Abstract
Background
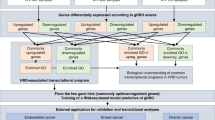
Approximately half of all high-grade serous ovarian carcinomas (HGSCs) have a therapeutically targetable defect in homologous recombination (HR) DNA repair. While there are genomic and transcriptomic methods, developed for other cancers, to identify HR deficient (HRD) samples, there are no gene expression-based tools to predict HR status in HGSC specifically. We have built a HGSC-specific model to predict HR status using gene expression.
Methods
We separated The Cancer Genome Atlas (TCGA) cohort of HGSCs into training (n = 288) and testing (n = 73) sets and labelled each case as HRD or HR proficient (HRP) based on the clinical standard for classification. Using the training set, we performed differential gene expression analysis between HRD and HRP cases. The 2604 significantly differentially expressed genes were used to train a penalised logistic regression model.
Results
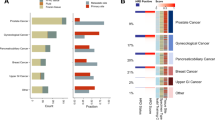
IdentifiHR uses the expression of 209 genes to predict HR status in HGSC. These genes preserve the genomic damage signal, capturing known regions of HR-specific copy number alteration which impact gene expression. IdentifiHR is 85% accurate in the TCGA test set and 86% accurate in an independent cohort of 99 samples, taken from primary tumours, ascites and normal fallopian tubes. Further, IdentifiHR is 84% accurate in pseudobulked single-cell HGSC sequencing from 37 patients and outperforms existing expression-based methods to predict HR status, being BRCAness, MutliscaleHRD and expHRD.
Conclusions
IdentifiHR is an accurate model to predict HR status in HGSC. It is available as an open source R package, empowering researchers to robustly classify HR status when only transcriptomic sequencing data is available.
Plain language summary
High-grade serous ovarian cancer (HGSC) is a type of ovarian cancer with very poor outcomes. However, half of HGSCs have faulty DNA repair that can be targeted for treatment if it is identified. Existing methods look at changes in DNA that arise when repair is faulty, but do not consider which genes are actively being used, or are “expressed”, by the cancer. We developed IdentifiHR, a machine learning method to predict DNA repair status using the expression of 209 genes. We tested IdentifiHR on 209 patient samples and found it correctly predicts repair status in about 85–86% of cases, performing better than existing tools on the same patient data. IdentifiHR is released as a software package for public use.
Similar content being viewed by others
Data availability
The results published here are in whole or part based upon data generated by The Cancer Genome Atlas, managed by the NCI and NHGRI. Information about TCGA can be found at http://cancergenome.nih.gov. RNA sequencing, gene-level copy number, methylation, SNP and structural variant data collected on the TCGA HGSC cohort, with associated clinical data, are available from the Genomic Data Commons (TCGA project) data portal (https://portal.gdc.cancer.gov/, https://www.cancer.gov/tcga, dbGaP Study Accession: phs000178.v11.p8). AOCS gene expression counts were accessed at the Gene Expression Omnibus (accession: GSE209964). Previously published WGS data are available from the European Genome-phenome Archive (accession: EGAD00001000877). MSKCC gene expression counts were available as a Seurat object on Synpase (SynID: syn51091849). The source data for Fig. 2A, D, E can be found in Supplementary Data 2, for Fig. 3A, B in Supplementary Data 5, for Fig. 3C in Supplementary Data 6, for Fig. 3D, E in supplementary data 7, for Fig. 3F in Supplementary Data 9 and for Fig. 4A–C in Supplementary Data 5. These data are available in the supplementary information and in the IdentifiHR repository, https://github.com/DavidsonGroup/IdentifiHR. All other data supporting the findings of this study, including the source data for all figures, are publicly available.
Code availability
All analyses were carried out in R v4.2.1. Code to reproduce the analysis can be found in the IdentifiHR repository, https://github.com/DavidsonGroup/IdentifiHR.
References
Moore, K. N. et al. Niraparib monotherapy for late-line treatment of ovarian cancer (QUADRA): a multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol. 20, 636–648 (2019).
Alsop, K. et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J. Clin. Oncol. 30, 2654–2663 (2012).
Miller, R. E. et al. ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann. Oncol. 31, 1606–1622 (2020).
Thorne, H. et al. BRCA1 and BRCA2 carriers with breast, ovarian and prostate cancer demonstrate a different pattern of metastatic disease compared with non-carriers: results from a rapid autopsy programme. Histopathology 83, 91–103 (2023).
Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Mafficini, A. et al. BRCA somatic and germline mutation detection in paraffin embedded ovarian cancers by next-generation sequencing. Oncotarget 7, 1076–1083 (2016).
Hennessy, B. T. et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J. Clin. Oncol. 28, 3570–3576 (2010).
Koczkowska, M. et al. Detection of somatic BRCA1/2 mutations in ovarian cancer - next-generation sequencing analysis of 100 cases. Cancer Med. 5, 1640–1646 (2016).
Vos, J. R. et al. Universal tumor DNA BRCA1/2 testing of ovarian cancer: prescreening PARPi treatment and genetic predisposition. J. Natl. Cancer Inst. 112, 161–169 (2020).
Song, H. et al. Contribution of germline mutations in the RAD51B, RAD51C, and RAD51D genes to ovarian cancer in the population. J. Clin. Oncol. 33, 2901–2907 (2015).
Popova, T. et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 72, 5454–5462 (2012).
Birkbak, N. J. et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2, 366–375 (2012).
Abkevich, V., Timms, K. M., Hennessy, B. T., Potter, J., Carey, M. S., Meyer, L. A. et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer 107, 1776–1782 (2012).
Marquard, A. M., Eklund, A. C., Joshi, T., Krzystanek, M., Favero, F., Wang, Z. C. et al. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark. Res. 3, 9 (2015).
Burdett, N. L., Willis, M. O., Alsop, K., Hunt, A. L., Pandey, A., Hamilton, P. T. et al. Multiomic analysis of homologous recombination-deficient end-stage high-grade serous ovarian cancer. Nat. Genet. 55, 437–450 (2023).
Macintyre, G., Goranova, T. E., De Silva, D., Ennis, D., Piskorz, A. M., Eldridge, M. et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat. Genet. 50, 1262–1270 (2018).
Drews, R. M., Hernando, B., Tarabichi, M., Haase, K., Lesluyes, T., Smith, P. S. et al. A pan-cancer compendium of chromosomal instability. Nature 606, 976–983 (2022).
Alexandrov, L. B., Kim, J., Haradhvala, N. J., Huang, M. N., Tian Ng, A. W., Wu, Y. et al. The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020).
Koskela, H., Li, Y., Joutsiniemi, T., Muranen, T., Isoviita, V. M., Huhtinen, K. et al. HRD related signature 3 predicts clinical outcome in advanced tubo-ovarian high-grade serous carcinoma. Gynecol. Oncol. 180, 91–98 (2024).
Steele, C. D., Abbasi, A., Islam, S. M. A., Bowes, A. L., Khandekar, A., Haase, K. et al. Signatures of copy number alterations in human cancer. Nature 606, 984–991 (2022).
Gulhan, D. C., Lee, J. J., Melloni, G. E. M., Cortes-Ciriano, I. & Park, P. J. Detecting the mutational signature of homologous recombination deficiency in clinical samples. Nat. Genet. 51, 912–919 (2019).
Nguyen, L., Van Hoeck, J. W. M. M. & Cuppen, A. E. Pan-cancer landscape of homologous recombination deficiency. Nat. Commun. 11, 5584 (2020).
Abbasi, A., Steele, C. D., Bergstrom, E. N., Khandekar, A., Farswan, A. & McKay, R. R. et al. HRProfiler detects homologous recombination deficiency in breast and ovarian cancers using whole-genome and whole-exome sequencing data. Cancer Res. 2504–2513 (2025).
Davies, H., Glodzik, D., Morganella, S., Yates, L. R., Staaf, J., Zou, X. et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med. 23, 517–525 (2017).
Sztupinszki, Z., Diossy, M., Krzystanek, M., Reiniger, L., Csabai, I., Favero, F. et al. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. npj Breast Cancer 4, 16 (2018).
Guo, M. & Wang, S. M. The BRCAness landscape of cancer. Cells 11 (2022).
Jacobson, D. H., Pan, S., Fisher, J. & Secrier, M. Multi-scale characterisation of homologous recombination deficiency in breast cancer. Genome Med. 15, 90 (2023).
Lee, J. J., Kang, H. J., Kim, D., Lim, S. O., Kim, S. S., Kim, G. et al. expHRD: an individualized, transcriptome-based prediction model for homologous recombination deficiency assessment in cancer. BMC Bioinformatics 25, 236 (2024).
Kang, J., Lee, J., Lee, A. & Lee, Y. S. Prediction of homologous recombination deficiency from cancer gene expression data. J. Int. Med. Res. 50, 3000605221133655 (2022).
Vazquez-Garcia, I., Uhlitz, F., Ceglia, N., Lim, J. L. P., Wu, M., Mohibullah, N. et al. Ovarian cancer mutational processes drive site-specific immune evasion. Nature 612, 778–786 (2022).
Zhou, W., Triche, T. J. Jr., Laird, P. W. & Shen, H. SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 46, e123 (2018).
Raine, K. M., Van Loo, P., Wedge, D. C., Jones, D., Menzies, A., Butler, A. P. et al. ascatNgs: identifying somatically acquired copy-number alterations from whole-genome sequencing data. Curr. Protoc. Bioinformatics 56, 1–9 7 (2016).
Aran, D., Sirota, M. & Butte, A. J. Systematic pan-cancer analysis of tumour purity. Nat. Commun. 6, 8971 (2015).
Goldman, M. J., Craft, B., Hastie, M., Repecka, K., McDade, F., Kamath, A. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38, 675–678 (2020).
Tothill, R. W., Tinker, A. V., George, J., Brown, R., Fox, S. B., Lade, S. et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res. 14, 5198–5208 (2008).
Knijnenburg, T. A., Wang, L., Zimmermann, M. T., Chambwe, N., Gao, G. F., Cherniack, A. D. et al. Genomic and molecular landscape of DNA damage repair deficiency across the Cancer Genome Atlas. Cell Rep. 23, 239–54.e6 (2018).
Carter, S. L., Cibulskis, K., Helman, E., McKenna, A., Shen, H., Zack, T. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Chen, X., Schulz-Trieglaff, O., Shaw, R., Barnes, B., Schlesinger, F., Kallberg, M. et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32, 1220–1222 (2016).
Van der Auwera GAOC, B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra 1st edn (O’Reilly Media, 2020).
Garsed, D. W., Pandey, A., Fereday, S., Kennedy, C. J., Takahashi, K., Alsop, K. et al. The genomic and immune landscape of long-term survivors of high-grade serous ovarian cancer. Nat. Genet. 54, 1853–1864 (2022).
Patch, A. M., Christie, E. L., Etemadmoghadam, D., Garsed, D. W., George, J., Fereday, S. et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 521, 489–494 (2015).
Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016).
Telli, M. L., Timms, K. M., Reid, J., Hennessy, B., Mills, G. B., Jensen, K. C. et al. Homologous Recombination Deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin. Cancer Res. 22, 3764–3773 (2016).
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22 (2010).
Lord, C. J. & Ashworth, A. BRCAness revisited. Nat. Rev. Cancer 16, 110–120 (2016).
Oshi, M., Gandhi, S., Wu, R., Asaoka, M., Yan, L., Yamada, A. et al. Development of a novel BRCAness score that predicts response to PARP inhibitors. Biomark. Res. 10, 80 (2022).
Zhang, M., Ma, S. C., Tan, J. L., Wang, J., Bai, X., Dong, Z. Y. et al. Inferring homologous recombination deficiency of ovarian cancer from the landscape of copy number variation at subchromosomal and genetic resolutions. Front. Oncol. 11, 772604 (2021).
Farrugia, D. J., Agarwal, M. K., Pankratz, V. S., Deffenbaugh, A. M., Pruss, D., Frye, C. et al. Functional assays for classification of BRCA2 variants of uncertain significance. Cancer Res. 68, 3523–3531 (2008).
Mesman, R. L. S., Calleja, F., Hendriks, G., Morolli, B., Misovic, B., Devilee, P. et al. The functional impact of variants of uncertain significance in BRCA2. Genet. Med. 21, 293–302 (2019).
Comitani, F., Nash, J. O., Cohen-Gogo, S., Chang, A. I., Wen, T. T., Maheshwari, A. et al. Diagnostic classification of childhood cancer using multiscale transcriptomics. Nat. Med. 29, 656–666 (2023).
Wong, M., Mayoh, C., Lau, L. M. S., Khuong-Quang, D. A., Pinese, M., Kumar, A. et al. Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat. Med. 26, 1742–1753 (2020).
Prat, A., Pineda, E., Adamo, B., Galvan, P., Fernandez, A., Gaba, L. et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 24, S26–S35 (2015).
Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Prat, A., Parker, J. S., Fan, C. & Perou, C. M. PAM50 assay and the three-gene model for identifying the major and clinically relevant molecular subtypes of breast cancer. Breast Cancer Res. Treat. 135, 301–306 (2012).
Acknowledgements
A.L.W. is supported by a Research Training Program scholarship and is partially funded by a CSL PhD top-up scholarship and a Tour De Cure PhD grant. N.M.D. is funded by NHMRC Investigator Grant [GNT2016547 to N.M.D.] and the Estate of Judith Corrie Philpots. S.J.R. is funded by NHMRC Investigator Grant [GNT2009840 to S.J.R]. We thank Dr Matthew Wakefield for offering insight and expertise in ovarian carcinoma biology. We acknowledge the contributions of Dr Ksenija Nesic and the entire laboratory of Professor Clare Scott at the Walter and Eliza Hall Institute for offering feedback on the complete IdentifiHR model. We offer thanks to Professor James Brenton and members of the Brenton laboratory for discussions surrounding HR and the training of our model. Figures 1 and 3 created, in part, in BioRender. Weir, A. (2025); https://BioRender.com/isms2aw. We thank the many patients who contributed to the data used in this research, and our cancer consumer advisers. We also acknowledge the Wurundjeri people of the Kulin nation as the traditional owners and guardians of the land on which the work was performed.
Author information
Authors and Affiliations
Contributions
A.L.W. and N.M.D. conceived and designed the study. A.L.W. collected, processed, and curated all data, developed the methodology, validated the method and results, wrote the original draft and all subsequent iterations, and produced all tables and visualisations in the study. N.M.D., S.J.R. and C.W.T. supervised the research and revised the manuscript. D.G and A.P. processed and analysed WGS data in the AOCS cohort. S.C.L. and M.L. advised on method development and analysis. All authors contributed to the review of the manuscript. All authors approved the manuscript for submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Michael Menzel and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Weir, A.L., Lee, S.C., Li, M. et al. IdentifiHR predicts homologous recombination deficiency in high-grade serous ovarian carcinoma using gene expression. Commun Med (2026). https://doi.org/10.1038/s43856-026-01387-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-026-01387-y