Abstract
Short and long sleep durations are associated with multiple physical, psychiatric and neurodegenerative diseases, yet their potentially shared and distinct biological mechanisms remain unclear. Here, using data from UK Biobank participants aged 38–73 years, we have characterized the in-depth genetic architecture of short (≤7 h) and long (≥7 h) sleep groups, along with their associations with behaviors, neuroimaging and blood biomarkers. The two sleep groups exhibited independent genetic architectures and distinct immunometabolic and proteomic profiles. Notably, long sleep showed more significant associations with cardiovascular-related biomarkers (for example, cholesterol), brain structures (for example, hippocampus) and plasma proteins (for example, GDF15), whereas short sleep demonstrated greater genetic overlap with psychiatric conditions, particularly depression. Mendelian randomization further supported this dissociation by showing that long sleep duration is probably a consequence of multiple brain disorders and cardiovascular diseases, whereas short sleep duration has a potential causal effect on various brain and physical illnesses. Our findings advance our understanding of the relationship between sleep and health conditions by revealing distinct biological origins and genetic mechanisms underlying short and long sleep duration.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$79.00 per year
only $6.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
This project corresponds to UK Biobank application ID 19542. Neuroimaging, genotype and behavioral data from UK Biobank dataset are available from https://biobank.ndph.ox.ac.uk/ by application. The variables utilized in this study are detailed in Supplementary Tables 1–3. Previous published GWASs of psychiatric disorders, including depression, bipolar disorder and schizophrenia, were provided by the Psychiatric Genomics Consortium, which can be downloaded from https://pgc.unc.edu/for-researchers/download-results/. GWAS summary statistics of immunometabolic phenotypes, obesity and aging disease are available in the MRC IEU OpenGWAS database (https://gwas.mrcieu.ac.uk), and the detailed PubMed identifiers (PMIDs) for the GWAS summary data are provided in Supplementary Table 10 (https://pubmed.ncbi.nlm.nih.gov/). European ancestral background LD scores from the 1000 Genomes Project were downloaded from https://alkesgroup.broadinstitute.org/LDSCORE/. The GRCh37 coordinates can be accessed via http://hgdownload.cse.ucsc.edu/goldenpath/hg19/database/.
Code availability
R version 4.2.0 was used to perform phenotype-wide association analysis. Matlab 2018b was used to perform linear association analysis. Freesurfer v6.0 was used to process the imaging data. PLINK 2.0 was used to perform GWAS analysis. R version 4.2.0 GenomicSEM version 0.0.3 was utilized to calculate heritability and genetic correlations. TwoSampleMR version 0.5.6 was utilized to measure the causal association. Scripts used to perform the analyses are available at https://github.com/yuzhulineu/UKB_short_longsleep/.
References
Mander, B. A., Winer, J. R. & Walker, M. P. Sleep and human aging. Neuron 94, 19–36 (2017).
Wang, C. et al. Association of estimated sleep duration and naps with mortality and cardiovascular events: a study of 116632 people from 21 countries. Eur. Heart J. 40, 1620–1629 (2018).
Hirshkowitz, M. et al. National Sleep Foundation’s sleep time duration recommendations: methodology and results summary. Sleep Health 1, 40–43 (2015).
Mutz, J., Roscoe, C. J. & Lewis, C. M. Exploring health in the UK Biobank: associations with sociodemographic characteristics, psychosocial factors, lifestyle and environmental exposures. BMC Med. 19, 240 (2021).
Li, Y. et al. The brain structure and genetic mechanisms underlying the nonlinear association between sleep duration, cognition and mental health. Nat. Aging 2, 425–437 (2022).
Huang, S.-Y. et al. Sleep, physical activity, sedentary behavior, and risk of incident dementia: a prospective cohort study of 431,924 UK Biobank participants. Mol. Psychiatry 27, 4343–4354 (2022).
Coutrot, A. et al. Reported sleep duration reveals segmentation of the adult life-course into three phases. Nat. Commun. 13, 7697 (2022).
Sabia, S. et al. Association of sleep duration at age 50, 60 and 70 years with risk of multimorbidity in the UK: 25-year follow-up of the Whitehall II cohort study. PLoS Med. 19, e1004109 (2022).
Itani, O. et al. Short sleep duration and health outcomes: a systematic review, meta-analysis and meta-regression. Sleep Med. 32, 246–256 (2017).
Jike, M. et al. Long sleep duration and health outcomes: a systematic review, meta-analysis and meta-regression. Sleep Med. Rev. 39, 25–36 (2018).
Buxton, O. M. et al. Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption. Sci. Transl. Med. 4, 129ra43 (2012).
Schmid, S. M., Hallschmid, M. & Schultes, B. The metabolic burden of sleep loss. Lancet Diabetes Endocrinol. 3, 52–62 (2015).
Reis, C. et al. Sleep duration, lifestyles and chronic diseases: a cross-sectional population-based study. Sleep Sci. 11, 217–230 (2018).
Doherty, A. et al. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nat. Commun. 9, 5257 (2018).
Gottlieb, D. J. et al. Novel loci associated with usual sleep duration: the CHARGE Consortium Genome-Wide Association Study. Mol. Psychiatry 20, 1232–1239 (2015).
Dashti, H. S. et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat. Commun. 10, 1100 (2019).
Jones, S. E. et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet. 12, e1006125 (2016).
Byrne, E. M. et al. Genetic correlation analysis suggests association between increased self-reported sleep duration in adults and schizophrenia and type 2 diabetes. Sleep 39, 1853–1857 (2016).
Lane, J. M. et al. Biological and clinical insights from genetics of insomnia symptoms. Nat. Genet. 51, 387–393 (2019).
Ai, S. et al. Causal associations of short and long sleep durations with 12 cardiovascular diseases: linear and nonlinear Mendelian randomization analyses in UK Biobank. Eur. Heart J. 42, 3349–3357 (2021).
Knutson, K. L. et al. The metabolic consequences of sleep deprivation. Sleep Med. Rev. 11, 163–178 (2007).
Tobaldini, E. et al. Short sleep duration and cardiometabolic risk: from pathophysiology to clinical evidence. Nat. Rev. Cardiol. 16, 213–224 (2019).
Furman, D. et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 25, 1822–1832 (2019).
Youngstedt, S. D. & Kripke, D. F. Long sleep and mortality: rationale for sleep restriction. Sleep Med. Rev. 8, 159–174 (2004).
Theorell-Haglöw, J. et al. Sleep duration is associated with protein biomarkers for cardiometabolic health: a large-scale population study. J. Sleep Res. 30, e13284 (2021).
Wojcik, G. L. et al. Opportunities and challenges for the use of common controls in sequencing studies. Nat. Rev. Genet. 23, 665–679 (2022).
Lukaski, H. C. Evolution of bioimpedance: a circuitous journey from estimation of physiological function to assessment of body composition and a return to clinical research. Eur. J. Clin. Nutr. 67, S2–S9 (2013).
Xiao, Q. et al. Relationship between sleep characteristics and measures of body size and composition in a nationally-representative sample. BMC Obesity 3, 48 (2016).
Zhang, Y.-R. et al. Peripheral immunity is associated with the risk of incident dementia. Mol. Psychiatry 27, 1956–1962 (2022).
Tian, Y. E. et al. Heterogeneous aging across multiple organ systems and prediction of chronic disease and mortality. Nat. Med. 29, 1221–1231 (2023).
Bhargava, S. et al. Lipids and lipoproteins in cardiovascular diseases: a classification. Trends Endocrinol. Metab. 33, 409–423 (2022).
Blake, G. J. et al. Blood pressure, C-reactive protein, and risk of future cardiovascular events. Circulation 108, 2993–2999 (2003).
Chiesa, S. T. et al. Glycoprotein acetyls: a novel inflammatory biomarker of early cardiovascular risk in the young. J. Am. Heart Assoc. 11, e024380 (2022).
Kettunen, J. et al. Biomarker glycoprotein acetyls is associated with the risk of a wide spectrum of incident diseases and stratifies mortality risk in angiography patients. Circ. Genom. Precis. Med. 11, e002234 (2018).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Jansen, P. R. et al. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat. Genet. 51, 394–403 (2019).
Morales-Muñoz, I. et al. Role of inflammation in short sleep duration across childhood and psychosis in young adulthood. JAMA Psychiatry 81, 825–833 (2024).
Lane, J. M. et al. Genetics of circadian rhythms and sleep in human health and disease. Nat. Rev. Genet. 24, 4–20 (2023).
Bhalla, S. et al. Protective role of IGF-1 and GLP-1 signaling activation in neurological dysfunctions. Neurosci. Biobehav. Rev. 142, 104896 (2022).
Plante, D. T. The evolving nexus of sleep and depression. Am. J. Psychiatry 178, 896–902 (2021).
Liew, S. C. & Aung, T. Sleep deprivation and its association with diseases—a review. Sleep Med. 77, 192–204 (2021).
Irwin, M. R., Olmstead, R. & Carroll, J. E. Sleep disturbance, sleep duration and inflammation: a systematic review and meta-analysis of cohort studies and experimental sleep deprivation. Biol. Psychiatry 80, 40–52 (2016).
Irwin, M. R. Sleep and inflammation: partners in sickness and in health. Nat. Rev. Immunol. 19, 702–715 (2019).
Smagula, S. F. et al. Risk factors for sleep disturbances in older adults: evidence from prospective studies. Sleep Med. Rev. 25, 21–30 (2016).
Garbarino, S. et al. Role of sleep deprivation in immune-related disease risk and outcomes. Commun. Biol. 4, 1304 (2021).
Besedovsky, L., Lange, T. & Haack, M. The sleep-immune crosstalk in health and disease. Physiol. Rev. 99, 1325–1380 (2019).
Stamatakis, K. A. & Punjabi, N. M. Long sleep duration: a risk to health or a marker of risk? Sleep Med. Rev. 11, 337–339 (2007).
Vyazovskiy, V. V. Sleep, recovery and metaregulation: explaining the benefits of sleep. Nat. Sci. Sleep 7, 171–184 (2015).
Rasmussen, M. K., Mestre, H. & Nedergaard, M. The glymphatic pathway in neurological disorders. Lancet Neurol. 17, 1016–1024 (2018).
Siow, T. Y. et al. Association of sleep, neuropsychological performance, and gray matter volume with glymphatic function in community-dwelling older adults. Neurology 98, e829–e838 (2022).
Xie, L. et al. Sleep drives metabolite clearance from the adult brain. Science 342, 373–377 (2013).
Walker, K. A. et al. Proteomics analysis of plasma from middle-aged adults identifies protein markers of dementia risk in later life. Sci. Transl. Med. 15, eadf5681 (2023).
Larsson, S. C. & Markus, H. S. Genetic liability to insomnia and cardiovascular disease risk. Circulation 140, 796–798 (2019).
Willoughby, A. R. et al. Country differences in nocturnal sleep variability: observations from a large-scale, long-term sleep wearable study. Sleep Med. 110, 155–165 (2023).
Wainberg, M. et al. Clinical laboratory tests and five-year incidence of major depressive disorder: a prospective cohort study of 433,890 participants from the UK Biobank. Transl. Psychiatry 11, 380 (2021).
Ahola-Olli, A. V. et al. Circulating metabolites and the risk of type 2 diabetes: a prospective study of 11,896 young adults from four Finnish cohorts. Diabetologia 62, 2298–2309 (2019).
Soininen, P. et al. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ. Cardiovasc. Genet. 8, 192–206 (2015).
Desikan, R. S. et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980 (2006).
Fischl, B. et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355 (2002).
Alfaro-Almagro, F. et al. Image processing and quality control for the first 10,000 brain imaging datasets from UK Biobank. NeuroImage 166, 400–424 (2018).
de Groot, M. et al. Improving alignment in tract-based spatial statistics: evaluation and optimization of image registration. NeuroImage 76, 400–411 (2013).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Kang, J. et al. Increased brain volume from higher cereal and lower coffee intake: shared genetic determinants and impacts on cognition and metabolism. Cereb. Cortex 32, 5163–5174 (2022).
Sun, B. B. et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature 622, 329–338 (2023).
Millard, L. A. C. et al. Software application profile: PHESANT: a tool for performing automated phenome scans in UK Biobank. Int. J. Epidemiol. 47, 29–35 (2018).
Lane, J. M. et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat. Genet. 49, 274–281 (2017).
Kocevska, D. et al. Sleep characteristics across the lifespan in 1.1 million people from the Netherlands, United Kingdom and United States: a systematic review and meta-analysis. Nat. Hum. Behav. 5, 113–122 (2021).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 7, e34408 (2018).
Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022).
Kunkle, B. W. et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430 (2019).
Nikpay, M. et al. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47, 1121–1130 (2015).
Willer, C. J. et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013).
Yasugaki, S. et al. Bidirectional relationship between sleep and depression. Neurosci. Res. 211, 57–64 (2025).
Liu, C. et al. Association between high-density lipoprotein cholesterol levels and adverse cardiovascular outcomes in high-risk populations. JAMA Cardiol. 7, 672–680 (2022).
Acknowledgements
This study utilized the UK Biobank resource under application no. 19542. We thank all the participants and researchers from UK Biobank. This work received support from the following sources: National Key R&D Program of China (2021YFC2501400 to T.J., 2019YFA0709502 to J.F., 2018YFC1312904 to J.F., 2022CSJGG1000 to T.J., 2019YFA0709501 to T.J., 2018YFC1312900 to T.J., 2023YFE0199700 to T.J. and 2023YFC3605400 to W.C.), the National Natural Science Foundation of China (T2122005 to T.J., 82472055 to W.C., 82071997 to W.C. and 81801773 to T.J.), the 111 Project (B18015 to J.F.), the key project of Shanghai Science and Technology (16JC1420402 to J.F.), Shanghai Municipal Science and Technology Major Project (2018SHZDZX01 to J.F.), Zhangjiang Lab (to J.F.), Shanghai Center for Brain Science and Brain-Inspired Technology (to J.F.), Shanghai Rising-Star Program (21QA1408700 to W.C.), Shanghai Pujiang Project (18PJ1400900 to T.J.), Shanghai Yangfan Project (24YF2738400 to Y.L.) and Postdoctoral Fellowship Program (Grade B) of China Postdoctoral Science Foundation (GZB20230164 to Y.L. and 2023M730697 to Y.L.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
J.F., W.C. and T.J. proposed the study. Y.L. and W.G. analyzed the data. W.Z. and S.H. preprocessed the data. T.J., S.N. and J.Y. contributed to interpretation of the results. Y.L. drafted the paper and T.J., S.N., B.J.S. and W.C. edited it. Y.L. and W.C. were responsible for visualization. Y.Z. and L.M. provided feedback to improve the paper. All authors contributed to discussions on data analysis and approved the final version of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Mental Health thanks Julian Mutz, Arezu Najafi, Masoud Tahmasian and Angeliki Tsapanou for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
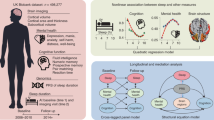
Extended Data Fig. 1 Guideline of the study.
Top, UK biobank data used in this study included environmental and behavioral measures, blood and imaging biomarkers, genomics, proteins, and sleep duration. Middle, A phenotype-wide association study (PheWAS) was performed to explore the associations of short and long sleep duration with these phenotypes. Biological blood biomarkers, neuroimaging biomarkers, and proteins were utilized to characterize distinct association profiles for short and long sleep duration. Bottom, Distinct genetic architectures and roles in association with health conditions were identified for short and long sleep duration. Finally, Mendelian randomization (MR) analysis showed distinct causal associations between short and long sleep duration and health-related phenotypes.
Supplementary information
Supplementary Information
Supplementary Tables 1–27 and Figs. 1–13.
Supplementary Data 1–6
Supplementary Data 1 Phenotype-wide associations study of sleep <= 7 h. Supplementary Data 2 Phenotype-wide associations study of sleep >= 7 h. Supplementary Data 3 Genetic correlations between sleep duration and health-related phenotypes. Supplementary Data 4 Phenotype-wide associations study of stratified short sleep (sleep < 7 versus sleep = 7/8 h). Supplementary Data 5 Phenotype-wide associations study of stratified long sleep (sleep > 8 versus sleep = 7/8 h). Supplementary Data 6 Genetic correlations between stratified sleep duration and health-related phenotypes.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Gong, W., Sahakian, B.J. et al. Divergent biological pathways linking short and long sleep durations to mental and physical health. Nat. Mental Health 3, 429–443 (2025). https://doi.org/10.1038/s44220-025-00395-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44220-025-00395-6
This article is cited by
-
The chain mediating effects of somatic pain and depression between chronic diseases and sleep in the older patients
Sleep and Breathing (2026)
-
Prevalence and correlates of perceived stress and psychological health among residents receiving standardized training: a multicenter cross-sectional survey study in China
BMC Public Health (2025)