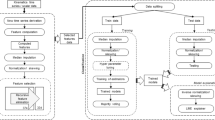
Abstract
Background
Gait disturbances are the clinical hallmark of ataxia. Their severity is assessed within a well-established clinical scale, which only allows coarse scoring and does not reflect the complexity of individual gait deterioration. We investigated whether sensor-free motion capture enables to replicate clinical scoring and improve the assessment of gait disturbances.
Methods
The normal walking task during clinical assessment was videotaped in 91 ataxia patients and 28 healthy controls. A full-body pose estimation model (AlphaPose) was used to extract positions, distances, and angles over time while walking. The resulting time series were analyzed with four machine learning (ML) models, which were combinations of feature extraction (tsfresh, ROCKET) and prediction methods (XGBoost, Ridge). First, in a regression and classification approach, we trained the ML models on reconstructing the clinical score. Second, we used explainable AI (SHAP) to identify the most important time series. Third, we investigated time series features to study longitudinal changes.
Results
Gait disturbances are assessed with high accuracy by ML models, slightly improving human rating (i) in the categorial prediction of the clinical score (F1-score best model: 63.99%, human: 60.57% F1-score), (ii) in the detection of subtle changes (pre-symptomatic patients, clinically rated unimpaired are differentiated from HC with a F1-score of 75.96%) and (iii) in the detection of longitudinal changes over time (Pearson’s correlation coefficient model: −0.626, p < 0.01; human: −0.060, not significant).
Conclusions
ML-based analysis shows improved sensitivity in assessing gait disturbances in ataxia. Subtle and longitudinal changes can be captured within this study. These findings suggest that such approaches may hold promise as potential outcome parameters for early interventions, therapy monitoring, and home-based assessments.
Plain language summary
This study explored a way to measure walking problems in people with ataxia, a condition that affects balance and movement. Researchers used video recordings of patients and healthy participants while walking and analyzed them with a machine learning model that tracks body movements without needing sensors. The model was used to predict clinical scores of walking difficulties and to detect subtle changes over time. The results showed that this approach can capture walking problems accurately and may help detect early changes before symptoms appear, as well as track changes over time. This method could support earlier interventions, improved therapy monitoring, and even enable home-based assessments for people with ataxia.
Similar content being viewed by others
Data availability
Data is made available upon reasonable request. The data is available in the form of videotaped assessments alongside a table providing the clinical ratings, as well as other characterizing information such as age. All requests shall be addressed to the corresponding author, Philipp Wegner (philipp.wegner@dzne.de). A source data file containing all numerical results underlying the graphs and charts presented in the main figures is available as Supplementary Data 1.
Code availability
This study did not use any custom code beyond scripts that run the models mentioned in the Methods. The software and the respective versions utilized were the following. The time series features were generated with the tsfresh (v. 0.20.0) framework implemented in Python https://tsfresh.readthedocs.io/en/latest/. The XGBoost models were taken from the Python implementations of XGBoost (v. 1.7.5) https://xgboost.readthedocs.io/en/stable/. The ridge regressor and classifier models were taken from scikit-learn (v. 1.2.2) https://scikit-learn.org/stable/api/ sklearn.linear_model.html. Hyperparameters for the XGBoost-based models were tuned using Optuna (v. 3.3.0) https://optuna.org/. SHAP values were calculated using the SHAP (v. 0.42.1) Python implementation https://SHAP.readthedocs.io/en/latest/index.html. We used the ROCKET implementation provided as a part of sktime (0.36.0) https://github.com/sktime/sktime. Statistical testing was performed using Scipy (v. 1.13.1) https://scipy.org/ Supplementary Table 3 lists the hyperparameter search spaces. Because all models were evaluated in a leave-one-out setting, no single final set of hyperparameters can be reported. To provide insight into typical configurations, we present the distribution of hyperparameters selected during cross-validation for one representative model (tsfresh+XGBoost) in the Supplementary (see Results section for the exact reference).
References
Klockgether, T. et al. The natural history of degenerative ataxia: a retrospective study in 466 patients. Brain 121, 589–600 (1998).
Mariotti, C., Fancellu, R. & Donato, S. An overview of the patient with ataxia. J. Neurol. 252, 511–518 (2025).
Diallo, A. et al. Survival in patients with spinocerebellar ataxia types 1, 2, 3, and 6 (EUROSCA): a longitudinal cohort study. en. Lancet Neurol. 17, 327–334 (2025).
Schmitz-Hubsch, T. et al. Scale for the assessment and rating of ataxia: development of a new clinical scale. Neurology 66, 1717–1720 (2023).
Serrao, M. & Conte, C. in Handbook of Human Motion 937–954. http://link.springer.com/10.1007/978-3-319-14418-4_46 (Springer International Publishing, 2018).
Ilg, W. & Timmann, D. Gait ataxia-specific cerebellar influences and their rehabilitation. Mov. Disord. 28, 1566–1575 (2023).
Buckley, E., Mazz`a, C. & McNeill, A. A systematic review of the gait characteristics associated with cerebellar Ataxia. Gait Posture 60, 154–163 (2023).
Kadirvelu, B. et al. A wearable motion capture suit and machine learning predict disease progression in Friedreich’s ataxia. Nat. Med. 29, 86–94 (2023).
Seemann, J. et al. Digital Gait measures capture 1-year progression in early-stage spinocerebellar ataxia type 2. Mov. Disord. 39, 788–797 (2024).
Jin, L. et al. Gait characteristics and clinical relevance of hereditary spinocerebellar ataxia on deep learning. Artif. Intell. Med. 103, 101794 (2025).
Lang, J. et al. Detecting and quantifying ataxia-related motor impairments in rodents using markerless motion tracking with deep neural networks. in Proc. 42nd Annual International Conference of the IEEE Engineering in Medicine and Biology Society, 3642–3648 (IEEE, 2020).
Summa, S. et al. Validation of low-cost system for gait assessment in children with ataxia. Comput. Methods Prog. Biomed. 196, 105705 (2025).
L’Italien, G. J. et al. Video-based kinematic analysis of movement quality in a phase 3 clinical trial of troriluzole in adults with spinocerebellar ataxia: a post hoc analysis. Neurol. Ther. 13, 1287–1301 (2025).
Eguchi, K. et al. Feasibility of differentiating gait in Parkinson’s disease and spinocerebellar degeneration using a pose estimation algorithm in two-dimensional video. J. Neurol. Sci. 464, 123158 (2025).
Ippisch, R., Jelusic, A., Bertram, J., Schniepp, R. & Wuehr, M. mVEGAS—mobile smartphone-based spatiotem- poral gait analysis in healthy and ataxic gait disorders. Gait Posture 97, 80–85 (2025).
Tsukagoshi, S. et al. Noninvasive and quantitative evaluation of movement disorder disability using an infrared depth sensor. J. Clin. Neurosci. 71, 135–140 (2025).
Ilg, W. et al. Quantitative Gait and balance outcomes for ataxia trials: consensus recommendations by the Ataxia Global Initiative Working Group on digital-motor biomarkers. Cerebellum 23, 1566–1592 (2024).
Reilly, M. M. et al. Trials for slowly progressive neurogenetic diseases need surrogate endpoints. Ann. Neurol. 93, 906–910 (2025).
Jacobi, H. et al. Inventory of Non-Ataxia Signs (INAS): validation of a new clinical assessment instrument. Cerebellum 12, 418–428 (2013).
Grobe-Einsler, M. et al. Scale for the Assessment and Rating of Ataxia (SARA): development of a training tool and certification program. Cerebellum 23, 877–880. https://doi.org/10.1007/s12311-023-01543-3 (2024).
Fang, H.-S. et al. AlphaPose: whole-body regional multi-person pose estimation and tracking in real-time. IEEE Trans. Pattern Anal. Mach. Intell. 45, 7157–7173 (2023).
Wei, S.-E., Ramakrishna, V., Kanade, T. & Sheikh, Y. Convolutional Pose Machines (IEEE, 2016).
Cao, Z., Simon, T., Wei, S.-E. & Sheikh, Y. Realtime Multi-Person 2D Pose Estimation using Part Affinity Fields (IEEE, 2017).
Cao, Z., Hidalgo Martinez, G., Simon, T., Wei, S. & Sheikh, Y. A. OpenPose: realtime multi-person 2D pose estimation using part affinity fields. in IEEE Transactions on Pattern Analysis and Machine Intelligence (IEEE, 2019).
Simon, T., Joo, H., Matthews, I. & Sheikh, Y. Hand Keypoint Detection in Single Images using Multiview Bootstrapping (IEEE, 2017).
Farhadi, A. & Redmon, J. Yolov3: An incremental improvement. Computer vision and pattern recognition. Vol. 1804 (Springer, Berlin/Heidelberg, Germany, 2018).
Tan, M., Pang, R. & Le, Q. V. EfficientDet: Scalable and Efficient Object Detection. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 10778–10787 (Seattle, WA, USA, 2020).
He, K. et al. Deep Residual Learning for Image Recognition. Proc. IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas. 770–778 (2016).
Christ, M., Braun, N., Neuffer, J. & Kempa-Liehr, A. W. Time Series FeatuRe Extraction on Basis of Scalable Hypothesis Tests (tsfresh—A Python package). Neurocomputing 307, 72–77 (2023).
Chen, T. & Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proc. 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD '16), 785–794 (Association for Computing Machinery, New York, NY, USA, 2016).
Dempster, A., Petitjean, F. & Webb, G. I. ROCKET: exceptionally fast and accurate time series classification using random convolutional kernels. Data Min. Knowl. Discov. 34, 1454–1495 (2023).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Fryer, D., Stru¨mke, I. & Nguyen, H. Shapley values for feature selection: the good, the bad, and the axioms. IEEE Access 9, 144352–144360 (2021).
Seabold, S. & Perktold, J. Statsmodels: Econometric and Statistical Modeling with Python in 9th Python in Science Conference (SCIPY, 2010).
Dean, C. B. & Nielsen, J. D. Generalized linear mixed models: a review and some extensions. Lifetime Data Anal. 13, 497–512. http://link.springer.com/10.1007/s10985-007-9065-x (2007).
Faber, J. et al. Stage-dependent biomarker changes in spinocerebellar ataxia type 3. Ann. Neurol. 95, 400–406 (2025).
Shah, V. V. et al. Gait variability in spinocerebellar ataxia assessed using wearable inertial sensors. Mov. Disord. 36, 2922–2931 (2021).
Ilg, W. et al. Digital gait biomarkers allow to capture 1-year longitudinal change in spinocerebellar ataxia type 3. Mov. Disord. 37, 2295–2301 (2024).
Shin, D. The effects of explainability and causability on perception, trust, and acceptance: implications for explainable AI. Int. J. Hum.-Comput. Stud. 146, 102551 (2024).
Weyer, A. et al. Reliability and validity of the scale for the assessment and rating of ataxia: a study in 64 ataxia patients. Mov. Disord. 22, 1633–1637 (2024).
Taheri Amin, A. et al. Comparison of live and remote video ratings of the scale for assessment and rating of ataxia. Mov. Disord. Clin. Pract. 10, 1404–1407 (2023).
Vizcarra, J. A., Casey, H. L., Hamedani, A. G. & Gomez, C. M. Reliability of remote video ratings of the scale for assessment and rating of ataxia. Park. Relat. Disord. 132, 107278 (2025).
Grobe-Einsler, M. et al. Development of SARA(home), a New video-based tool for the assessment of ataxia at home. Mov. Disord. 36, 1242–1246. https://www.ncbi.nlm.nih.gov/pubmed/33433030 (2021).
Acknowledgements
This study was funded by the iBehave Network, sponsored by the Ministry of Culture and Science of the State of North Rhine-Westphalia. J.F. received funding from the Advanced Clinician Scientist Programme (ACCENT, funding code 01EO2107). The ACCENT Program is funded by the German Federal Ministry of Education and Research (BMBF). This publication is an outcome of ESMI, an EU Joint Programme - Neurodegenerative Disease Research (JPND) project (see www.jpnd.eu). The project is supported through the following funding organizations under the aegis of JPND: Germany, Federal Ministry of Education and Research (BMBF; funding codes 01ED1602A/B). J.F. received funding of the National Ataxia Foundation (NAF) and consultancy honoraria from Vico Therapeutics and Biogen, unrelated to the present manuscript. Several authors are members of the European Reference Network for Rare Neurological Diseases (ERN-RND). M.G.E. received research support from the German Ministry of Education and Research (BMBF) within the European Joint Program for Rare Diseases (EJP-RD) 2021 Transnational Call for Rare Disease Research Projects (funding number 01GM2110), from the National Ataxia Foundation (NAF), and from Ataxia UK, and received honoraria from Biogen and Healthcare Manufaktur, Germany, all unrelated to this study. We would like to acknowledge Matthis Synofzik, (Division Translational Genomics of Neurodegenerative Diseases, Hertie Institute for Clinical Brain Research and Center of Neurology, University of Tu¨bingen, Germany & German Center for Neurodegenerative Diseases (DZNE), Tu¨bingen, Germany) who contributed to the retrospective consensus video ratings.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
M.G.E., J.F., T.K., B.K., O.K., D.S., F.H., and S.B. conducted the clinical part of this study. T.E., A.L., and P.W. prepared and updated clinical information about the study participants. L.R., F.K., and M.F. consulted this work and contributed to conceptualizing the project. J.F. supervised this work and contributed to the final paper writing. PW implemented the models, performed the analysis, and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wegner, P., Grobe-Einsler, M., Reimer, L. et al. Leveraging machine learning for digital gait analysis in ataxia using sensor-free motion capture. Commun Med (2026). https://doi.org/10.1038/s43856-025-01258-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-025-01258-y