Abstract
Background
Gastric neuroendocrine carcinoma (G-NEC) presents with clinical and pathological features that closely resemble those of gastric adenocarcinoma (GC), often complicating differential diagnosis. However, G-NEC is markedly more aggressive and associated with a significantly poorer prognosis, necessitating accurate and timely identification to guide appropriate therapeutic interventions.
Methods
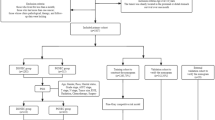
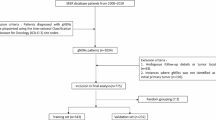
In response to this clinical need, we developed G-NECNet, a deep convolutional neural network tailored to detect G-NEC from histopathological whole-slide images.
Results
The model demonstrates excellent diagnostic performance, yielding an average area under the receiver operating curve (AUROC) of 0.993 in the internal validation cohort, 0.985 on an external single-institutional dataset, and 1.000 on an external multi-institutional consultation dataset. These consistently high AUROC values highlight the robustness, accuracy, and generalizability of G-NECNet across diverse clinical settings.
Conclusions
The integration of G-NECNet into routine diagnostic workflows may not only improve the precision of G-NEC classification but also reduce misdiagnosis-related healthcare costs, offering a practical and scalable solution for clinical application.
Plain language summary
This study aims to improve the diagnosis of gastric neuroendocrine carcinoma (G-NEC), a rare but aggressive stomach cancer often mistaken for the more common stomach cancer called gastric adenocarcinoma. Since accurate diagnosis typically necessitates time-consuming and resource-intensive staining of sections from the tumor, we developed G-NECNet, a computational model that analyzes routine tumor images to distinguish G-NEC precisely. The model works well across multiple datasets, demonstrating high reliability and generalizability in different clinical settings. These findings suggest that G-NECNet could assist pathologists in making faster and more accurate diagnoses of G-NEC. Integrating this tool into routine diagnostic workflows may help reduce errors, improve patient outcomes, and lower healthcare costs, providing a practical and scalable solution for clinical application.
Similar content being viewed by others
Code availability
Source code is at https://github.com/Kepler1647b/G-NECNet (https://doi.org/10.5281/zenodo.18059065)33.
Data availability
All relevant data are available upon request, but cannot be shared publicly. Restrictions are applied to the whole slide images and annotation data of Internal cohort, External-cohort, and Consultation-cohort, which are used with institutional permission via IRB approval for the current study, and thus are not publicly available due to patient privacy obligations. All data supporting the findings of this study are available on request for non-commercial and academic purposes from the corresponding author, M.C. (caimy@sysucc.org.cn) within 10 working days. We do not require you to sign a data use agreement. Processed data can be reproduced stably by the source code. Supplementary Data are provided with this paper (Supplementary Data 1). The source data for Fig. 2 and Table 1 are in Supplementary Data 1.
References
Cives, M. & Strosberg, J. R. Gastroenteropancreatic neuroendocrine tumors. CA Cancer J. Clin. 68, 471–487 (2018).
Fang, J. M., Li, J. & Shi, J. An update on the diagnosis of gastroenteropancreatic neuroendocrine neoplasms. World J. Gastroenterol. 28, 1009–1023 (2022).
Dasari, A. et al. Trends in the incidence, prevalence, and survival outcomes in patients with neuroendocrine tumors in the United States. JAMA Oncol. 3, 1335–1342 (2017).
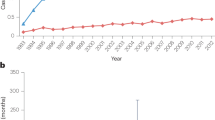
Hu, P. et al. Trends of incidence and prognosis of gastric neuroendocrine neoplasms: a study based on SEER and our multicenter research. Gastric Cancer 23, 591–599 (2020).
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Lin, J. et al. Comparison of survival and patterns of recurrence in gastric neuroendocrine carcinoma, mixed adenoneuroendocrine carcinoma, and adenocarcinoma. JAMA Netw Open 4, e2114180 (2021).
Yamagata, Y. et al. Is lymph node dissection for neuroendocrine carcinoma of the stomach effective as it is for adenocarcinoma? Eur. J. Surg. Oncol. 47, 2004–2009 (2021).
White, B. E. et al. Incidence and survival of neuroendocrine neoplasia in England 1995–2018: a retrospective, population-based study. Lancet Reg. Health Eur. 23, 100510 (2022).
Ishida, M. et al. Neuroendocrine carcinoma of the stomach: morphologic and immunohistochemical characteristics and prognosis. Am. J. Surg. Pathol. 37, 949–959 (2013).
Kawasaki, K. et al. Neuroendocrine neoplasms of the lung and gastrointestinal system: convergent biology and a path to better therapies. Nat. Rev. Clin. Oncol. https://doi.org/10.1038/s41571-022-00696-0 (2022).
Jiang, Y. et al. Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun. (Lond.) 40, 154–166 (2020).
Krittanawong, C. et al. Deep learning for cardiovascular medicine: a practical primer. Eur. Heart J. 40, 2058–2073 (2019).
Chiu, Y. C. et al. Deep learning of pharmacogenomics resources: moving towards precision oncology. Brief. Bioinform. 21, 2066–2083 (2020).
Xu, J., Xue, K. & Zhang, K. Current status and future trends of clinical diagnoses via image-based deep learning. Theranostics 9, 7556–7565 (2019).
Zheng, K. et al. Deep learning model with pathological knowledge for detection of colorectal neuroendocrine tumor. Cell Rep. Med. 5, 101785 (2024).
van der Laak, J., Litjens, G. & Ciompi, F. Deep learning in histopathology: the path to the clinic. Nat. Med. 27, 775–784 (2021).
Zhang, X. et al. A multicenter proof-of-concept study on deep learning-based intraoperative discrimination of primary central nervous system lymphoma. Nat. Commun. 15, 3768 (2024).
Naik, N. et al. Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains. Nat. Commun. 11, 5727 (2020).
Coudray, N. et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018).
Yamashita, R. et al. Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study. Lancet Oncol. 22, 132–141 (2021).
Zheng, X. et al. A deep learning model and human-machine fusion for prediction of EBV-associated gastric cancer from histopathology. Nat. Commun. 13, 2790 (2022).
Iabichino, G. et al. Diagnosis, treatment, and current concepts in the endoscopic management of gastroenteropancreatic neuroendocrine neoplasms. World J. Gastroenterol. 28, 4943–4958 (2022).
Sigel, C. S. Advances in the cytologic diagnosis of gastroenteropancreatic neuroendocrine neoplasms. Cancer Cytopathol. 126, 980–991 (2018).
Pavel, M. et al. Gastroenteropancreatic neuroendocrine neoplasms: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 31, 844–860 (2020).
Board, W.C.o.T.E. Digestive System Tumours (IARC Press, 2022).
Griger, J. et al. An integrated cellular and molecular model of gastric neuroendocrine cancer evolution highlights therapeutic targets. Cancer Cell 41, 1327–1344.e10 (2023).
Liao, Y. et al. Artificial intelligence for predicting HER2 status of gastric cancer based on whole-slide histopathology images: a Retrospective Multicenter Study. Adv. Sci. (Weinh.) 12, e2408451 (2025).
Xia, L. et al. Lymph node metastasis prediction from in situ lung squamous cell carcinoma histopathology images using deep learning. Lab. Investig. 105, 102187 (2025).
Saillard, C. et al. Validation of MSIntuit as an AI-based pre-screening tool for MSI detection from colorectal cancer histology slides. Nat. Commun. 14, 6695 (2023).
Huang, B. et al. Accurate diagnosis and prognosis prediction of gastric cancer using deep learning on digital pathological images: a retrospective multicentre study. EBioMedicine 73, 103631 (2021).
Li, C. et al. Deep learning-based AI model for signet-ring cell carcinoma diagnosis and chemotherapy response prediction in gastric cancer. Med. Phys. 49, 1535–1546 (2022).
Vorontsov, E. et al. A foundation model for clinical-grade computational pathology and rare cancers detection. Nat. Med. https://doi.org/10.1038/s41591-024-03141-0 (2024).
Tianchen Zhu, Z. Z. et al. G-NECNet. Zenodo https://doi.org/10.5281/zenodo.18059065 (2025).
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (No. 2021YFA1300201), the National Natural Science Foundation of China (Grant Nos. 81972227, 82172646, 81872001, and 82073189), and the Guangdong Esophageal Cancer Institute Science and Technology Program (No. M202108).
Author information
Authors and Affiliations
Contributions
Z.L., Y.H., M.C., and J.L. conceived and designed the study. T.Z., C.W., and X.Z. collected the samples and acquired the image data. Z. Zhao performed the machine learning. M.C., L.Z., and W.C. conducted the reader study. T.C. and Z. Zhou did the statistical analyses. All authors vouch for the data, analyses, and interpretations. T.Z. and M.C. wrote the first draft of the manuscript, and all authors reviewed, contributed to, and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Wataru Uegami and Bing Ren for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhu, T., Zhao, Z., Wang, C. et al. A deep learning model for the diagnosis of gastric neuroendocrine carcinoma. Commun Med (2026). https://doi.org/10.1038/s43856-026-01382-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-026-01382-3