Abstract
Background
Next-generation sequencing (NGS) tests are integral to oncology care. To address the need for clinical and NGS data management, interpretation, and reporting, we developed iCatalog for the multi-institutional Individualized Cancer Therapy 2/Genomic Assessment Informs Novel Therapy Consortium (GAIN) pediatric precision oncology (PO) study.
Methods
We designed iCatalog as a secure, web-based clinical decision support application that stores and integrates clinical, specimen, and molecular data from multiple sources at the patient level. The knowledge base (KB) and centralized patient/test database are intended to manage information for the 825 patients expected to enroll in the GAIN study. User permissions and access are controlled. Gene- and variant-level interpretation is facilitated through linked external resources and an internal KB that can be updated during application use. iCatalog generates editable, study-specific patient reports for each molecular test.
Results
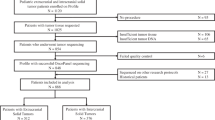
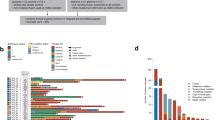
Launched to support the GAIN study, iCatalog integrates genomic data from eight NGS platforms, generates 1002 clinical interpretation reports, and stores data for 1194 tests involving 777 patients with pediatric solid tumors across 133 diagnoses. The KB contains pediatric cancer-specific curations, authored by the research team, spanning 581 genes and 2659 variants (including 2146 single-nucleotide variants and insertions-deletions, 235 copy-number variants, 278 structural variants).
Conclusions
iCatalog is a robust tool designed and proven to support a PO study. It integrates clinical and genomic data to facilitate the clinical interpretation and reporting of variants identified through NGS testing while maintaining a pediatric-specific KB generated during the study. As a scalable, modular platform, iCatalog can accelerate clinical decision-making and elevate PO insights across studies.
Plain language summary
iCatalog is a web-based application developed to support a study evaluating the benefit of tumor profiling in the care of young cancer patients. iCatalog combines information used to determine patient care implications of tumor profiling. Patient (e.g., age, cancer diagnosis), sample (e.g., identification number, date), and genomic data (e.g., mutations, deletions, amplifications) are merged. iCatalog links genomic results to knowledge repositories and auto-generates reports, allowing experts to easily create patient-specific reports for use in the clinic. On this platform, 1002 reports were generated detailing the therapeutic and diagnostic implications of tumor profiling. Pediatric-specific knowledge on 581 genes was generated. As tumor sequencing becomes standard of care, tools reducing barriers to interpreting results for patient care become more important.
Similar content being viewed by others
Data availability
Data and additional supporting information are available in supplemental tables. The source data for Fig. 4A-B is in Supplementary Data 7 and 8, and Fig. 4C is in Supplementary Data 9. The source data for Fig. 5A-B is in Supplementary Data 10, and Fig. 5C is in Supplementary Data 11. Data not included in the supplemental files are available in a prior publication by Church et al.8 or can be provided upon request to Katherine Janeway (katherine_janeway@dfci.harvard.edu).
Code availability
iCatalog was developed through a collaborative effort between the Dana-Farber Cancer Institute (DFCI) and the University of Chicago, with the University of Chicago retaining administrative control over the release of the source code. The codes for the original iCatalog and research versions are hosted in a private Bitbucket repository and available upon request. Interested parties may request access by contacting the co-first author and software developer at the University of Chicago (Wenjun Kang, wkang2@bsd.uchicago.edu) or the corresponding author (Katherine Janeway, katherine_janeway@dfci.harvard.edu).
References
Jiang, W. et al. Tri(c)DB: an integrated platform of knowledgebase and reporting system for cancer precision medicine. J. Transl. Med. 21, 885 (2023).
Yu, Y. et al. PreMedKB: an integrated precision medicine knowledgebase for interpreting relationships between diseases, genes, variants and drugs. Nucleic Acids Res. 47, D1090–D1101 (2019).
Dahary, D. et al. Genome analysis and knowledge-driven variant interpretation with TGex. BMC Med Genomics 12, 200 (2019).
de Bruijn, I. et al. Genome nexus: a comprehensive resource for the annotation and interpretation of genomic variants in cancer. JCO Clin. Cancer Inf. 6, e2100144 (2022).
Borchert, F. et al. Knowledge bases and software support for variant interpretation in precision oncology. Brief Bioinform. https://doi.org/10.1093/bib/bbab134 (2021).
Tamborero, D. et al. The Molecular Tumor Board Portal supports clinical decisions and automated reporting for precision oncology. Nat. Cancer 3, 251–261 (2022).
Reisle, C. et al. A platform for oncogenomic reporting and interpretation. Nat. Commun. 13, 756 (2022).
Church, A. J. et al. Molecular profiling identifies targeted therapy opportunities in pediatric solid cancer. Nat. Med. 28, 1581–1589 (2022).
Harris, M. H. et al. Multicenter feasibility study of tumor molecular profiling to inform therapeutic decisions in advanced pediatric solid tumors: the individualized cancer therapy (iCat) study. JAMA Oncol. 2, 608–615 (2016).
Kang, W. et al. System for informatics in the molecular pathology laboratory: an open-source end-to-end solution for next-generation sequencing clinical data management. J. Mol. Diagn. 20, 522–532 (2018).
Suehnholz, S. P. et al. Quantifying the expanding landscape of clinical actionability for patients with cancer. Cancer Discov. 14, 49–65 (2024).
Chakravarty, D. et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. https://doi.org/10.1200/PO.17.00011 (2017).
Ioannidis, N. M. et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 99, 877–885 (2016).
Tian, Y. et al. REVEL and BayesDel outperform other in silico meta-predictors for clinical variant classification. Sci. Rep. 9, 12752 (2019).
Feng, B. J. PERCH: a unified framework for disease gene prioritization. Hum. Mutat. 38, 243–251 (2017).
Hashem, O. et al. An overview of RAF kinases and their inhibitors (2019-2023). Eur. J. Med Chem. 275, 116631 (2024).
Povedano, J. M. et al. TK216 targets microtubules in Ewing sarcoma cells. Cell Chem. Biol. 29, 1325–1332 e4 (2022).
Meyers, P. A. et al. Open-label, multicenter, phase I/II, first-in-human trial of TK216: a first-generation EWS::FLI1 fusion protein antagonist in ewing sarcoma. J. Clin. Oncol. 42, 3725–3734 (2024).
Gazola, A. A., Lautert-Dutra, W., Archangelo, L. F., Reis, R. B. D. & Squire, J. A. Precision oncology platforms: practical strategies for genomic database utilization in cancer treatment. Mol. Cytogenet. 17, 28 (2024).
Nakken, S. et al. Personal cancer genome reporter: variant interpretation report for precision oncology. Bioinformatics 34, 1778–1780 (2018).
Acknowledgements
Funding for this study was provided by the Precision For Kids Pan Mass Challenge Team, the 4 C’s Fund, Lamb Family Fund, C&S Wholesale Grocers, and C&S Charities.
Author information
Authors and Affiliations
Contributions
W.K.: validation, software development, resources, visualization; L.L.D.L.V.: data curation, writing—original draft, visualization, formal analysis of knowledge base, refinement of user interface; H.C.: resources, visualization, refinement of user interface, project administration; E.A.: data curation, visualization, refinement of user interface; L.D.M.: investigation, data curation; A.K.: investigation, data curation; E.S.: visualization, refinement of user interface; E.C.: resources, supervision, project administration; L.C.: conception, refinement of user interface, data curation; J.W.: resources, refinement of user interface; D.A.W.: investigation, data curation; M.A.A.: investigation, data curation; S.I.C.: investigation, data curation; M.C.: resources, supervision; J.A.J.: resources, supervision; S.V.: conception, funding acquisition; N.R.P.: investigation, data curation, refinement of user interface; A.J.C.: conception; K.A.J.: investigation, conception and refinement of user interface visualization, validation, data curation, funding acquisition; All authors contributed to revisions and approved the final manuscript for publication.
Corresponding author
Ethics declarations
Competing interests
W.K., L.L.D.L.V., H.C., E.A., L.D.M., A.K., E.S., E.C., L.C., J.W., D.A.W., M.A.A., S.I.C., S.V., N.R.P., A.J.C. have no conflicts. M.C. consults for Tellic and Impetus. J.A.J. has received travel support from MDClone to attend scientific meetings. K.A.J. consults for Recordati and receives research funding from AstraZeneca.
Peer review
Peer review information
Communications Medicine thanks Yaqiong Jin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kang, W., Lazo de la Vega, L., Comeau, H. et al. Introducing iCatalog as a clinical decision support tool for collaborative pediatric precision oncology studies. Commun Med (2026). https://doi.org/10.1038/s43856-025-01351-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-025-01351-2