample size, which has limited age variability. It is also notable that outliers demonstrated epigenetic age deceleration and not acceleration and that the most extreme values were found in the double-exposed group. The Horvath clock had a more reasonable range (23.0, 38.3), with a slightly longer right tail to the distribution, reflecting more acceleration than deceleration results. It is worth noting that all the included epigenetic clocks are trained on reference datasets that are not representative of the Rwandan population in terms of ancestry and that were not exposed to such extreme prenatal and postnatal stress17. As such, it is possible that these clocks are less accurate at estimating biological age in this population, potentially due to confounding by cell type30,46,47. For example, the Hannum clock was validated using a sample of 426 “Caucasian” and 230 “Hispanic” individuals, and the authors found a correlation of 96% between chronological and epigenetic age15. In our sample, this correlation was 3.9%. This finding highlights the limitations of creating epigenetic (or genetic, i.e., polygenic risk scores) algorithms based on non-fully representative datasets, reflecting broader anthropological critiques of biological normativity48.
First generation clocks, trained only on chronological age, may be particularly vulnerable to unmeasured confounding (e.g., population structure). The other clocks tested were also trained on data not representative of our sample (see Supplementary Data 1), but each include longitudinal measures of age-related decline, with the Ying clocks additionally constraining CpGs in its algorithm to those most causally related to age-related traits. Our findings suggest that these clocks are potentially more generalizable to other populations17.
We note several limitations to our study. The relatively small sample size may limit the generalizability of our findings. The majority of our participants had some college education which may not be representative of the socioeconomic status of our target population, but which we hypothesize would bias our results towards the null. There is also the possibility for misclassification, with individuals conceived through genocidal rape potentially being unaware of their birth origins and therefore being included in the single-exposure group. We note that this misclassification would also reduce group differences and bias our results toward the null. Further, we utilized a cross-sectional study design with self-report on ACEs in adulthood, which introduces some degree of measurement error into our measure of ELA and limits causal inferences about the effect of ELA on biological aging. Our study sample, which had a very constrained age range, may have led to errors in the epigenetic age calculations. While we included ACEs in our analysis, other unmeasured confounders, such as nutritional status, other environmental exposures, and genetic factors may have influenced the results. Finally, we did not evaluate preconception exposure in our sample. To our knowledge, there has been limited research on preconception exposures—only among Holocaust survivors49 and more recently survivors of the Syrian war50, and no similar studies have been conducted in Africa. This is an important area that future research should address. Despite these limitations, the shared directionality of effect between the multiple second-generation clocks tested, the highly impacted study population, and the use of a control group lend strength to our study findings.
Conclusions
In sum, we found a relationship between maternal exposure to genocide-related trauma during pregnancy and epigenetic age acceleration in young adult offspring using third- and fourth- generation epigenetic age estimators. Adjusting for adverse childhood experiences does not attenuate most of these results, and in the case of YingAdaptAge increased the coefficient. Future longitudinal research on larger samples should evaluate the potential interactive or moderating effects of both positive and negative environmental experiences later in the life course on epigenetic age acceleration.
Code availability
All code to reproduce analyses and figures is available at https://github.com/luisamariariverarivera/Rwanda_Aging_Public33.
References
Ministere de l’Administration Locale, de l’Information et des Affaires Sociales . Denombrement des Victimes du Genocide. Rwanda Ministry of, 1–145 (2002).
Uwizeye, G. et al. Double Jeopardy: Young adult mental and physical health outcomes following conception via genocidal rape during the 1994 genocide against the Tutsi in Rwanda. Soc. Sci. Med. 278, 113938 (2021).
Musanabaganwa, C. No title. Intergenerational and epigenetic effects of trauma and PTSD following exposure to the 1994 genocide against the Tutsi in Rwanda (2023).
Perroud, N. et al. The Tutsi genocide and transgenerational transmission of maternal stress: epigenetics and biology of the HPA axis. The World Journal of Biological Psychiatry 15, 334–345 (2014).
Nowrojee, B. in Shattered lives: Sexual violence during the Rwandan genocide and its aftermath (Human Rights Watch, 1996).
Bijleveld, C., Morssinkhof, A. & Smeulers, A. Counting the countless: Rape victimization during the Rwandan genocide. International Criminal Justice Review 19, 208–224 (2009).
Uwizeye, G., DeVon, A. H., McCreary, L. L., Patil, L. C. & Rutherford, N. J. Children born of genocidal rape: What do we know about their experiences and needs?
Mukamana, D. & Brysiewicz, P. The lived experience of genocide rape survivors in Rwanda. Journal of Nursing Scholarship 40, 379–384 (2008).
Nyirandamutsa, F. et al. Are the Offspring Still Affected by their Mothers’ Genocidal Rape 28 Years Ago? Thematic Analysis of Offspring Experience. Rwanda J. Med. Health Sci. 6, 251–263 (2023).
Uwizeye, G., Rutherford, J. N. & Thayer, Z. M. Associations between duration of first trimester intrauterine exposure to genocide against the Tutsi and health outcomes in adulthood. Am. J. Biol. Anthropol. (2023).
Rivera, L. et al. Prenatal Exposure to the Genocide against the Tutsi in Rwanda is associated with DNA methylation at candidate genes in early adulthood: the role of trauma severity and postnatal adversity. medRxiv, 2024.06. 12.24308615 (2024).
Sullivan, A. D. et al. Intervening after trauma: child–parent psychotherapy treatment is associated with lower pediatric epigenetic age acceleration. Psychol. Sci. 09567976241260247 (2024).
Zheng, C., Berger, N. A., Li, L. & Xu, R. Epigenetic age acceleration and clinical outcomes in gliomas. PLoS ONE 15, e0236045 (2020).
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14, 1–20 (2013).
Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367 (2013).
Higgins-Chen, A. T. et al. A computational solution for bolstering reliability of epigenetic clocks: Implications for clinical trials and longitudinal tracking. Nat. Aging 2, 644–661 (2022).
Watkins, S. H. et al. Epigenetic clocks and research implications of the lack of data on whom they have been developed: a review of reported and missing sociodemographic characteristics. Environ. Epigenet. 9, dvad005 (2023).
Ryan, C. P. Epigenetic clocks”: Theory and applications in human biology. Am. J. Hum. Biol. 33, e23488 (2021).
Parrott, B. B. & Bertucci, E. M. Epigenetic aging clocks in ecology and evolution. Trends Ecol. Evolut. 34, 767–770 (2019).
Joshi, D., Gonzalez, A., Lin, D. & Raina, P. The association between adverse childhood experiences and epigenetic age acceleration in the Canadian longitudinal study on aging (CLSA). Aging Cell 22, e13779 (2023).
Colich, N. L., Rosen, M. L., Williams, E. S. & McLaughlin, K. A. Biological aging in childhood and adolescence following experiences of threat and deprivation: a systematic review and meta-analysis. Psychol. Bull. 146, 721 (2020).
Quinn, E. B., Hsiao, C. J., Maisha, F. M. & Mulligan, C. J. Prenatal maternal stress is associated with site-specific and age acceleration changes in maternal and newborn DNA methylation. Epigenetics 18, 2222473 (2023).
Kuzawa, C. W. et al. Birth weight and maternal energy status during pregnancy as predictors of epigenetic age acceleration in young adults from metropolitan Cebu, Philippines. Epigenetics 17, 1535–1545 (2022).
Faul, F., Erdfelder, E., Buchner, A. & Lang, A. Statistical power analyses using G* Power 3.1: Tests for correlation and regression analyses. Behav. Res. Methods 41, 1149–1160 (2009).
World Health Organization. Adverse childhood experiences international questionnaire. Adverse childhood experiences international questionnaire (ACE-IQ), 245–258 (2018).
Kazeem, O. T. A validation of the adverse childhood experiences scale in Nigeria. Res. Humanit. Soc. Sci. 5, 18–23 (2015).
McClendon-Weary, B., Putnick, D. L., Robinson, S. & Yeung, E. Little to give, much to gain—what can you do with a dried blood spot? Curr. Environ. Health Rep. 7, 211–221 (2020).
Sasaki, A., Kim, B., Murphy, K. E. & Matthews, S. G. Impact of ex vivo sample handling on DNA methylation profiles in human cord blood and neonatal dried blood spots. Front. Genet. 11, 224 (2020).
Aryee, M. J. et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369 (2014).
Salas, L. A. et al. Enhanced cell deconvolution of peripheral blood using DNA methylation for high-resolution immune profiling. Nat. Commun. 13, 1–13 (2022).
Ying, K. et al. Biolearn, an open-source library for biomarkers of aging. bioRxiv, 2023.12. 02.569722 (2023).
McEwen, L. M. et al. Systematic evaluation of DNA methylation age estimation with common preprocessing methods and the Infinium MethylationEPIC BeadChip array. Clin. Epigenet. 10, 1–9 (2018).
Rivera, L. Code to reproduce Uwizeye et al. 2025, “Prenatal exposure to genocide accelerates aging across third and fourth generation clocks in young adults in Rwanda”. Zenodo https://doi.org/10.5281/zenodo.15730937 (2025).
Habiyaremye, F., Rwunganira, S., Musanabaganwa, C., Muhimpundu, M. A. & Omolo, J. Tobacco use and associated factors among Rwandan youth aged 15-34 years: Findings from a nationwide survey, 2013. PLoS ONE 14, e0212601 (2019).
Bollepalli, S., Korhonen, T., Kaprio, J., Anders, S. & Ollikainen, M. EpiSmokEr: a robust classifier to determine smoking status from DNA methylation data. Epigenomics 11, 1469–1486 (2019).
Musanabaganwa, C. et al. Leukocyte methylomic imprints of exposure to the genocide against the Tutsi in Rwanda: a pilot epigenome-wide analysis. Epigenomics 14, 11–25 (2022).
Kertes, D. A. et al. BNDF methylation in mothers and newborns is associated with maternal exposure to war trauma. Clin. Epigenet. 9, 1–12 (2017).
Feltes, B. C., de Faria Poloni, J. & Bonatto, D. The developmental aging and origins of health and disease hypotheses explained by different protein networks. Biogerontology 12, 293–308 (2011).
Thayer, Z. & Gildner, T. in The Routledge Handbook of Anthropology and Reproduction 36–51 (Routledge, 2021).
Hoffman, D. J., Powell, T. L., Barrett, E. S. & Hardy, D. B. Developmental origins of metabolic diseases. Physiol. Rev. 101, 739–795 (2021).
Wang, N. et al. Exposure to severe famine in the prenatal or postnatal period and the development of diabetes in adulthood: an observational study. Diabetologia 60, 262–269 (2017).
Schmitz, L. L. & Duque, V. In utero exposure to the Great Depression is reflected in late-life epigenetic aging signatures. Proc. Natl. Acad. Sci. USA 119, e2208530119 (2022).
Cheng, M. et al. Accelerated biological aging six decades after prenatal famine exposure. Proc. Natl. Acad. Sci. USA 121, e2319179121 (2024).
Ellison, P. T. & Jasienska, G. Constraint, pathology, and adaptation: how can we tell them apart? Am. J. Hum. Biol. 19, 622–630 (2007).
Bell, C. G. et al. DNA methylation aging clocks: challenges and recommendations. Genome Biol. 20, 1–24 (2019).
Zhang, Z. et al. Deciphering the role of immune cell composition in epigenetic age acceleration: Insights from cell-type deconvolution applied to human blood epigenetic clocks. Aging Cell 23, e14071 (2024).
Nissen, E. et al. Assessment of immune cell profiles among post-menopausal women in the Women’s Health Initiative using DNA methylation-based methods. Clin. Epigenet. 15, 69 (2023).
Wiley, A. S. Biological normalcy. Annu. Rev. Anthropol. 52, 223–238 (2023).
Bierer, L. M. et al. Intergenerational effects of maternal holocaust exposure on FKBP5 methylation. Am. J. Psychiatry 177, 744–753 (2020).
Mulligan, J. C. et al. Epigenetic signatures of intergenerational exposure to violence in three generations of Syrian refugees. Sci. Rep. 15, 5945 (2025).
Acknowledgements
GU was supported by the University of Illinois Chicago, Dartmouth College Society of Fellows, Margaret McNamara Education Grants, and the University of Western Ontario during the study duration. This study was funded by Western University Faculty of Health Sciences; Dartmouth College—Goodman Faculty Research Grant, Society of Fellows Venture Funding, Neukom Institute for Computational Science and Christensen laboratory; and the University of Illinois Chicago—College of Nursing & Graduate College. All funders, except the Christensen laboratory, were uninvolved in data analysis and interpretation. Special thanks to the study participants, SEVOTA, and AVEGA Agahozo. We thank Dr. Donatilla Mukamana for her insightful contributions to the design and execution of the study, as well as for supervising the fieldwork. The authors thank members of the Christensen lab for their contribution to data analysis and their comments on the manuscript that led to improvements of the manuscript.
Author information
Authors and Affiliations
Contributions
G.U. designed the study, collected the data, interpreted the results, and led the writing of the manuscript. L.M.R. conducted statistical analyses, interpreted results and co-wrote the manuscript. H.S. conducted laboratory and statistical analyses and edited the manuscript. B.C. and J.N.R. contributed to the study design, interpreted results, and edited the manuscript. Z.M.T. contributed to the study design, interpreted results, and co-wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Charlotte A. M. Cecil and Carlos Guerrero-Bosagna for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Uwizeye, G., Rivera, L.M., Stolrow, H.G. et al. Prenatal exposure to genocide accelerates epigenetic aging in third- and fourth-generation clocks among young adults in Rwanda. Commun Med 5, 346 (2025). https://doi.org/10.1038/s43856-025-01065-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43856-025-01065-5